Abstract
Quorum sensing (QS) in vitro controls production of plant cell wall degrading enzymes (PCWDEs) and other virulence factors in the soft rotting enterobacterial plant pathogen Pectobacterium atrosepticum (Pba). Here, we demonstrate the genome-wide regulatory role of QS in vivo during the Pba–potato interaction, using a Pba-specific microarray. We show that 26% of the Pba genome exhibited differential transcription in a QS (expI-) mutant, compared to the wild-type, suggesting that QS may make a greater contribution to pathogenesis than previously thought. We identify novel components of the QS regulon, including the Type I and II secretion systems, which are involved in the secretion of PCWDEs; a novel Type VI secretion system (T6SS) and its predicted substrates Hcp and VgrG; more than 70 known or putative regulators, some of which have been demonstrated to control pathogenesis and, remarkably, the Type III secretion system and associated effector proteins, and coronafacoyl-amide conjugates, both of which play roles in the manipulation of plant defences. We show that the T6SS and a novel potential regulator, VirS, are required for full virulence in Pba, and propose a model placing QS at the apex of a regulatory hierarchy controlling the later stages of disease progression in Pba. Our findings indicate that QS is a master regulator of phytopathogenesis, controlling multiple other regulators that, in turn, co-ordinately regulate genes associated with manipulation of host defences in concert with the destructive arsenal of PCWDEs that manifest the soft rot disease phenotype.
Author Summary
Many Gram-negative bacteria use a population density-dependent regulatory mechanism called quorum sensing (QS) to control the production of virulence factors during infection. In the bacterial plant pathogen Pectobacterium atrosepticum (formerly Erwinia carotovora subsp. atroseptica), an important model for QS, this mechanism regulates production of enzymes that physically attack the host plant cell wall. This study used a whole genome microarray-based approach to investigate the entire QS regulon during plant infection. Results demonstrate that QS regulates a much wider set of essential virulence factors than was previously appreciated. These include virulence factors similar to those in other plant and animal pathogens that have not previously been associated with QS, e.g., a Type VI secretion system (and its potential substrates), shown for the first time to be required for virulence in a plant pathogen; and the plant toxin coronafacic acid, known in other pathogens to play a role in manipulating plant defences. This study provides the first evidence that Pectobacterium may target host defences simultaneously with a physical attack on the plant cell wall. Moreover, the study demonstrates that a wide range of previously known and unknown virulence regulators lie within the QS regulon, revealing it to be the master regulator of virulence.
Introduction
Quorum sensing (QS) is a population density-dependent regulatory mechanism, utilising freely diffusible chemical signal molecules, which controls a wide range of phenotypes in many different bacteria [1]. The best-studied QS systems are those utilising N-acyl-homoserine lactone (AHL) signal molecules, synthesised by LuxI homologues. AHL concentration increases with bacterial population growth until, at high cell density, a threshold level of signal is reached. This is detected by AHL binding to receptor proteins, LuxR-family transcriptional regulators, resulting in altered gene expression [2]. QS plays an essential role in the pathogenesis of many bacterial pathogens of both plants and animals. Amongst the best studied AHL QS systems are those of the soft rotting enterobacterial plant pathogens Pectobacterium atrosepticum (Pba) and Pectobacterium carotovorum subsp. carotovorum (Pcc; formerly Erwinia carotovora subsp. atroseptica and E. c. subsp. carotovorum respectively) [3]. These pathogens cause disease primarily through the coordinate and prolific production of a variety of plant cell wall degrading enzymes (PCWDEs), which are secreted to the extracellular environment through the Type I (protease) and Type II (pectinases and cellulases) secretion systems [4]. However, they also possess a Type III secretion system (T3SS) with cognate effector (DspA/E) and helper/harpin proteins (HrpN/HrpW), which is required for full virulence [5]. While the role of the T3SS in the soft rotting pathogens remains to be elucidated, in the closely-related E. amylovora, DspA/E has been reported to interact with leucine-rich repeat receptor-like protein kinases (LLR-RLKs) of apple plants, implying a role in the manipulation of host defences [6]. QS in pectobacteria has been reported to regulate PCWDEs [7], the Type III secreted harpin HrpN [8], and other virulence factors, including Nip and Svx [9]–[11], a very small number of virulence regulators (expR, rsmA and virR) [12]–[14], and the antibiotic carbapenem [15]. These are controlled by the AHL, N-(3-oxohexanoyl)-L-homoserine lactone (OHHL), synthesised by ExpI. Different strains of pectobacteria possess up to three homologues of LuxR [16] including: VirR, which plays a central role in the repression of QS-regulated virulence factors [12]; CarR, which regulates the production of carbapenem [15]; and ExpR, which activates transcription of the global repressor, rsmA, in the absence of AHL [13].
Until now, studies on QS in pectobacteria have largely been in vitro and have examined its role in the regulation of targeted virulence factors, particularly PCWDEs. Such virulence factors are thought to operate as part of a necrotrophic mode of action (where the invading organism causes death of host tissue and colonises dead substrate). As a consequence, this group of pathogens have been termed “brute force” in line with this physical attack on plant cell walls. This is in contrast to pathogens such as Pseudomonas syringae, which are hemibiotrophic (requiring living host tissue as part of the infection process, during which they actively manipulate host defences) and, due to their ability to manipulate plant defences as part of the infection process, have been termed “stealth” pathogens. In the pectobacteria, it has been hypothesised that QS acts to delay the onset of PCWDE production until sufficient numbers of cells are present to overcome plant defences, which are induced by the formation of cell wall breakdown products [17],[18]. However, in previous work we showed that premature addition of OHHL to potato plants infected with low numbers of Pba induced early disease development [19], suggesting that this hypothesis may be an over-simplification of a more complex process. In addition, the full genome sequence of Pba strain SCRI1043 (Pba1043) has revealed many additional putative virulence determinants, including coronafacoyl-amide conjugates and homologues of the hemolysin-co-regulated protein (Hcp) and Rhs accessory element VgrG [20]. In Pseudomonas syringae, coronafacoyl-amide conjugates promote disease development and, together with the T3SS, may act to suppress salicylic acid-based defences as part of this process [21],[22]. Hcp and VgrG have been associated with virulence in animal pathogens and are potential effector proteins delivered through a Type VI secretion system (T6SS) [23]–[26]. Hcp and VgrG homologues were recently detected in the secretome of Pba1043 and over-expression of hcp1 increased Pba virulence, suggesting that this and other hcp family members are virulence determinants [27]. The presence of such determinants in Pba suggests that the pectobacteria may also act in a stealth-like manner by manipulating resistance during the infection process. However, whether these determinants are produced and act independently, or together with PCWDEs as part of a coordinated assault on the plant, is unknown. We developed a whole genome microarray for Pba1043 and report its use to study gene expression from an expI mutant of Pba1043 grown in planta, to determine global effects of QS on gene regulation during potato infection, with particular emphasis on the relationship between PCWDEs and possible stealth mechanisms.
Results/Discussion
A Mutation in expI Reduces Virulence and OHHL Production
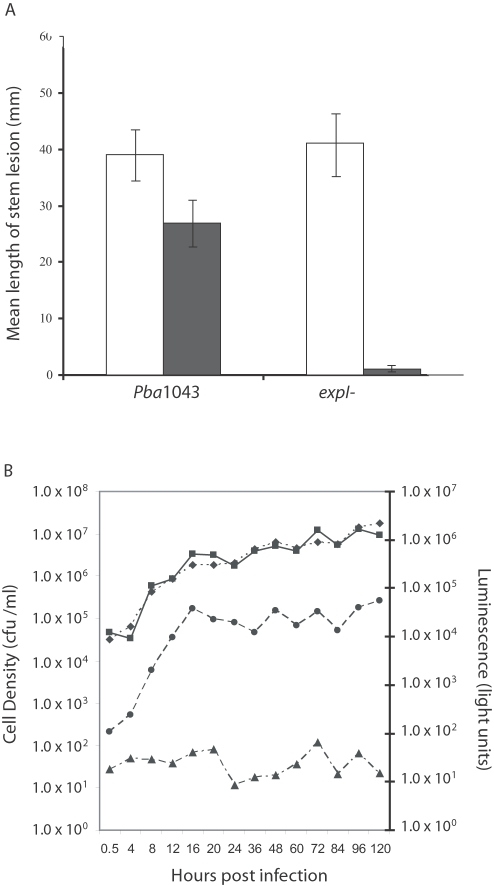
The expI gene and ExpI product, OHHL, are required for full virulence in Pba and Pcc [7],[12],[28]. The virulence of an expI (ECA0105) mutant was significantly reduced on both potato stems and tubers and was restored following complementation with the expI gene in trans (Fig. S1). To confirm that virulence could be restored in planta by the presence of OHHL, the expI mutant strain was inoculated at low cell densities into an OHHL-producing transgenic potato plant [19], where virulence was restored compared to inoculation on a non-transgenic control plant (Fig. 1A). This supports previous work where the presence of OHHL in these transgenic plants induced early disease development from low cell densities (102 cells per inoculation site) of both WT and expI mutant strains [19].
