Abstract
Src family tyrosine kinases are involved in modulating various signal transduction pathways leading to the induction of DNA synthesis and cytoskeletal reorganization in response to cell-cell or cell-matrix adhesion. The critical role of these kinases in regulating cellular signaling pathways requires that their activity be tightly controlled. Src family proteins are regulated through reversible phosphorylation and dephosphorylation events that alter the conformation of the kinase. We have found evidence that Src also is regulated by ubiquitination. Activated forms of Src are less stable than either wild-type or kinase-inactive Src mutants and can be stabilized by proteasome inhibitors. In addition, poly-ubiquitinated forms of active Src have been detected in vivo. Taken together, our results establish ubiquitin-mediated proteolysis as a previously unidentified mechanism for irreversibly attenuating the effects of active Src kinase.
The Src family kinases are nonreceptor tyrosine kinases that mediate a variety of cellular signaling pathways. Nine vertebrate members of the Src kinase family are known: some are expressed in a cell-type specific manner and others are expressed more broadly. For example, Blk and Lck expression is limited to lymphoid lineages. In contrast, Src, Fyn, and Yes are more broadly expressed although their levels of expression do vary among cell types. For instance, Src is abundant in brain and platelets, whereas Fyn is abundant in brain, platelets, and T lymphocytes (for review see ref. 1).
All Src family members share the same basic structural features. Each possesses an amino-terminal Src homology (SH) 4 domain that contains a myristylation signal sequence required for membrane localization. Adjacent to the SH4 domain is a unique region followed by conserved SH3 and SH2 domains, which mediate protein–protein interactions. The catalytic kinase domain resides in the carboxyl-terminal half of the protein. Finally, there is a short carboxyl-terminal tail domain that functions to negatively regulate the kinase activity (for reviews see refs. 1 and 2). An intramolecular interaction between the SH2 domain and a phosphorylated tyrosine residue (amino acid 527 in chicken c-Src) within this tail domain alters the conformation of Src such that the catalytic domain is confined to an inactive state (closed conformation; ref. 3). The dephosphorylation of tyrosine 527 results in a release of the intramolecular interaction with the SH2 domain relaxing conformational constraints. The molecule takes on an “open conformation” that facilitates catalytic activation (3–5).
Src family kinases function in numerous signaling pathways including those mediating DNA synthesis and proliferation. Activated cell surface receptors interact with and signal through the Src family kinases. For example, binding of ligand to the platelet-derived growth factor receptor results in its association with and activation of the Src family kinases Src, Fyn, and Yes (6). In turn, the activated Src kinases trigger a cascade of events, ultimately leading to entry into S phase and subsequent DNA replication (7–9). Src kinases also have a role in progression through the G2/M transition of the cell cycle, suggesting that they function in both the G1 and mitotic checkpoints (10–12).
Several Src family members (Src, Fyn, and Yes) are also important for transducing signals in response to cell-cell or cell-matrix adhesion (13). Adherence of cells to the extracellular matrix via integrin receptors results in the assembly of protein complexes required to modify the cellular cytoskeleton (for reviews see refs. 1, 14, and 15). One of the early responses to integrin receptor engagement is the activation of Src family kinases that phosphorylate substrates such as focal adhesion kinase, paxillin, and p130cas. These Src-mediated phosphorylation events have been associated with changes in cell adhesion, motility, and shape (13, 16).
Because the Src family kinases affect both cell cycle progression and cytoskeletal organization, dysregulation may lead to constitutive activation and cellular transformation (2). For example, the viral derivative of Src, v-Src, and the point mutant SrcY527F both lack the negative regulatory Tyr-527, leaving them in the “open,” active conformation (17–21). Additional Src variants bearing mutations in the SH2, SH3, and kinase domains are also constitutively active presumably because of disruption of the “closed” conformation of the protein (22, 23).
Although the reversible phosphorylation of the regulatory tyrosine within the carboxyl terminus plays an important regulatory function, additional mechanisms may exist for controlling the activity of Src family kinases. Indeed, our laboratory recently has demonstrated that the Src family member Blk is regulated by ubiquitin-mediated degradation (24). Specifically, the active form of Blk is recognized by E6AP, an E3 ubiquitin-protein ligase that promotes its ubiquitination and subsequent degradation (24–26). In this study, we show that Src itself is degraded in a ubiquitin-dependent manner and that the active form is specifically targeted for degradation. Taken together, these results suggest that targeted degradation of active forms of the Src family of tyrosine kinases may constitute an additional mechanism for restricting the activity of these important signaling proteins.
Materials and Methods
Plasmid Constructs.