Figure 1. OHHL is Required for Virulence in Potato Stems and Peaks at 16 hpi in P. atrosepticum-infected Potato Tubers.
(A) Lesion development on potato stems following inoculation of the wild type Pba1043 and expI mutant strains. Filled bars = potato cv Desiree control; open bars = OHHL-producing transgenic Desiree (YI5A) [19]. (B) Cell density and OHHL production (in light units) of wild type P. atrosepticum and expI mutant strains in potato tubers over 120 h. Wild type cell density (square); expI mutant cell density (diamond); wild type OHHL production (circle); mutant OHHL production (triangle). Bars show mean +/− standard error of the mean.
A luminescence-based assay was used to monitor OHHL production during growth of the expI mutant and wild type strains in potato tubers (Fig. 1B). Both strains grew at comparable rates over a 120 h infection time course and reached similar population levels. Although the wild type and expI mutant strains would be expected to show differences in growth in natural condition during the course of disease development, the relatively short infection time (120 h) and the method of inoculation (onto the cut tuber surface), may account for the results observed. In the wild type, the level of OHHL rose sharply over the first 16 hours in line with log phase growth, before reaching a plateau at a concentration of approximately 80 µg/ml. At this plateau, bacterial cell density was approximately 5.0×106 cfu/ml, which was similar to previous reports [15]. In the expI mutant, OHHL production remained at background levels. Based on the above data, time points at 12 and 20 hours post inoculation (hpi), i.e. just prior to and just following maximum OHHL synthesis in planta, were selected to study transcriptional changes during QS (Fig. 1B).
Comparison of Pba Wild Type and expI Mutant Transcriptomes in planta
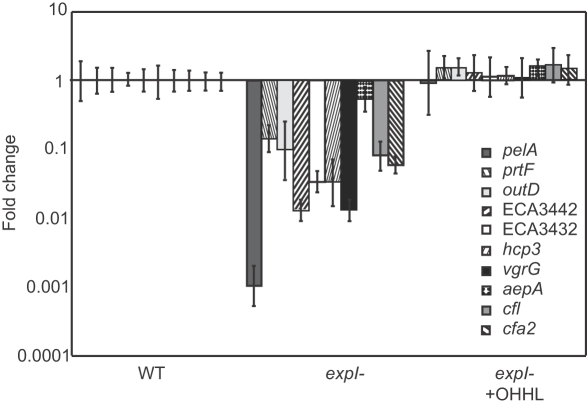
Differential expression of genes (pelA [ECA4067], pelC [ECA4069], celV [ECA1981], prtW [ECA2785], pehA [ECA1095], ECA2220, svx [ECA0931] and nip [ECA3087]) previously shown to be under QS control [9],[10],[12] was investigated using quantitative real-time PCR (qRT-PCR) at 12 and 20 hpi in the expI mutant and wild type strains. In all cases, significant up-regulation of these genes was observed in the wild type only (Table S1). cDNA from the wild type and expI mutant at 12 and 20 hpi was hybridised to the Pba microarray. 1167 coding sequences (CDSs) (approx. 26% of the genome) showed statistically significant differences (P≤0.05) in expression between the expI mutant and wild type (Table S2). 498 CDSs showed reduced transcript abundance (421 at 12 hpi, 169 at 20 hpi, 92 at both time points) and 687 CDSs exhibited increased transcript abundance (551 at 12 hpi, 180 at 20 hpi, 44 at both time points) in the expI mutant compared to the wild type. Microarray comparison of mutant and wild type cDNAs from cells in buffer solution prepared for tuber inoculation following overnight growth in LB to stationary phase (zero time-point), was consistent with there being no overall transcriptional difference (P≤0.05) between the strains prior to plant inoculation (data not shown). Only 16% of CDSs within the horizontally-acquired islands [20] showed differential gene expression, suggesting that such CDSs are less likely to have been incorporated into the QS regulon than those on the chromosome backbone. qRT-PCR was used to study a number of genes in the expI mutant and wild type to examine differential gene expression, either to verify changes observed in the microarray or to examine the effects of a mutation in expI on additional genes (Table S1). Importantly, qRT-PCR analysis of selected genes after growth of the expI mutant and wild type in vitro revealed the same pattern of expI-dependence as observed in vivo, and these changes could be fully complemented by the addition of exogenous OHHL (Fig. 2).
Figure 2. OHHL Complementation of Selected Quorum Sensing-regulated Genes.
RT-PCR analysis of selected genes after 18h growth of wild type P. atrosepticum in Pel Minimal Medium (PMM), and growth of the expI mutant in PMM with and without the addition of OHHL (5 µM final concentration). vgrG = ECA2867. Bars show mean +/− standard deviation.
The role of QS in pathogenesis of pectobacteria has been intensively studied in vitro, particularly for its ability to co-ordinately up-regulate PCWDEs [7]–[10],[12],[14],[28]. Previous work based on enzyme plate assays observed that all major groups of PCWDEs, including pectate lyases (Pel), cellulases (Cel), protease (Prt), pectin lyase (Pnl) polygalacturonase (Peh) and pectin methyl esterase (Pme) were under QS control [8]. In this study, we found that genes encoding all these groups showed lower transcript abundance in the expI mutant compared to the wild type at both 12 and 20 hpi (Table 1, Table S1). The major pectate lyases PelA, PelB (ECA4068) and PelC (ECA4069), as well as CelV (ECA1981), a putative cellulase ECA2220, PrtW (ECA2785) and PehA (ECA1095) have previously been associated with QS in pectobacteria, either through transcriptional or proteomic analyses [7],[10],[12],[28]. However, in addition the transcription (using microarray and/or qRT-PCR analyses) of genes encoding other PCWDEs and their isoforms, including “minor” pectate lyases (PelZ [ECA4070], Pel-3 [ECA1094], PelB and PelW [ECA2402]), CelB (ECA2827) and CelH (ECA3646), PehN (ECA1190), PmeB (ECA0107) and Pnl (ECA1499) was found to be expI-dependent. These results confirm previous observations of QS regulatory control in vitro and validate our in planta approach.
Table 1. Microarray Analysis of Selected P. atrosepticum Genes in planta.