The pLNCX vectors encoding the chicken c-Src, v-Src, c-Src(Y416F), and c-Src(K295R) were kind gifts of Joan Brugge, Harvard Medical School (18, 20, 21, 27). pLNCX vectors encoding c-Src(Y527F) and c-Src(E378G) were generated by subcloning ClaI fragments into pLNCX from plasmids also given by Dr. Brugge (19, 21–23). pCMV4c-Src and pCMV4-c-Src(E378G) were generated by subcloning KpnI–HindIII fragments into pCMV4.
Cell Culture and Transfection.
All cell lines were maintained in DMEM (GIBCO/BRL), supplemented with 100 units/ml of penicillin, 100 μg/ml of streptomycin, and 10% FBS, at 37° in a 5% CO2 incubator. csk+/+, csk−/−, src+/+, and src−/− mouse embryo fibroblasts (MEFs; refs. 28 and 31) were kind gifts of Sheila Thomas, Harvard Medical School. Transfections were performed by using standard electroporation procedures at 0.25 V for Cos-7 cells and 0.30 V for src−/− MEFs.
Antibodies.
mAb 327 (anti-Src antibody; Oncogene Science and generous gift of Dr. Thomas), polyclonal antibody PAb N16 (anti-Src antibody; Santa Cruz Biotechnology), polyclonal antibody PAb C11 (anti-actin antibody; Santa Cruz Biotechnology), mAb 9E10 (anti-Myc antibody; Oncogene Science), and mAb Ubi-1 (anti-ubiquitin antibody; Zymed) were used.
Cellular Lysate Preparation.
Forty eight hours posttransfection, cells were washed twice with PBS and incubated in 1 ml of ice-cold RIPA buffer (150 mM NaCl/1% NP40/0.5% deoxycholate/0.1% SDS/50 mM Tris⋅HCl, pH 7.5/5 μg/ml PMSF/5 μg/ml aprotinin/5 μg/ml leupeptin) or NP-40 lysis buffer (100 mM Tris⋅HCl, pH 7.4/120 mM NaCl/1% NP-40/5 μg/ml PMSF/5 μg/ml aprotinin/5 μg/ml leupeptin) as indicated. Lysates were sonicated and cleared of debris by centrifuging for 5 min at 4°C. For experiments requiring proteasome inhibitor treatment, 48 hr posttransfection, cells were treated with 50 μM Z-L3VS. (generous gift of Hidde Ploegh, Harvard Medical School; ref. 30) for 8–14 hr. Control cells were treated with an equal amount of DMSO and harvested as described above.
Immunoblotting.
For each experiment, equal amounts of total protein were resolved by 8% SDS/PAGE. After SDS/PAGE, proteins were transferred to poly(vinylidene difluoride) (PVDF) membrane in 12.5 mM Tris⋅HCl-100 mM glycine for 2 hr at 60 V. The membrane then was incubated in a blocking solution of 5% nonfat dried milk in TNET (10 mM Tris⋅HCl/2.5 mM EDTA/50 mM NaCl/0.1% Tween20) with rocking. After washing three times in TNET, the membrane was incubated in primary antibody for at least 2 hr, washed three times, and incubated in secondary antibody (horseradish peroxidase linked; Amersham Pharmacia) for 1 hr. Membranes then were washed three times in TNET and developed with enhanced chemiluminescence reagents (NEN).
Pulse–Chase Experiments.
src−/− cells were transiently transfected with the pLNCX-Src constructs described above. After plasmid transfection, the cells were pooled and divided equally among five 6-cm2 dishes. Forty eight hours posttransfection, cells were washed twice in PBS and then incubated in DMEM lacking methionine and cysteine for 1 hr at 37°C. Each 6-cm2 dish was labeled with 100 μCi of 35S methionine/cysteine (Express Protein Labeling Mix; NEN) for a 45-min pulse, washed three times with PBS, and chased with DMEM containing 10% FCS and 100-fold excess l-methionine. At each time point, cells were harvested in 0.5 ml of NP-40 lysis buffer as described above, and equivalent amounts of cell lysate were immunoprecipitated by using mAb 327. Immune complexes bound to the Sepharose beads were washed three times in NP-40 lysis buffer and released into gel loading buffer (2% SDS/60 mM Tris⋅HCl, pH 6.8/10% glycerol/0.1% bromophenol blue/292 mM β-mercaptoethanol) by boiling for 3 min. Samples were resolved by 8% SDS/PAGE, dried, and exposed to film. Results were quantitated by using a Molecular Dynamics PhosphorImager (Storm860).
In Vivo Ubiquitination Assay.
Cos-7 cells were cotransfected with pCMV4 expressing either c-Src or c-SrcE378G and pCMV4Myc-Ub. Where indicated, cells were treated with proteasome inhibitors as described above. Lysates were harvested in 1 ml of RIPA buffer and immunoprecipitated with N16 anti-Src antibodies. Immunoprecipitates were separated by SDS/PAGE, transferred to PVDF membrane, and immunoblotted as described above by using mAb 9E10. For csk−/− cells, lysates were harvested and immunoprecipitated as described above. Immunoprecipitates were separated and transferred as above and immunoblotted with mAb Ubi-1.
Results
Previous work from our laboratory has demonstrated that the Src family tyrosine kinase Blk is degraded via the ubiquitin pathway (24). Specifically, we found that activated Blk is ubiquitinated and is a substrate for E6AP, an E3 ubiquitin-protein ligase. Our studies with E6AP and Blk suggested to us a model in which the ubiquitin-mediated degradation of activated forms of the Src family kinase might be a more general mechanism by which this family of signaling proteins is regulated. To test this model, we chose to investigate whether Src itself was regulated by ubiquitination and proteolysis.
Activated Forms of Src Have Reduced Steady-State Protein Levels.
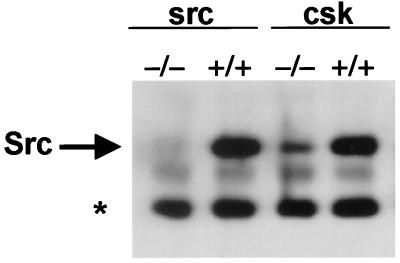
To test whether activated Src is less stable, steady-state Src levels were analyzed in csk+/+ and csk−/− MEF cells (29, 31). Csk (C-terminal Src kinase) is responsible for the negative regulatory tyrosine phosphorylation of Src at Tyr-527 (32). In cells from Csk-deficient mice, Src activity is increased 11-fold as compared with cells from a wild-type littermate (31). However, the steady-state levels of Src were reduced 5.4-fold from the levels observed in wild-type cells (Fig. 1), suggesting that steady-state levels of Src are inversely proportional to their activity (29, 31). Half-life experiments using cyclohexamide treatment to block protein synthesis demonstrated that whereas Src protein from csk+/+ cells remained stable for the duration of the experiment (2 hr), Src protein levels from csk−/− cells were undetectable after 90 min (data not shown). These results demonstrated that the difference in steady-state levels was caused by a decrease in the protein stability of endogenous Src in csk−/− cells, presumably because it is activated.
Figure 1.
Steady-state levels of endogenous Src protein. Equivalent amounts of whole-cell lysate from src−/− and csk−/− MEFs and matched wild-type MEFs were immunoprecipitated with mAb 327, separated by SDS/PAGE, transferred to PVDF membrane, and probed with mAb 327. * indicates the antibody heavy chain.
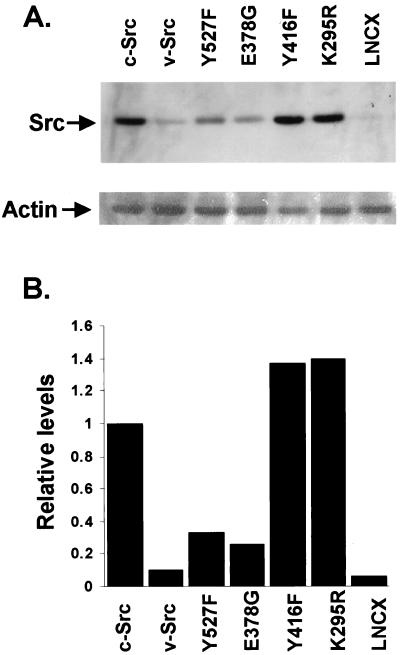
To test this hypothesis further, we analyzed the steady-state levels of a series of active and inactive mutants of Src kinase transfected into src−/− MEFs (28). The Rous sarcoma virus derivative of Src, v-Src, encodes a protein with 10–20 times the activity of c-Src (18, 20). c-SrcY527F lacks the negative regulatory Tyr-527 and exhibits 5–10 times the kinase activity of c-Src (19, 21). c-SrcE378G contains a mutation in the kinase domain and is 20 times more active than c-Src (22, 23). We also characterized the steady-state levels of two inactive mutants of c-Src: c-SrcY416F, which lacks a positive regulatory tyrosine required for maximal kinase activity, and c-SrcK295R, which lacks the critical lysine in the ATP-binding pocket, rendering it completely inactive (20, 21, 27).
Western blot analysis performed on src−/− cells transfected with the above constructs demonstrated that the levels of the activated forms of Src, namely v-Src, c-SrcY527F, and c-SrcE378G, were much lower than the levels of wild-type c-Src protein (Fig. 2A, compare lanes 1–4). Quantitation of the Src levels indicated a reduction in the steady-state protein by 3-fold, 4-fold, and 10-fold for c-SrcY527F, c-SrcE378G, and v-Src, respectively, compared with c-Src levels. This inverse correlation of protein levels to kinase activity has been noted previously for v-Src and c-SrcY527F (20). The less active c-SrcY416F and the completely inactive c-SrcK295R both showed steady-state levels equivalent to or greater than c-Src (Fig. 2A, lanes 5 and 6). There was a 1.4-fold increase in the steady-state levels of c-SrcY416F and a 1.3-fold increase in the levels of c-SrcK295R compared with c-Src (Fig. 2B). These results were confirmed in multiple separate experiments using either RIPA or NP-40 lysis buffers (data not shown). Each blot was probed for actin to control for protein loading. The comparable levels of c-Src, Y416F, and K295R forms of Src are consistent with our model because c-Src is found predominantly in the inactive form (20, 21).