| Gene ID | Gene name | Predicted function | F/C 12h | F/C 20h |
| Plant cell wall degrading enzymes | ||||
| ECA4067* | pelA | Pectate lyase I | −15.9 | −12.5 |
| ECA1094* | pel-3 | Pectate lyase | −54.2 | −43.2 |
| ECA2553 | - | Pectate lyase | −11.2 | −7.4 |
| ECA4068* | pelB | Pectate lyase | −28.6 | −11.4 |
| ECA4070* | pelZ | Pectate lyase | −4.1 | −6.3 |
| ECA2827* | celB | Beta(1,4)-glucan glucanohydrolase | −9.2 | −3.8 |
| ECA1499* | pnl | Pectin lyase | −5.5 | −7.8 |
| Type I secretion | ||||
| ECA2781* | prtF | Protease secretion | −3.1 | −2.8 |
| ECA2782* | prtE | Protease secretion | - | - |
| ECA2783* | prtD | Protease secretion | - | −1.9 |
| Type II secretion | ||||
| ECA3100* | outM | Secretion prot. M | −2.7 | −2.2 |
| ECA3101* | outL | Secretion prot. L | −3.0 | −2.3 |
| ECA3105* | outH | Secretion prot. H | −3.8 | −2.4 |
| ECA3106* | outG | Secretion prot. G | −7.5 | −3.5 |
| ECA3107* | outF | Secretion prot. F | −1.8 | −1.8 |
| ECA3109* | outD | Secretion prot. D | −1.7 | −1.5 |
| Type III secretion | ||||
| ECA2097* | hrpE | Secretion prot. | - | −1.6 |
| ECA2108* | - | Putative lipoprot. | −1.7 | - |
| Type VI secretion and putative substrates | ||||
| ECA3444* | - | Hypothetical prot. | −82.0 | −50.0 |
| ECA3445* | - | Hypothetical prot. | −2.5 | −2.0 |
| ECA2867* | vgrG | VgrG homologues | −2.9 | −4.3 |
| ECA3427* | - | VgrG homologues | −4.8 | −3.6 |
| ECA3420 | - | Hypothetical prot. | −7.7 | −2.6 |
| ECA3430* | - | Putative phospholipase | −3.8 | - |
| ECA3432* | vasK | IcmF-like prot. | −5.0 | −2.5 |
| ECA3433 | - | Hypothetical prot. | −2.6 | −1.8 |
| ECA3436* | vasG | ClpB-like prot. | −4.4 | −3.2 |
| ECA3440 | - | Hypothetical prot. | −11.1 | −5.6 |
| ECA3442* | vasA | Hypothetical prot. | −3.7 | −4.2 |
| ECA3443 | - | Hypothetical prot. | −5.0 | −3.4 |
| ECA4275 | hcp1 | hcpA homologues | −50.0 | −50.0 |
| ECA3428* | hcp2 | |||
| ECA2866* | hcp3 | hcpA homologue | −20.0 | −20.0 |
| ECA0456* | hcp4 | hcpA homologue | −14.3 | −5.9 |
| ECA3672* | - | hcpA homologue | −6.9 | −4.1 |
| ECA0176* | - | hcpA homologue | −21.3 | −12.7 |
| ECA4277 | - | hcpA homologue | −20.0 | −16.7 |
| Regulators | ||||
| ECA0105* | § expI | AHL synthesis | −28.2 | −18.3 |
| ECA0106* | § expR | QS regulator | −9.4 | −8.5 |
| ECA0809 | hexY | Global regulator | −2.4 | −1.5 |
| ECA1022* | aepA | Virulence regulator | −10.5 | −3.7 |
| ECA1561 | § virR | QS regulator | −1.6 | −1.5 |
| ECA1931* | hor | Global regulator | −4.2 | −2.5 |
| ECA2882* | expA | Two component regulator | −2.3 | −1.8 |
| ECA4123 | rexZ | PCWDE regulator | −3.3 | - |
| ECA1562* | virS | TetR regulator | 3.7 | 1.8 |
| ECA2425* | kdgR | Pectin degradation repressor | 2.1 | - |
| ECA2445 | pehR (phoP) | Two component regulator | 2.3 | - |
| ECA3030* | hexA | LysR regulator | 3.5 | 2.1 |
| ECA3366* | § rsmA | Global regulator | 1.9 | - |
| ECA2724* | rscR | LysR regulator | 3.3 | 2.9 |
| ECA3168* | ohrR | Regulator hydro-peroxide resistance | 2.5 | 4.2 |
| ECA1740* | fliZ | Alternative sigma factor regulator | −2.0 | −2.9 |
| ECA2435* | rdgA | Regulator of pectin lyase production | 1.6 | 1.5 |
| Toxin/virulence associated proteins | ||||
| ECA0601 | cfa8A | Putative oxidoreductase | - | −1.8 |
| ECA0607* | cfa2 | Coronafacic acid dehydratase | 1.7 | −2.0 |
| ECA0931* | svx | Putative virulence-associated prot. | −6.9 | −3.3 |
| ECA3087* | nip | Putative virulence-associated prot. | −34.5 | −11.0 |
| ECA3946* | - | Putative exported prot. | −4.6 | −2.0 |
Showing differences in gene transcript abundance in the expI mutant compared with the wild type P. atrosepticum strain at 12 and 20 hours post inoculation. Based on a 1.5 fold statistically significant difference [P value <0.05] in expression. F/C = Fold change; negative value for fold change = genes showing reduced transcript abundance in the expI mutant; * = qRT-PCR data available (see Table S1); § = Regulators previously shown to be quorum sensing controlled in Pectobacterium; - = no observed change in expression by microarray analysis. Prot. = protein.
Other genes previously shown to fall under QS control in vitro, including svx, nip and a gene of unknown function (ECA3946), as well as three regulators (expR [ECA0106], rsmA [ECA3366] and virR [ECA1561]) involved in the production of PCWDEs [9], [10], [12]–[14], also showed reduced transcript abundance in the expI mutant compared to the wild type strain. This again justifies our approach in assessing the genome-wide effects of QS regulation during the potato interaction. While 1167 genes, representing a variety of processes, were found to be differentially expressed in the microarray experiment (Table S2), we focus predominantly on those that display reduced transcript levels in the mutant (as these are presumably induced directly or indirectly by QS), and which also have a known or putative role in virulence (Table 1).
Secretion Systems
To successfully cause disease Pba must secrete a multitude of PCWDEs and other proteins, many of which are under QS control. We observed that both Type I and Type II secretion systems (T1SS and T2SS, respectively), which can be considered as ‘accessory virulence factors’, are modulated by QS (Table 1). Prior to this study the secretion systems responsible for the delivery of these virulence factors had not been reported as QS-regulated, and this observation indicates a novel facet to QS control of pathogenesis in pectobacteria.
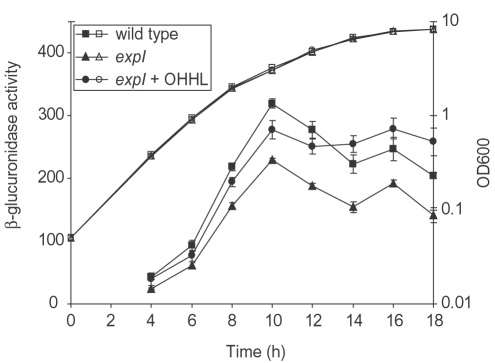
The T2SS is well characterised in pectobacteria and is responsible for secretion of many key virulence factors, e.g. Pel, Cel and Svx [10]. The T2SS of pectobacteria is encoded by a cluster of 15 out genes (ECA3098-3110 and ECA3113-3114) [20],[29], of which six (outMLHGFD) (by microarray analysis) exhibited reduced transcript abundance levels in the expI mutant (Table 1). Analysis of these and seven other out genes by qRT-PCR confirmed that all were expressed at a lower level in the expI mutant (Table S1), implying that expression of the Out T2SS is up-regulated by QS in vivo. Similar QS modulation of out expression was also demonstrated by both qRT-PCR (Table S1) and the use of an outD-gusA reporter fusion in vitro (Fig. 3). In the latter experiment, expression of outD (ECA3109) was reduced in the expI mutant and restored to wild type levels by the exogenous addition of OHHL, confirming QS modulation of out gene expression.
Figure 3. Expression of outD-gusA in vitro is QS-dependent.
β-glucuronidase activity from an outD-gusA reporter fusion was measured in a wild type P. atrosepticum background (wild type, squares), in an expI mutant (expI, triangles) and in an expI mutant with the addition of exogenous 1 µg/ml OHHL (expI+OHHL, circles) throughout growth in PMB. β-glucuronidase activity (solid symbols) is expressed as A405/min/ml/OD600 and growth was measured as OD600 (open symbols). Bars show mean +/− standard error of the mean.
Regulation of the major secreted protease, PrtW, is QS-dependent in Pba [10]. Secretion of Prt by the PrtDEF T1SS is well-characterised in Dickeya dadantii (formerly Erwinia chrysanthemi) [30] and, by analogy, PrtW is expected to be secreted by the T1SS encoded by the neighbouring prtDEF (ECA 2781-2783) genes in Pba. To support this, the microarray data indicated that transcription of the T1SS genes prtDF was reduced in the expI mutant. Use of qRT-PCR confirmed that expression of all three T1SS genes, prtDEF, was significantly reduced in the expI mutant compared to the wild type (Table S1). QS-dependence of the T1SS and T2SS is a logical accompaniment to the simultaneous QS-dependent induction of their substrates, presumably allowing the systems to cope efficiently with the greatly increased quantity of these substrates. Examples of QS-modulated secretion have been reported previously in other pathogens, e.g. the Xcp T2SS of Pseudomonas aeruginosa and the Lip T1SS of Serratia marcescens [31],[32], although this is the first time that QS-dependant secretion systems have been described in pectobacteria.
As well as physically attacking the plant cell wall through the action of PCWDEs, in Pba1043 the Type III secretion system (T3SS) is also necessary for full virulence [5]. The T3SS is found in many Gram-negative pathogens of both animals and plants and is used to translocate effector proteins into host cells, where they manipulate host defences. Helper proteins (or harpins) are secreted to the extracellular environment, and may assist in effector translocation [33]. We observed that expression of the T3SS structural, putative effector and helper genes, and Type III-associated regulators were all modulated by QS. In Pba1043, and other pectobacteria, the T3SS is encoded by the hrp cluster, composed of around 40 CDSs. These CDSs encode components of the structural apparatus, as well as the putative effector DspA/E [ECA2113], and helpers HrpN [ECA2103] and HrpW [ECA2112]. The Pba1043 hrp cluster also contains a group of CDSs (ECA2104-ECA2110), which includes a number of lipoproteins, that appear to be absent in closely-related species [5]. ECA2104 shows homology to vgrG and is described below. In the microarray experiment, two CDSs hrpE [ECA2097], associated with the Type III structural apparatus, and a putative lipoprotein (ECA2108) exhibited decreased transcript abundance in the expI mutant (Table 1). qRT-PCR analysis of these and an additional 17 CDSs subsequently confirmed that CDSs encoding the Type III structural apparatus, the putative effector dspE, helpers hrpN and hrpW, regulators hrpL, hrpS and hrpY, and all CDSs between ECA2104 and ECA2110 were significantly reduced in the expI mutant compared to the wild type, predominantly at 12 h (Table S1). Either positive or negative QS regulation of the T3SS has been observed in other pathogens, e.g. Pseudomonas aeruginosa [34], Vibrio harveyi [35], enteropathogenic E. coli [36], Ralstonia solanacearum [37], and QS regulation of hrpN has been shown in Pba [8]. However, this is the first published evidence that QS plays a role in regulating the entire T3SS and its effectors in the enterobacterial plant pathogens, indicating that co-ordinated physical (PCWDEs) and stealth (T3SS) attacks may be necessary for successful disease development.