Figure 2.
Steady-state levels of wild-type and mutant Src proteins. (A) (Upper) Src−/− MEFs were transfected with pLNCX vector expressing c-Src, v-Src, Y527F, E378G, Y416F, or K295R forms of Src, or pLNCX alone. Cells were harvested after 48 hr, and 50 μg of whole-cell lysate from each transfection was separated by SDS/PAGE, transferred to PVDF membrane, and probed with mAb 327. (Lower) The same membrane reprobed with anti-actin antibody C11. (B) Quantitation of the results shown in A by using densitometry.
Activated Forms of Src Kinase Are Rapidly Degraded.
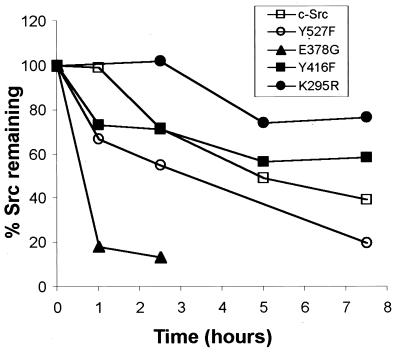
We next tested whether the decreased steady-state levels of the activated forms of Src were caused by degradation and a shortened protein half-life. Pulse–chase analyses therefore were performed on individual Src proteins. The half-life of wild-type c-Src was approximately 5–6 hr. Previous reports also have indicated that c-Src is a relatively stable protein (33, 34). In contrast to the stability of c-Src, we found that the half-lives of the active forms of the kinase, c-SrcE378G and c-SrcY527F, were 1 hr and 3 hr, respectively. The decreased stability observed for the active forms of Src is consistent with the reduction in steady-state levels being caused by increased protein turnover. Furthermore, the less active and inactive forms of Src were more stable than the wild-type c-Src. Both c-SrcY416F and c-SrcK295R proteins had half-lives greater than 7.5 hr (Fig. 3).
Figure 3.
Pulse–chase analysis of wild-type and mutant Src proteins. Src−/− MEFs were transfected with pLNCX vector expressing c-Src, or the following Src mutants: Y527F, E378G, Y416F, or K295R. Forty eight hours posttransfection, cells were pulse-labeled with 100 μCi 35S Met/Cys for 45 min. Plates were incubated in “chase” media with 100-fold excess methionine and then harvested at the indicated times. Lysates were immunoprecipitated by using mAb 327 and separated by SDS/PAGE. Gels were quantitated with a PhosphorImager, and each time point was normalized to the 0-hr time point.
Active Src Kinase Is a Target for the Ubiquitin Degradation Machinery.
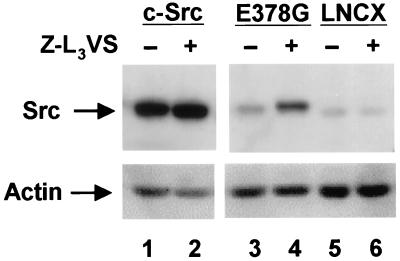
We next examined whether the differential stabilities of the various forms of the Src kinase were the result of the susceptibility of the activated forms to ubiquitination. Our previous results with Blk indicated that the preferential proteolysis of the activated forms of the kinase involved ubiquitination and subsequent degradation by the proteasome (24). To determine whether activated Src was being degraded by the proteasome, we examined the effect of the proteasome inhibitor, Z-L3VS, on the levels of Src protein (30). We compared the effects of proteasome inhibitor on the steady-state levels of c-Src (predominantly inactive) and the active form c-SrcE378G. As shown in Fig. 4, there was a minimal 1.2-fold increase in c-Src protein level in the presence of proteasome inhibitor; however, there was a dramatic 3.3-fold increase in the c-SrcE378G protein levels. This result, demonstrating the stabilization of active Src by a proteasome inhibitor, confirmed the involvement of the proteasome in the degradation of activated Src. The slight stabilization of c-Src might be caused by a low level of active wild-type c-Src or the normal turnover of inactive c-Src. The proteasome inhibitor had no effect on actin levels.
Figure 4.
Steady-state levels of Src in the presence of the proteasome inhibitor Z-L3VS. (Upper) Src−/− MEFs were transfected with pLNCX vector expressing c-Src, the active E378G mutant, or vector alone. Forty eight hours after transfection, cells were treated with either 50 μM Z-L3VS in DMSO or DMSO alone as a negative control. Eight hours after treatment, cells were harvested, separated by SDS/PAGE, transferred to PVDF membrane, and probed with mAb 327. The faint band in the LNCX lane is a background band. (Lower) The same membrane reprobed with anti-actin antibody C11.
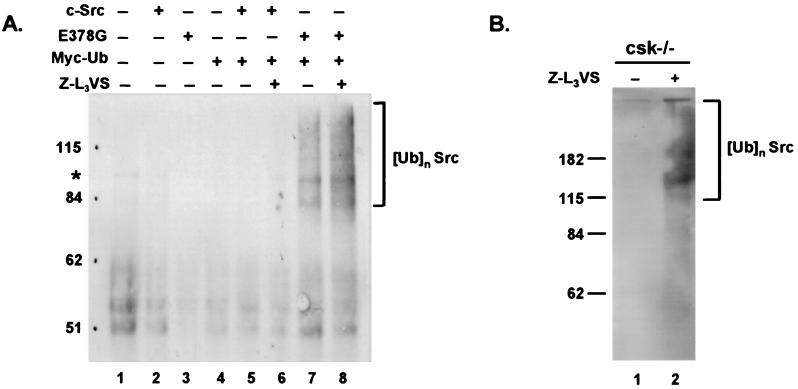
To investigate further the role of the ubiquitin pathway in Src proteolysis, we examined whether ubiquitinated forms of Src could be detected in vivo. For these experiments c-Src or c-SrcE378G were transfected into Cos-7 cells together with a plasmid expressing Myc-tagged ubiquitin. Ubiquitinated forms of c-SrcE378G were readily detected as a smear of higher molecular weight bands in the cells coexpressing c-SrcE378G and Myc-tagged ubiquitin (Fig. 5A, lanes 7 and 8). Although only a faint ladder of multiubiquitinated Src was detected, the high molecular weight smear is characteristic of multiubiquitinated proteins and is similar to those found in previous reports using Myc-tagged ubiquitin (35, 36). The addition of proteasome inhibitor did not greatly stabilize the ubiquitinated forms of c-SrcE378G, which is consistent with previous reports demonstrating the overall stability of Myc-tagged ubiquitin protein conjugates (37). We next examined the ubiquitination state of endogenous Src in csk−/− cells where Src is predominantly active. After treatment with proteasome inhibitor for 14 hr, cell lysates were immunoprecipitated with anti-Src antibodies and then analyzed by immunoblotting with anti-ubiquitin antibodies. As shown in Fig. 5B, we could detect the characteristic high molecular weight smear indicative of ubiquitination. In longer exposures of the transfection experiment (Fig. 5A) and in experiments using csk+/+ cells we also were able to detect evidence of high molecular bands indicative of ubiquitinated c-Src (data not shown). Because the half-life of c-Src is 5–6 hr (Fig. 3), the 14-hr proteasome inhibitor treatment period would account for at least two half-lives of c-Src. This result suggests that normal turnover of c-Src also may be regulated by ubiquitination. Nonetheless, these results demonstrate the in vivo ubiquitination of endogenous Src. Taken together with the stabilization of Src observed with proteasome inhibitors, these data confirm a role for ubiquitination in the regulation of Src through targeted proteolysis of its activated forms (Fig. 6).
Figure 5.
In vivo ubiquitination of active c-Src. (A) Cos-7 cells were transiently transfected with pCMV4 vector alone (lane 1) or pCMV4 expressing c-Src (lane 2), E378G (lane 3), Myc-tagged ubiquitin (lane 4) alone, or in the combinations indicated (lanes 5–8). For each transfection, 1 mg of lysate was immunoprecipitated with polyclonal antibody (PAb) N16 (anti-Src), separated by SDS/PAGE, transferred to PVDF membrane, and probed with mAb 9E10 (anti-myc). Cells used for lanes 6 and 8 were treated with 50 μM Z-L3VS for 14 hr before harvesting. * indicates a background band. (B) csk−/− cells were treated with either 50 μM Z-L3VS in DMSO or DMSO alone as a negative control. One milligram of lysate was immunoprecipitated with PAb N16 (anti-Src), separated by SDS/PAGE, transferred to PVDF membrane, and probed with mAb Ubi-1 (anti-ubiquitin).
Figure 6.
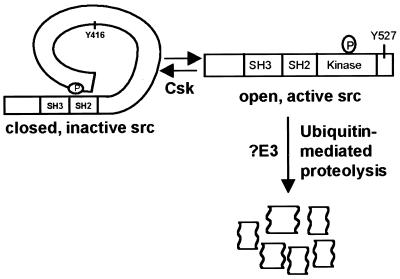
Proposed model for the targeted degradation of active Src. Phosphorylation of Tyr-527 maintains Src in a “closed” inactive conformation. Dephosphorylation of pTyr-527 and autophosphorylation of Tyr-416 releases the kinase into an open, fully active state. The active form of Src is specifically targeted for ubiquitin-mediated proteolysis. The E3 ubiquitin-protein ligase that recognizes active Src has not yet been identified.