Recently, a novel T6SS was described and implicated in pathogenicity in Vibrio cholerae and P. aeruginosa [23],[38]. In V. cholerae, the system is encoded by the VAS locus, genes VCA0107-VCA0123. This locus is one member of a group of conserved gene clusters that are conserved in several pathogens. In both V. cholerae and P. aeruginosa, the T6SS is required for secretion of HcpA and VgrG proteins, although whether these represent putative effectors or simply secreted components of the secretion machinery is not yet clear [23],[38]. In Pba1043, the locus ECA3445–ECA3427 is predicted to encode a VAS-like T6SS and its putative substrates, since these genes encode proteins very similar to those encoded by VCA0107-VCA0123 and is similarly arranged on the chromosome. Microarray analysis indicated that 11 of the 18 genes were expressed at significantly lower levels in the expI mutant (Table 1), and so transcription of the T6SS also appears to be QS-dependent. The modulated genes included Pba homologues of VCA0120, VCA0116 and VCA0110, which are required for Type VI secretion in V. cholerae and/or P. aeruginosa [23],[38]. Moreover, the expression of several predicted T6SS substrates, i.e. encoded by hcpA and vgrG-like genes, was also found to be QS-dependent (Table 1). There are seven hcpA homologues in Pba, three of which (ECA4275[hcp1], ECA3428[hcp2] and ECA2866[hcp3]) are highly similar [20],[27]. ECA3428 and ECA4275 are sufficiently similar that it was not possible to design probes specific to each locus. Nevertheless, the probe detecting expression of both these genes showed decreased transcript abundance in expression in the expI mutant, indicating QS-dependent regulation. Expression of ECA2866 and four other homologues (ECA0456[hcp4], ECA3672, ECA0176 and ECA4277) was also decreased (Table 1). A combination of microarray analysis and qRT-PCR indicated reduced transcript abundance in the expI mutant of all five vgrG homologues, ECA2867, ECA3427, ECA2104, ECA4142 and ECA4276 in the Pba1043 genome (Table S1). Previous work showed that Hcp1-4 and a VgrG homologue (ECA3427) were found in the secretome of Pba1043. Over-expression of Hcp1 increased virulence, suggesting that this and related proteins are virulence factors in pectobacteria [27]. The VAS-like T6SS genes, ECA3445-ECA3427, appear to constitute an operon that may extend for a further seven CDSs (ECA3426-ECA3420). Of these, expression of six was reduced in the expI mutant (Table S2), raising the possibility that they may encode T6SS-dependent effectors.
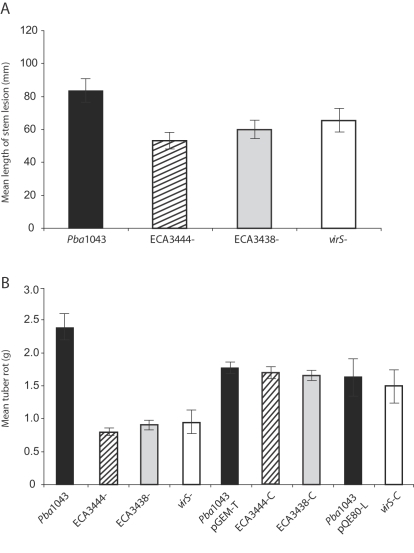
As the T6SS is clearly important for virulence in other pathogens, and a predicted substrate (Hcp1) affects virulence in Pba, we investigated whether the putative T6SS plays a role in virulence in Pba. Mutants in ECA3438 and ECA3444, when tested in potato stem and tuber virulence assays, both showed significantly reduced virulence compared with the wild type (Fig. 4). In tuber tests, complementation of the mutants in trans was shown to return virulence to wild type levels (Fig. 4B). Our results indicate, for the first time in any pathogen, a role for QS in the regulation of the T6SS and its putative substrates. It also demonstrates that the T6SS in Pba plays a role in pathogenesis, which appears to act in conjunction with PCWDE, the T3SS and other virulence determinants during the QS process.
Figure 4. The Regulator virS and Components of the Type VI Secretion System are Involved in Virulence.
Virulence assays in potato stems (A) and potato tubers (B), following inoculation of wild type P. atrosepticum and mutants affected in virS and Type VI secretion (ECA3444 and ECA3438). Complementation of tuber rotting phenotype using plasmids pGEM-T (ECA3444 and ECA3438) and pQE80-L (virS) (B). “C” indicates complemented. Bars show mean +/- standard error of the mean.
Regulators
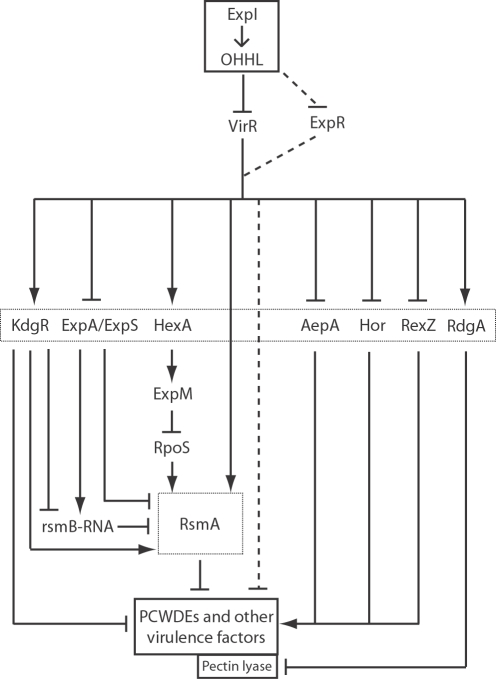
Microarray analysis revealed the QS-dependent differential expression of at least 79 CDSs with either known or putative regulatory functions (Table S2). Twelve CDSs, five of which showed enhanced (hexA [ECA3030], kdgR [ECA2425], phoP[pehR] [ECA2445], rdgA [ECA2435] and rsmA) and seven of which showed reduced (aepA [ECA1022], expA [ECA2882], expR, hexY [ECA0809], hor [ECA1931], rexZ [ECA4123] and virR) transcript abundance in the expI mutant, are known to regulate PCWDEs production and are required for full virulence in pectobacteria [4], [12]–[14],[39] (Table 1). However, only three (expR, rsmA and virR) have previously been shown, in vitro, to fall under QS control [12]–[14]. Three CDSs (hrpL [ECA2087], hrpY [ECA2089] and hrpS [ECA2090]) involved in the regulation of the T3SS in pectobacteria and other phytopathogens [40] also showed decreased transcript abundance in the expI mutant. As all 15 of these CDSs are QS-dependent, this places QS at the apex of a regulatory hierarchy controlling both PCWDEs and the T3SS with its cognate effector proteins. Other QS-controlled regulators are also likely to be important during interaction with the plant (see below). Although QS is central to pathogenesis, elucidating the hierarchical relationships between “subordinate” regulators presents a particular challenge due to the lack of data on such relationships in this particular strain. Several virulence regulators in pectobacteria are known to operate though the Rsm system, which plays a major role in controlling virulence [14]. While not investigated as part of this work, it is highly likely that at least some of the regulators identified in this study operate through this system. Nevertheless, we have still been able to add considerable new information to existing regulatory models [41] and propose an extended model for virulence in the pectobacteria (Fig. 5).
Figure 5. Schematic Model of the Hierarchical Relationships Between ExpI, Other Regulators and Virulence Factors in Pectobacterium.
OHHL is believed to act via antagonism of the LuxR-family AHL-responsive transcriptional regulator, VirR, in turn activating or repressing numerous other regulators that control virulence factor gene expression. A second LuxR-type repressor, ExpR, may also contribute to OHHL regulation of downstream genes. This model represents an integrated summary of regulatory data derived from the study of multiple Pectobacterium strains, and is unlikely to apply in every aspect to all such strains. Adapted from von Bodman et al., [41] using additional information from Barnard and Salmond [16], and this study. Solid arrowheads indicate activation and bars indicate repression. Dotted lines indicate uncertain and/or strain dependent effects.
In addition to regulators previously characterised in pectobacteria, differential expression of 18 further CDSs were found that are similar to a diverse range of transcriptional regulators in other bacteria (Table S2). These include CDSs with putative regulatory functions in nitrogen signal transduction and assimilation (citB [ECA2578], glnB [ECA3254], nac [ECA4483]), hydrogenase activity (hypA [ECA1235]), oxygen sensing (fnr [ECA2207]), defence against superoxides and other stress responses (ohrR [ECA3168], phoB [ECA1110], recX [ECA3368], rseB [ECA3282], rseC [ECA3281]), motility (flgM [ECA1700], fliZ [ECA1740]) and survival in soil (sftR [ECA4305]) (Table S2). Three of these additional regulators (fliZ, ohrR and rscR) have been implicated in virulence in other bacterial pathogens (Table 1) [42]–[44]. However, it does not necessarily follow that homologous regulatory proteins in bacteria are responsible for regulation of homologous processes [45].
Many CDSs encoding putative regulators of unknown function were shown to be regulated by QS. These CDSs thus represent novel candidates for virulence factors. Expression of one such CDS, ECA1562, subsequently named virS, was enhanced in the expI mutant at 12 and 20 hpi and is thus proposed to be repressed by QS (Table 1). VirS is a predicted TetR-family transcriptional regulator whose target(s) is unknown, although its closest reported homologue is a TetR family regulator, TvrR, implicated in virulence in the plant pathogen Pseudomonas syringae pv. tomato [46]. virS is located adjacent to the gene encoding a key QS-controlled regulator, VirR (ECA1561, [12]). However, inactivation of virS does not affect transcription of virR (data not shown). In order to determine whether virS plays a role in virulence, a defined virS mutant was constructed and tested in stem and tubers virulence assay. The virS mutant showed significantly reduced lesion formation compared with the wild type (Fig. 4) and is thus a novel virulence factor in Pba. In tuber tests, complementation of the mutant in trans returned virulence to wild type levels (Fig. 4B). The precise role of virS in planta is under investigation.
Phytotoxins
The microarray data revealed a small reduction in expression of genes cfa2 (ECA0607) and cfa8A (ECA0601) in the expI mutant compared to the wild type (Table 1). These genes are of particular interest as they are part of a cluster responsible for the synthesis of coronafacic acid (CFA) which, in Pseudomonas syringae, is a component of the phytotoxin coronatine [47]. We showed previously that mutations in this cluster (cfa6 [ECA0603] and cfa7 [ECA0602]) significantly reduce pathogenicity of Pba1043 on potato stems [20]. Transcriptional changes in cfa2, cfa6 and cfa7, compared to a QS up-regulated (pelA) control, were thus examined at 12 and 20 hpi using qRT-PCR. At both time-points, pelA, the cfa genes, and the cfl (ECA0609) gene (involved in the formation of coronafacoyl conjugates by ligation of amino acids to CFA) showed reduced expression in the expI mutant, indicating that they are all under QS control (Table S1).
Salicylic acid (SA) and jasmonic acid (JA) are signalling molecules that play major roles in the activation of plant defences against pathogen attack [48]. CFA and its amino acid conjugates appear to act as structural and functional analogues of JA and its conjugates [49]. Recent work by Uppalapati et al. [21] showed that Pseudomonas syringae DC3000 mutants lacking CFA and/or coronatine were impaired in their ability to persist in tomato plants at the later stages of infection, and that the ability to persist coincided with the activation of JA-based, and concomitant suppression of SA-based, defences. It is hypothesised that, through this suppression of SA-mediated defences, coronafacoyl conjugates may aid P. syringae to enter the necrotrophic phase of infection and promote disease symptoms. It would appear therefore that Pba, through QS, synthesises CFA and coronafacoyl conjugates co-ordinately with multiple PCWDEs, the T3SS and T6SS in a synchronised assault on the plant as it progresses from biotrophy to necrotrophy. Although the effect of Pba-encoded CFA conjugates on plant defences has yet to be determined, such a two-pronged attack may be necessary for Pba to establish disease. It will be interesting to determine whether QS plays a similar role in P. syringae and related pathogens.
Conclusions
QS regulation in pectobacteria was observed originally in vitro through dramatic impacts on PCWDE production, pathogenesis and (in Pcc) carbapenem antibiotic production [16]. The microarray analysis of global gene expression in planta presented here indicates a far broader physiological impact of QS, uncovering effects on the expression of many other genes associated with pathogenesis, and on other physiological processes not necessarily connected to plant pathogenesis. As QS is AHL concentration-dependent, its impact is likely to be greatest towards the latter stages of infection, where large quantities of PCWDEs are induced to attack plant cells and the characteristic soft rot disease symptoms occur [4]. Correspondingly, we find that production of the T1SS and T2SS, which are involved in the secretion of PCWDEs to the extracellular environment, are also under QS control.
A very small number of virulence regulators have previously been shown to fall under QS control. Our study has added over 70 other regulators to this list, including the major known virulence regulators associated with PCWDE production in pectobacteria. An important inference from these microarray analyses is that the QS control system occupies a critical position in the regulatory hierarchy and that multiple downstream regulators, some which may operate through the Rsm system [14], are under QS control. Furthermore, QS is seen to have both positive (activation) and negative (repression) effects on its downstream targets. Our knowledge of the hierarchical chain of command in control of the complex regulatory systems of PCWDE and other virulence factors is fragmentary, in part because the current literature describes experimental data derived from multiple strains of Pba, Pcc, and Dickeya spp. It may not therefore be completely legitimate to assume that the identified regulators play conserved roles in these different bacterial strains [45]. Nevertheless, while accepting this caveat, our results are consistent with the notion that, within the infected potato plant, QS acts as a key “master regulator” sensory system in this phytopathogen (Fig. 5). Additionally, regulators associated with virulence in other bacteria, and many novel putative regulators have also been identified; including VirS, which has been associated with virulence in this study.
In addition to the T1SS and T2SS, we have identified a T6SS in Pba and shown, for the first time in a plant pathogen, that it has a role in virulence. Moreover, we have described the first example of a QS-controlled T6SS in any pathogen. The precise functional roles of Hcp, VgrG and other possible Type VI substrates is unknown, but their proposed functions as effector proteins may be important for manipulating host defences whilst PCWDEs mount a simultaneous physical attack on plant cell walls. This does appear to be the case for both the T3SS and associated effectors, and coronafacoyl-amide conjugates, which are similarly QS-dependent, and consequently may suppress or otherwise manipulate defences. This has important implications for the infection process in pectobacteria, as it suggests that these pathogens do not infect merely by “brute force”, where the action of PCWDEs alone is sufficient to overwhelm plant defences and break down plant cell walls towards the end of infection. It seems increasingly likely that, in conjunction with PCWDEs, the production of virulence determinants that actively suppress plant defences, may be necessary to facilitate the transition from biotrophy to necrotrophy during disease development.
Materials and Methods
Bacterial strains, media and pathogenicity assays
Pba1043 [20], and strains with mutations in expI, ECA3438, ECA3444 and virS were used in this study. The expI mutant was derived from phage M1-mediated transduction of expI::mTn5gusAgfp from mutant MC3 into the wild type strain [10]. Mutants ECA3438 and ECA3444 were isolated from a mutation library of Pba1043 [5]. For inactivation of virS, 1085 bp of virS and surrounding regions were PCR-amplified using primers SC51 (ATTTGGATCCGTTGTTCCTGTTCTGTCG) and SC52 (TATATCTAGAGTTTACTGAGCAAGCGACG) and cloned into pBluescript-II KS+ using BamHI-XbaI sites. The KnR cassette from pACYC177 (NEB) was cloned into the NsiI site in the middle of virS. The resulting virS::KnR fragment was then cloned into the suicide vector, pKNG101 [50], generating the marker-exchange plasmid. The plasmid was introduced into Pba1043 by conjugation and transconjugants, resulting from integration of the suicide plasmid into the chromosome by homologous recombination, were selected by ability to grow on minimal medium containing 0.2% glucose+streptomycin. Following overnight growth in the absence of antibiotic selection, exconjugants, in which resolution of the plasmid from the chromosome leaving only the disrupted allele had occurred, were selected by ability to grow on minimal medium containing kanamycin+10% sucrose as sole carbon source and inability to grow on streptomycin. The disruption of the locus was confirmed by PCR analysis and DNA sequencing. All strains were maintained on Luria Bertani (LB) agar supplemented with kanamycin (50 µg/ml) and, unless stated otherwise, were cultured in 10 ml LB broth at 27°C overnight with aeration. Mutations were transduced into a clean Pba1043 background using phage M1 [51]. Pathogenicity tests were performed both on potato stems and tubers [19]. Approx. 102 and 104 cells per inoculation site were used for stems and tubers, respectively. Complementation of mutant strains was carried out in trans following cloning of ECA3444, ECA3438 and expI into plasmid pGEM-T (Promega, Southampton, UK) and virS into pQE80-L (Qiagen, Crawley, UK) with their own ribosome binding sites. OHHL-producing transgenic potato plants used for in planta OHHL complementation are as described [19]. GENSTAT for Windows was used for statistical analyses [20].