Discussion
Our laboratory recently has shown that activated forms of the Src family member Blk are specifically degraded by ubiquitin-mediated proteolysis (24). In the present study we have extended this observation to Src itself and have examined whether this mechanism of regulation is a property of other members of this kinase family. Using csk−/− cells, which lack the negative regulator of Src, and mutant forms of Src as sources of active kinase, we demonstrated a decrease in the steady-state levels of the active forms of Src. Pulse–chase experiments revealed shorter half-lives for activated forms of c-Src, suggesting that the decreased protein levels were the result of increased protein turnover. In contrast, Src mutants fully or partially impaired in their catalytic activity were quite stable and were present at higher steady-state levels. Proteasome inhibitors stabilized only the active forms of Src, implicating the proteasome machinery in the proteolysis of the active forms of Src. We also demonstrated the in vivo ubiquitination of kinase active Src, thus establishing the involvement of ubiquitination in the regulation of Src.
Regulation of Src by ubiquitin-mediated degradation provides a previously unrecognized mechanism for limiting Src activity. A major switch for the activation and inactivation of Src involves the phosphorylation and dephosphorylation of tyrosine residues within the C terminus and kinase domains of the protein. Regulation by reversible phosphorylation allows the kinase to alternate between active and inactive states. The specific proteolysis of the kinase-active forms of Src by ubiquitination as demonstrated in this manuscript provides an alternative mechanism for restricting Src kinase activity. The irreversible nature of ubiquitin-mediated proteolysis allows the cell to control and attenuate the activity of a mitogenic protein.
Although ubiquitin-mediated degradation has not previously been shown to be a mechanism for regulation of Src activity, the correlation between increased activity and decreased protein stability has been noted previously. Using NIH 3T3 cells stably expressing various Src constructs, Kmiecik and Shalloway (20) demonstrated that v-Src and c-SrcY527F have higher specific kinase activities and significantly lower steady-state protein levels than c-Src. In addition, other studies have shown that v-Src has a short-half life that varies among different viral strains, suggesting that the stability of v-Src depends on mutations unique to each strain. Protein synthesis inhibitors greatly increased the half-life of v-Src in these studies, indicating that additional proteins are required for v-Src degradation (38, 39).
Unregulated, constitutive activation of Src induces cellular transformation presumably through activation of mitogenic and cellular adhesion pathways. In most cases, transformation by Src occurs only when activated forms of the kinase are present; however, there are data suggesting that elevated levels of c-Src are transforming. Johnson et al. (40) found that c-Src transfected into NIH 3T3 cells could induce low levels of foci. When these transformed foci were examined, Src levels were consistently higher than those found in transfected, nontransformed NIH 3T3 cells (average ratio of 5:1). Confirmation of the wild-type status of the c-Src in these foci led these researchers to propose that a level of c-Src above a certain threshold was oncogenic.
Defects in Src ubiquitination could cause Src protein levels to exceed a “threshold” level and result in cellular transformation. In fact, various human cancers, including breast cancer, exhibit elevated Src levels despite the absence of any detectable Src mutations (41). Elevated levels of Src in these cancers possibly could result from mutations in genes that regulate Src levels, possibly in those that participate in the proper targeting of Src to the ubiquitin proteasome pathway.
Although we have established that the ubiquitin/proteasome pathway is involved in degrading activated Src, the specific enzymes involved in the process have yet to be determined. The E3 ubiquitin-protein ligases are the critical enzymes responsible for substrate recognition by the ubiquitin machinery (for review see ref. 42). E6AP was the first mammalian E3 enzyme to be identified (26). In cooperation with the human papillomavirus E6 protein, E6AP mediates the ubiquitination of p53 (25, 26). E6AP is the prototype of a family of E3 proteins known as HECT domain proteins (homology to E6AP carboxyl terminus), all of which share similarities in their catalytic domains (43, 44). Based on our previous results demonstrating a role for E6AP in targeting Blk for degradation, E6AP, or perhaps a related HECT family member, is a strong candidate for the E3 responsible for recognizing activated forms of Src. In preliminary coimmunoprecipitation experiments, we have detected an interaction between E6AP and Src (K.F.H., E.M.C., and P.M.H., unpublished results). Additional experiments, however, will need to be performed to establish the specific ubiquitin protein ligase that is involved in the degradation of Src.