In planta RNA preparation and microarrays
Wild type and expI mutant strains were grown in 10 ml LB broth at 27°C with aeration to stationary phase (approx. 1.0×109 cells/ml) and re-suspended in 10 mM Mg SO4 prior to inoculation into sterilized potato tubers (2×107 cells/ml into cv Maris Piper). The tubers were then wrapped in cling film and placed in a tray with wetted tissue to retain high humidity before incubation at 19°C in the dark. At 12 and 20 hours post inoculation (hpi), the bacterial cells were isolated from the tuber by scraping infected tissue into sterilised water. Starch was removed by centrifugation twice at 1000 rpm for 1 min. The bacterial cells in the supernatant were transferred to RNA stabilization buffer containing 1% phenol (pH 4.3, v/v) and 20% ethanol (v/v) and incubated on ice for at least 30 min. Total RNA was isolated using the SV Total RNA Isolation System (Promega) as described by the manufacturer and quantified using a NanoDrop ND-100 spectrophotometer (NanoDrop Technologies, Wilminton, DE). The quality of RNA was analyzed using an Agilent Bioanalyzer 2100 electrophoresis system (Agilent Technologies Inc., West Lothian, UK). In total 12 µg RNA was reverse transcribed and cDNA labelled [52]. 60-mer oligonucleotide probes were designed to Pba CDSs and used, together with controls, to generate 11K custom arrays with 99.5% genome coverage (Agilent, Inc., Santa Clara, CA, USA) [53]. Microarrays were carried out in triplicate for each time point [52],[53]. All microarray images were visually assessed for quality prior to feature extraction, whereby standard probe QC standards were applied (see further information in ArrayExpress-http://www.ebi.ac.uk/microarray-as/aer/). Features flagged as poor were removed prior to importing into Genespring software. Box plots and principle components analysis of whole datasets were used to assess array to array variation. Any outlying microarrays were repeated as necessary.
Data analysis
Microarray data were analysed using GeneSpring software (version 7.2) and normalized using the Lowess algorithm (Agilent Technologies Inc.). Gene expression was considered to be different between the wild type and expI mutant strains for a probe if there was at least 1.5 fold change in normalised hybridisation score, and that change showed a statistically significant (Student's t test: P value <0.05) difference in their normalized data. Microarray data were submitted to the ArrayExpress repository (http://www.ebi.ac.uk/microarray-as/aer/), submission E-TABM-384, including details of the SCRI Pectobacterium atrosepticum 11k array (submission A-MEXP-942).
RT-PCR and Gus expression
qRT-PCR was performed using recA as an endogenous control to validate differences in expression of genes identified from the microarray experiment, and to test additional genes in the Pba genome. RNA samples were analysed in triplicate. 5 µg total RNA was used to synthesize cDNA and 1 µl diluted template DNA (1:10) was used in a reaction of 25 µl containing 1x SYBR Green PCR Master Mix (Qiagen) and 10 pmol of the appropriate primers. qRT-PCR data were analysed using the Relative Expression Software Tool [REST] 2005 (Corbett Life Science, Cambridge, UK). Selected mutants were complemented by the addition of OHHL (Sigma) in DMSO (or with DMSO alone as a control) to PMM media at a final concentration of 5 µM, and strains grown to a final cell density of 5×107 cfu/ml. After 18h incubation, bacteria were harvested and RNA extracted, purified and quantified as previously described. Differential expression was considered statistically significant if the t-test P-value was <0.05. To analyse gene expression of the out gene cluster in vitro, cultures were grown in Pel Minimal Medium (PMM) at 27 °C, RNA samples were prepared and qRT-PCR analysis performed as described in Burr et al., [12]. The outD-gusA strain, MC4, was as described by Corbett et al., [10], an outD-gusA/expI double mutant was generated by generalised transduction (data not shown), and β-glucuronidase (GusA) activity was measured throughout growth in Pel minimal broth (PMB) as described [10]. All primers used are described in Table S1.
Bacterial counts and measurement of OHHL
Bacterial cells for counting were collected in 10 ml sterile water prior to dilution and plating as described [54]. Bacterial cells used for measuring OHHL levels were taken from these samples prior to dilution. OHHL levels were analysed using E. coli JM109 carrying a bioluminescence reporter vector (pSB401) [55]. Each sample of 100 µl was aliquoted into three wells of a sterile black 96-well microtitre plate, and 100 µl of the sensor strain (grown to an OD600 of 1.0) was added to each well. The microtitre plate was incubated at 37°C for 3 hours and the luminescence from each well was measured using a SpectraMax M5 luminescence plate reader at the default setting with an integration time of 1 second (Molecular Devices Corp., Sunnyvale, CA). A series of OHHL standards was used both as a positive control and to determine the level of OHHL.
Identifiers, names and accessions numbers of genes referred to in the text
ECA0105 (expI), YP048233; ECA0106 (expR), YP048234; ECA0176, YP048830; ECA0456 (hcp4), YP048574; ECA0601 (cfa8A), YP048718; ECA0602 (cfa7), YP048719; ECA0603 (cfa6), YP048720; ECA0607 (cfa2), YP048724; ECA0609 (cfl), YP048726; ECA0809 (hexY), YP048920; ECA0931 (svx), YP049040; ECA1017 (pmeB), YP049124; ECA1022 (aepA), YP049129; ECA1094 (pel-3), YP049200; ECA1095 (pehA), YP049201; ECA1110 (phoB), YP049216; ECA1190 (pehN), YP049296; ECA1235 (hypA), YP049341; ECA1499 (pnl), YP049604; ECA1561 (virR), YP049663; ECA1562 (virS), YP049664; ECA1700 (flgM), YP049801; ECA1740 (fliZ), YP049840; ECA1931 (hor), YP050028; ECA1981 (celV), YP 050075; ECA2087 (hrpL), YP050182; ECA2089 (hrpY), YP050184; ECA2090 (hrpS), YP050185; ECA2097 (hrpE), YP050792; ECA2103 (hrpN), YP050198; ECA2104 (vgrG), YP050199; ECA2108, YP050203; ECA2105-2110, YP050200-050205; ECA2112 (hrpW), YP050207; ECA2113 (dpsA/E), YP050208; ECA2207 (fnr), YP050300; ECA2220, YP050313; ECA2402 (pelW), YP050497; ECA2425 (kdgR), YP050520; ECA2435 (rdgA), YP050530; ECA2445 (pehR), YP050539; ECA2553, YP050644; ECA2578 (citB), YP050669; ECA2724 (rscR), YP050815; ECA2781 (prtF), YP050872; ECA2782 (prtE), YP050873; ECA2783 (prtD), YP050874; ECA2785 (prtW), YP050876; ECA2827 (celB), YP050918; ECA2866 (hcp3), YP050957; ECA2867 (vgrG), YP050958; ECA2882 (expA), YP050973; ECA3030 (hexA), YP051120; ECA3087 (nip), YP051177; ECA3098-3114, YP051188-051204; ECA3100 (outM), YP051190; ECA3101 (outL), YP051191; ECA3105 (outH), YP051195; ECA3106 (outG), YP051196; ECA3107 (outF), YP051197; ECA3109 (outD), YP051199; ECA3168 (ohrR), YP051257; ECA3254 (glnB), YP051343; ECA3281 (rseC), YP051370; ECA3282 (rseB), YP051371; ECA3366 (rsmA), YP051455; ECA3368 (recX), YP051457; ECA3420, YP051511; ECA3421-3426, YP051512-051517; ECA3427, YP051518; ECA3428 (hcp), YP051519; ECA3430, YP051520; ECA3432 (vasK), YP051522; ECA3433, YP051523; ECA3436 (vasG), YP051526; ECA3438, YP051528; ECA3440, YP051530; ECA3442 (vasA), YP051532; ECA3443, YP051533; ECA3444, YP051534; ECA3445, YP051535; ECA3427-3445, YP051518-051535; ECA3646 (celH), YP051234; ECA3672, YP051760; ECA3946, YP052033; ECA4067 (pelA), YP052154; ECA4068 (pelB), YP052155; ECA4069 (pelC), YP052156; ECA4070 (pelZ), YP052157; ECA4123 (rexZ), YP052210; ECA4142 (vgrG), YP052229; ECA4275, YP052362; ECA4276 (vgrG), YP052363; ECA4277, YP052364; ECA4305 (sftR), YP052392; ECA4483 (nac), YP052566.
Supporting Information
ExpI is Involved in Virulence. Virulence assays in potato tubers following inoculation of wild type P. atrosepticum and the expI mutant, and complementation of the expI mutant by plasmid pGEM-T containing the expI coding region against the wild type containing empty vector. “C” indicates complemented. Bars show mean +/− standard error of the mean.
(0.39 MB EPS)
qRT-PCR analysis of selected genes from Pectobacterium atrosepticum during potato infection. qRT-PCR data and primers of QS regulated genes.
(0.23 MB DOC)
Microarray analysis of genes from Pectobacterium atrosepticum during potato infection. Expression values for microarray data of QS regulated genes.
(2.22 MB DOC)
Acknowledgments
The authors wish to acknowledge the financial support of the Scottish Government Rural and Environment Research and Analysis Directorate (RERAD), the BBSRC, and the Research Council of Norway.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by the Scottish Government Rural and Environment Research and Analysis Directorate (RERAD) and the Biotechnology and Biological Sciences Research Council (BBSRC). G.T. was funded by a studentship from the Research Council of Norway.
References
- 1.Whitehead NA, Barnard AM, Slater H, Simpson NJ, Salmond GP. Quorum-sensing in Gram-negative bacteria. FEMS Microbiol Rev. 2001;25:365–404. doi: 10.1111/j.1574-6976.2001.tb00583.x. [DOI] [PubMed] [Google Scholar]
- 2.Engebrecht J, Silverman M. Identification of genes and gene products necessary for bacterial bioluminescence. Proc Natl Acad Sci USA. 1984;81:4154–4158. doi: 10.1073/pnas.81.13.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gardan L, Gouy C, Christen R, Samson R. Elevation of three subspecies of Pectobacterium carotovorum to species level: Pectobacterium atrosepticum sp. nov., Pectobacterium betavasculorum sp. nov. and Pectobacterium wasabiae sp. nov. Int J Syst Evol Microbiol. 2003;53:381–391. doi: 10.1099/ijs.0.02423-0. [DOI] [PubMed] [Google Scholar]
- 4.Toth IK, Bell KS, Holeva MC, Birch PRJ. Soft rot erwiniae: from genes to genomes. Molecular Plant Pathology. 2003;4:17–30. doi: 10.1046/j.1364-3703.2003.00149.x. [DOI] [PubMed] [Google Scholar]
- 5.Holeva MC, Bell KS, Hyman LJ, Avrova AO, Whisson SC, et al. Use of a pooled transposon mutation grid to demonstrate roles in disease development for Erwinia carotovora subsp. atroseptica putative type III secreted effector (DspE/A) and helper (HrpN) proteins. Mol Plant Microbe Interact. 2004;17:943–950. doi: 10.1094/MPMI.2004.17.9.943. [DOI] [PubMed] [Google Scholar]
- 6.Meng X, Bonasera JM, Kim JF, Nissinen RM, Beer SV. Apple proteins that interact with DspA/E, a pathogenicity effector of Erwinia amylovora, the fire blight pathogen. Mol Plant Microbe Interact. 2006;19:53–61. doi: 10.1094/MPMI-19-0053. [DOI] [PubMed] [Google Scholar]
- 7.Jones S, Yu B, Bainton NJ, Birdsall M, Bycroft BW, et al. The lux autoinducer regulates the production of exoenzyme virulence determinants in Erwinia carotovora and Pseudomonas aeruginosa. Embo J. 1993;12:2477–2482. doi: 10.1002/j.1460-2075.1993.tb05902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smadja B, Latour X, Faure D, Chevalier S, Dessaux Y, et al. Involvement of N-acylhomoserine lactones throughout plant infection by Erwinia carotovora subsp. atroseptica (Pectobacterium atrosepticum). Mol Plant Microbe Interact. 2004;17:1269–1278. doi: 10.1094/MPMI.2004.17.11.1269. [DOI] [PubMed] [Google Scholar]
- 9.Pemberton CL, Whitehead NA, Sebaihia M, Bell KS, Hyman LJ, et al. Novel quorum-sensing-controlled genes in Erwinia carotovora subsp. carotovora: identification of a fungal elicitor homologue in a soft-rotting bacterium. Mol Plant Microbe Interact. 2005;18:343–353. doi: 10.1094/MPMI-18-0343. [DOI] [PubMed] [Google Scholar]
- 10.Corbett M, Virtue S, Bell K, Birch P, Burr T, et al. Identification of a new quorum-sensing-controlled virulence factor in Erwinia carotovora subsp. atroseptica secreted via the type II targeting pathway. Mol Plant Microbe Interact. 2005;18:334–342. doi: 10.1094/MPMI-18-0334. [DOI] [PubMed] [Google Scholar]
- 11.Mattinen L, Tshuikina M, Mae A, Pirhonen M. Identification and characterization of Nip, necrosis-inducing virulence protein of Erwinia carotovora subsp. carotovora. Mol Plant Microbe Interact. 2004;17:1366–1375. doi: 10.1094/MPMI.2004.17.12.1366. [DOI] [PubMed] [Google Scholar]
- 12.Burr T, Barnard AM, Corbett MJ, Pemberton CL, Simpson NJ, et al. Identification of the central quorum sensing regulator of virulence in the enteric phytopathogen, Erwinia carotovora: the VirR repressor. Mol Microbiol. 2006;59:113–125. doi: 10.1111/j.1365-2958.2005.04939.x. [DOI] [PubMed] [Google Scholar]
- 13.Cui Y, Chatterjee A, Hasegawa H, Dixit V, Leigh N, et al. ExpR, a LuxR homolog of Erwinia carotovora subsp. carotovora, activates transcription of rsmA, which specifies a global regulatory RNA-binding protein. J Bacteriol. 2005;187:4792–4803. doi: 10.1128/JB.187.14.4792-4803.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chatterjee A, Cui Y, Liu Y, Dumenyo CK, Chatterjee AK. Inactivation of rsmA leads to overproduction of extracellular pectinases, cellulases, and proteases in Erwinia carotovora subsp. carotovora in the absence of the starvation/cell density-sensing signal, N-(3-oxohexanoyl)-L-homoserine lactone. Appl Environ Microbiol. 1995;61:1959–1967. doi: 10.1128/aem.61.5.1959-1967.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGowan SJ, Barnard AM, Bosgelmez G, Sebaihia M, Simpson NJ, et al. Carbapenem antibiotic biosynthesis in Erwinia carotovora is regulated by physiological and genetic factors modulating the quorum sensing-dependent control pathway. Mol Microbiol. 2005;55:526–545. doi: 10.1111/j.1365-2958.2004.04397.x. [DOI] [PubMed] [Google Scholar]
- 16.Barnard AM, Salmond GP. Quorum sensing in Erwinia species. Anal Bioanal Chem. 2007;387:415–423. doi: 10.1007/s00216-006-0701-1. [DOI] [PubMed] [Google Scholar]
- 17.Mae A, Montesano M, Koiv V, Palva ET. Transgenic plants producing the bacterial pheromone N-acyl-homoserine lactone exhibit enhanced resistance to the bacterial phytopathogen Erwinia carotovora. Mol Plant Microbe Interact. 2001;14:1035–1042. doi: 10.1094/MPMI.2001.14.9.1035. [DOI] [PubMed] [Google Scholar]
- 18.Salmond GP, Bycroft BW, Stewart GS, Williams P. The bacterial ‘enigma’: cracking the code of cell-cell communication. Mol Microbiol. 1995;16:615–624. doi: 10.1111/j.1365-2958.1995.tb02424.x. [DOI] [PubMed] [Google Scholar]
- 19.Toth IK, Newton JA, Hyman LJ, Lees AK, Daykin M, et al. Potato plants genetically modified to produce N-acylhomoserine lactones increase susceptibility to soft rot erwiniae. Mol Plant Microbe Interact. 2004;17:880–887. doi: 10.1094/MPMI.2004.17.8.880. [DOI] [PubMed] [Google Scholar]
- 20.Bell KS, Sebaihia M, Pritchard L, Holden MT, Hyman LJ, et al. Genome sequence of the enterobacterial phytopathogen Erwinia carotovora subsp. atroseptica and characterization of virulence factors. Proc Natl Acad Sci USA. 2004;101:11105–11110. doi: 10.1073/pnas.0402424101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uppalapati SR, Ishiga Y, Wangdi T, Kunkel BN, Anand A, et al. The phytotoxin coronatine contributes to pathogen fitness and is required for suppression of salicylic acid accumulation in tomato inoculated with Pseudomonas syringae pv. tomato DC3000. Mol Plant Microbe Interact. 2007;20:955–965. doi: 10.1094/MPMI-20-8-0955. [DOI] [PubMed] [Google Scholar]
- 22.Elizabeth SV, Bender CL. The phytotoxin coronatine from Pseudomonas syringae pv. tomato DC3000 functions as a virulence factor and influences defence pathways in edible brassicas. Mol. Plant Pathol. 2007;8:83–92. doi: 10.1111/j.1364-3703.2006.00372.x. [DOI] [PubMed] [Google Scholar]
- 23.Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, et al. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci USA. 2006;103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wechter WP, Glandorf DCM, Derrick WC, Leverentz B, Kluepfel DA. Identification of genetic loci in a rhizosphere-inhabiting species of Pseudomonas involved in expression of a phytoparasitic nematode ovicidal factor. Soil Biol. Biochem. 2001;33:1749–1758. [Google Scholar]
- 25.Schell MA, Ulrich RL, Ribot WJ, Brueggemann EE, Hines HB, et al. Type VI secretion is a major virulence determinant in Burkholderia mallei. Mol Microbiol. 2007;64:1466–1485. doi: 10.1111/j.1365-2958.2007.05734.x. [DOI] [PubMed] [Google Scholar]
- 26.Williams SG, Varcoe LT, Attridge SR, Manning PA. Vibrio cholerae Hcp, a secreted protein coregulated with HlyA. Infect Immun. 1996;64:283–289. doi: 10.1128/iai.64.1.283-289.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mattinen L, Nissinen R, Riipi T, Kalkkinen N, Pirhonen M. Host-extract induced changes in the secretome of the plant pathogenic bacterium Pectobacterium atrosepticum. Proteomics. 2007;7:3527–3537. doi: 10.1002/pmic.200600759. [DOI] [PubMed] [Google Scholar]