In addition to identifying the E3 enzyme involved, it will be important to determine the specific degradation signal(s) in the active form of Src that is recognized by the ubiquitin machinery. Activated Src adopts an open conformation in which tyrosine 416 becomes phosphorylated (2). It is possible that either the change in conformation status or the phosphorylation of specific residues such as tyrosine 416 serves as the degradation signal. The stability of c-Src K295R argues for the latter possibility. This mutant can adopt the open conformation, yet remains stable, suggesting that conformation alone is not sufficient to target Src for degradation. There also may be a requirement for interactions with additional cellular proteins before recognition of Src as a substrate for the ubiquitination machinery. These mechanisms are not mutually exclusive and must be carefully explored.
The work presented in this report demonstrates the targeted degradation of active forms of Src by the ubiquitin/proteasome pathway. These results reveal a previously unidentified cellular mechanism for irreversibly limiting Src kinase activity. Further investigation and elucidation of this pathway is necessary to gain a general understanding of the cellular regulation of Src activity and how this regulation may be disrupted in cancer cells.
Acknowledgments
We thank J. Brugge for plasmids expressing wild-type and mutant Src kinases. We thank S. Thomas for anti-Src antibodies and src+/+, src−/−, csk+/+, and csk−/− MEFs and H. Ploegh for Z-L3VS proteasome inhibitor. We are grateful to J. Brugge, A. Hudson, L. Decker, and W. Kao for a critical review of this manuscript. This work was supported by a grant from the National Institutes of Health (R01-CA64888). K.F.H. was supported by a postdoctoral fellowship from the National Institutes of Health/National Cancer Institute (award number 1 F32 CA81727–01). S.K. was supported by a postdoctoral fellowship from the American Cancer Society (award number PF-4309).
Abbreviations
- SH
Src homology
- MEF
mouse embryo fibroblast
- PVDF
poly(vinylidene difluoride)
References
- 1.Thomas S M, Brugge J S. Annu Rev Cell Dev Biol. 1997;13:513–609. doi: 10.1146/annurev.cellbio.13.1.513. [DOI] [PubMed] [Google Scholar]
- 2.Brown M T, Cooper J A. Biochim Biophys Acta. 1996;1287:121–149. doi: 10.1016/0304-419x(96)00003-0. [DOI] [PubMed] [Google Scholar]
- 3.Xu W, Doshi A, Lei M, Eck M J, Harrison S C. Mol Cell. 1999;3:629–638. doi: 10.1016/s1097-2765(00)80356-1. [DOI] [PubMed] [Google Scholar]
- 4.Taylor S J, Shalloway D. Curr Opin Genet Dev. 1993;3:26–34. doi: 10.1016/s0959-437x(05)80337-5. [DOI] [PubMed] [Google Scholar]
- 5.Xu W, Harrison S C, Eck M J. Nature (London) 1997;385:595–602. doi: 10.1038/385595a0. [DOI] [PubMed] [Google Scholar]
- 6.Ralston R, Bishop J M. Proc Natl Acad Sci USA. 1985;82:7845–7849. doi: 10.1073/pnas.82.23.7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Twamley-Stein G M, Pepperkok R, Ansorge W, Courtneidge S A. Proc Natl Acad Sci USA. 1993;90:7696–7700. doi: 10.1073/pnas.90.16.7696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roche S, Koegl M, Barone M V, Roussel M F, Courtneidge S A. Mol Cell Biol. 1995;15:1102–1109. doi: 10.1128/mcb.15.2.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barone M V, Courtneidge S A. Nature (London) 1995;378:509–512. doi: 10.1038/378509a0. [DOI] [PubMed] [Google Scholar]
- 10.Chackalaparampil I, Shalloway D. Cell. 1988;52:801–810. doi: 10.1016/0092-8674(88)90422-9. [DOI] [PubMed] [Google Scholar]
- 11.Fumagalli S, Totty N F, Hsuan J J, Courtneidge S A. Nature (London) 1994;368:871–874. doi: 10.1038/368871a0. [DOI] [PubMed] [Google Scholar]
- 12.Roche S, Fumagalli S, Courtneidge S A. Science. 1995;269:1567–1569. doi: 10.1126/science.7545311. [DOI] [PubMed] [Google Scholar]
- 13.Klinghoffer R A, Sachsenmaier C, Cooper J A, Soriano P. EMBO J. 1999;18:2459–2471. doi: 10.1093/emboj/18.9.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Juliano R L, Haskill S. J Cell Biol. 1993;120:577–585. doi: 10.1083/jcb.120.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwartz M A. Cancer Res. 1993;53:1503–1506. [PubMed] [Google Scholar]
- 16.Hanks S K, Polte T R. BioEssays. 1997;19:137–145. doi: 10.1002/bies.950190208. [DOI] [PubMed] [Google Scholar]
- 17.Takeya T, Hanafusa H. Cell. 1983;32:881–890. doi: 10.1016/0092-8674(83)90073-9. [DOI] [PubMed] [Google Scholar]
- 18.Hunter T. Cell. 1987;49:1–4. doi: 10.1016/0092-8674(87)90745-8. [DOI] [PubMed] [Google Scholar]
- 19.Cartwright C A, Eckhart W, Simon S, Kaplan P L. Cell. 1987;49:83–91. doi: 10.1016/0092-8674(87)90758-6. [DOI] [PubMed] [Google Scholar]
- 20.Kmiecik T E, Shalloway D. Cell. 1987;49:65–73. doi: 10.1016/0092-8674(87)90756-2. [DOI] [PubMed] [Google Scholar]
- 21.Piwnica-Worms H, Saunders K B, Roberts T M, Smith A E, Cheng S H. Cell. 1987;49:75–82. doi: 10.1016/0092-8674(87)90757-4. [DOI] [PubMed] [Google Scholar]
- 22.Levy J B, Iba H, Hanafusa H. Proc Natl Acad Sci USA. 1986;83:4228–4232. doi: 10.1073/pnas.83.12.4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bjorge J D, Bellagamba C, Cheng H C, Tanaka A, Wang J H, Fujita D J. J Biol Chem. 1995;270:24222–24228. doi: 10.1074/jbc.270.41.24222. [DOI] [PubMed] [Google Scholar]
- 24.Oda H, Kumar S, Howley P M. Proc Nat Acad Sci USA. 1999;96:9557–9562. doi: 10.1073/pnas.96.17.9557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huibregtse J M, Scheffner M, Howley P M. Mol Cell Biol. 1993;13:775–784. doi: 10.1128/mcb.13.2.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scheffner M, Huibregtse J M, Vierstra R D, Howley P M. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 27.Snyder M A, Bishop J M, McGrath J P, Levinson A D. Mol Cell Biol. 1985;5:1772–1779. doi: 10.1128/mcb.5.7.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soriano P, Montgomery C, Geske R, Bradley A. Cell. 1991;64:693–702. doi: 10.1016/0092-8674(91)90499-o. [DOI] [PubMed] [Google Scholar]
- 29.Nada S, Yagi T, Takeda H, Tokunaga T, Nakagawa H, Ikawa Y, Okada M, Aizawa S. Cell. 1993;73:1125–1135. doi: 10.1016/0092-8674(93)90642-4. [DOI] [PubMed] [Google Scholar]
- 30.Bogyo M, McMaster J S, Gaczynska M, Tortorella D, Goldberg A L, Ploegh H. Proc Natl Acad Sci USA. 1997;94:6629–6634. doi: 10.1073/pnas.94.13.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Imamoto A, Soriano P. Cell. 1993;73:1117–1124. doi: 10.1016/0092-8674(93)90641-3. [DOI] [PubMed] [Google Scholar]
- 32.Nada S, Okada M, MacAuley A, Cooper J A, Nakagawa H. Nature (London) 1991;351:69–72. doi: 10.1038/351069a0. [DOI] [PubMed] [Google Scholar]
- 33.Iba H, Cross F R, Garber E A, Hanafusa H. Mol Cell Biol. 1985;5:1058–1066. doi: 10.1128/mcb.5.5.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirai H, Varmus H E. Mol Cell Biol. 1990;10:1307–1318. doi: 10.1128/mcb.10.4.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ward C L, Omura S, Kopito R R. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- 36.Hofmann F, Martelli F, Livingston D M, Wang Z. Genes Dev. 1996;10:2949–2959. doi: 10.1101/gad.10.23.2949. [DOI] [PubMed] [Google Scholar]
- 37.Ellison M J, Hochstrasser M. J Biol Chem. 1991;266:21150–21157. [PubMed] [Google Scholar]
- 38.Ziemiecki A, Friis R R, Bauer H. Mol Cell Biol. 1982;2:355–360. doi: 10.1128/mcb.2.4.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sefton B M, Patschinsky T, Berdot C, Hunter T, Elliott T. J Virol. 1982;41:813–820. doi: 10.1128/jvi.41.3.813-820.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson P J, Coussens P M, Danko A V, Shalloway D. Mol Cell Biol. 1985;5:1073–1083. doi: 10.1128/mcb.5.5.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kolibaba K S, Druker B J. Biochim Biophys Acta. 1997;1333:F217–F248. doi: 10.1016/s0304-419x(97)00022-x. [DOI] [PubMed] [Google Scholar]
- 42.Ciechanover A, Schwartz A L. FASEB J. 1994;8:182–191. doi: 10.1096/fasebj.8.2.8119489. [DOI] [PubMed] [Google Scholar]
- 43.Huibregtse J M, Scheffner M, Beaudenon S, Howley P. Proc Natl Acad Sci USA. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huibregtse J M, Scheffner M, Howley P M. Cold Spring Harbor Symposia on Quantitative Biology. Vol. 59. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. pp. 237–245. [DOI] [PubMed] [Google Scholar]