- 28.Pirhonen M, Flego D, Heikinheimo R, Palva ET. A small diffusible signal molecule is responsible for the global control of virulence and exoenzyme production in the plant pathogen Erwinia carotovora. Embo J. 1993;12:2467–2476. doi: 10.1002/j.1460-2075.1993.tb05901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reeves PJ, Whitcombe D, Wharam S, Gibson M, Allison G, et al. Molecular cloning and characterization of 13 out genes from Erwinia carotovora subspecies carotovora: genes encoding members of a general secretion pathway (GSP) widespread in gram-negative bacteria. Mol Microbiol. 1993;8:443–456. doi: 10.1111/j.1365-2958.1993.tb01589.x. [DOI] [PubMed] [Google Scholar]
- 30.Binet R, Letoffe S, Ghigo JM, Delepelaire P, Wandersman C. Protein secretion by Gram-negative bacterial ABC exporters–a review. Gene. 1997;192:7–11. doi: 10.1016/s0378-1119(96)00829-3. [DOI] [PubMed] [Google Scholar]
- 31.Chapon-Herve V, Akrim M, Latifi A, Williams P, Lazdunski A, et al. Regulation of the xcp secretion pathway by multiple quorum-sensing modulons in Pseudomonas aeruginosa. Mol Microbiol. 1997;24:1169–1178. doi: 10.1046/j.1365-2958.1997.4271794.x. [DOI] [PubMed] [Google Scholar]
- 32.Riedel K, Ohnesorg T, Krogfelt KA, Hansen TS, Omori K, et al. N-acyl-L-homoserine lactone-mediated regulation of the lip secretion system in Serratia liquefaciens MG1. J Bacteriol. 2001;183:1805–1809. doi: 10.1128/JB.183.5.1805-1809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Collmer A, Lindeberg M, Petnicki-Ocwieja T, Schneider DJ, Alfano JR. Genomic mining type III secretion system effectors in Pseudomonas syringae yields new picks for all TTSS prospectors. Trends Microbiol. 2002;10:462–469. doi: 10.1016/s0966-842x(02)02451-4. [DOI] [PubMed] [Google Scholar]
- 34.Bleves S, Soscia C, Nogueira-Orlandi P, Lazdunski A, Filloux A. Quorum sensing negatively controls type III secretion regulon expression in Pseudomonas aeruginosa PAO1. J Bacteriol. 2005;187:3898–3902. doi: 10.1128/JB.187.11.3898-3902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henke JM, Bassler BL. Quorum sensing regulates type III secretion in Vibrio harveyi and Vibrio parahaemolyticus. J Bacteriol. 2004;186:3794–3805. doi: 10.1128/JB.186.12.3794-3805.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sircili MP, Walters M, Trabulsi LR, Sperandio V. Modulation of enteropathogenic Escherichia coli virulence by quorum sensing. Infect Immun. 2004;72:2329–2337. doi: 10.1128/IAI.72.4.2329-2337.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Genin S, Brito B, Denny TP, Boucher C. Control of the Ralstonia solanacearum Type III secretion system (Hrp) genes by the global virulence regulator PhcA. FEBS Lett. 2005;579:2077–2081. doi: 10.1016/j.febslet.2005.02.058. [DOI] [PubMed] [Google Scholar]
- 38.Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312:1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y, Wang X, Mukherjee A, Chatterjee AK. RecA relieves negative autoregulation of rdgA, which specifies a component of the RecA-Rdg regulatory circuit controlling pectin lyase production in Erwinia carotovora ssp. carotovora. Mol Microbiol. 1996;22:909–918. doi: 10.1046/j.1365-2958.1996.01537.x. [DOI] [PubMed] [Google Scholar]
- 40.Tang X, Xiao Y, Zhou JM. Regulation of the type III secretion system in phytopathogenic bacteria. Mol Plant Microbe Interact. 2006;19:1159–1166. doi: 10.1094/MPMI-19-1159. [DOI] [PubMed] [Google Scholar]
- 41.Von Bodman SB, Bauer WD, Coplin DL. Quorum sensing in plant-pathogenic bacteria. Annu Rev Phytopathol. 2003;41:455–482. doi: 10.1146/annurev.phyto.41.052002.095652. [DOI] [PubMed] [Google Scholar]
- 42.Iyoda S, Kamidoi T, Hirose K, Kutsukake K, Watanabe H. A flagellar gene fliZ regulates the expression of invasion genes and virulence phenotype in Salmonella enterica serovar Typhimurium. Microb Pathog. 2001;30:81–90. doi: 10.1006/mpat.2000.0409. [DOI] [PubMed] [Google Scholar]
- 43.Nelson KM, Young GM, Miller VL. Identification of a locus involved in systemic dissemination of Yersinia enterocolitica. Infect Immun. 2001;69:6201–6208. doi: 10.1128/IAI.69.10.6201-6208.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chuchue T, Tanboon W, Prapagdee B, Dubbs JM, Vattanaviboon P, et al. ohrR and ohr are the primary sensor/regulator and protective genes against organic hydroperoxide stress in Agrobacterium tumefaciens. J Bacteriol. 2006;188:842–851. doi: 10.1128/JB.188.3.842-851.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Price MN, Dehal PS, Arkin AP. Orthologous transcription factors in bacteria have different functions and regulate different genes. PLoS Comput Biol. 2007;3:1739–1750. doi: 10.1371/journal.pcbi.0030175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Preiter K, Brooks DM, Penaloza-Vazquez A, Sreedharan A, Bender CL, et al. Novel virulence gene of Pseudomonas syringae pv. tomato strain DC3000. J Bacteriol. 2005;187:7805–7814. doi: 10.1128/JB.187.22.7805-7814.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bender CL, Alarcon-Chaidez F, Gross DC. Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol Mol Biol Rev. 1999;63:266–292. doi: 10.1128/mmbr.63.2.266-292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dong X. SA, JA, ethylene, and disease resistance in plants. Curr Opin Plant Biol. 1998;1:316–323. doi: 10.1016/1369-5266(88)80053-0. [DOI] [PubMed] [Google Scholar]
- 49.Weiler EW, Kutchan TM, Gorba T, Brodschelm W, Niesel U, et al. The Pseudomonas phytotoxin coronatine mimics octadecanoid signalling molecules of higher plants. FEBS Lett. 1994;345:9–13. doi: 10.1016/0014-5793(94)00411-0. [DOI] [PubMed] [Google Scholar]
- 50.Kaniga K, Delor I, Cornelis GR. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene. 1991;109:137–141. doi: 10.1016/0378-1119(91)90599-7. [DOI] [PubMed] [Google Scholar]
- 51.Toth IK, Mulholland V, Cooper V, Bentley S, Shih YL, et al. Generalized transduction in the potato blackleg pathogen Erwinia carotovora subsp. atroseptica by bacteriophage phi M1. Microbiol. 1997;143:2433–2438. doi: 10.1099/00221287-143-7-2433. [DOI] [PubMed] [Google Scholar]
- 52.Venkatesh B, Babujee L, Liu H, Hedley P, Fujikawa T, et al. The Erwinia chrysanthemi 3937 PhoQ sensor kinase regulates several virulence determinants. J Bacteriol. 2006;188:3088–3098. doi: 10.1128/JB.188.8.3088-3098.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hyman LJ, Sullivan L, Toth IK, Perombelon MCM. Modified crystal violet pectate medium (CVP) based on a new polypectate source (Slendid) for the detection and isolation of soft rot erwinias. Potato Res. 2001;44:265–270. [Google Scholar]
- 55.Winson MK, Swift S, Hill PJ, Sims CM, Griesmayr G, et al. Engineering the luxCDABE genes from Photorhabdus luminescens to provide a bioluminescent reporter for constitutive and promoter probe plasmids and mini-Tn5 constructs. FEMS Microbiol Lett. 1998;163:193–202. doi: 10.1111/j.1574-6968.1998.tb13045.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ExpI is Involved in Virulence. Virulence assays in potato tubers following inoculation of wild type P. atrosepticum and the expI mutant, and complementation of the expI mutant by plasmid pGEM-T containing the expI coding region against the wild type containing empty vector. “C” indicates complemented. Bars show mean +/− standard error of the mean.
(0.39 MB EPS)
qRT-PCR analysis of selected genes from Pectobacterium atrosepticum during potato infection. qRT-PCR data and primers of QS regulated genes.
(0.23 MB DOC)
Microarray analysis of genes from Pectobacterium atrosepticum during potato infection. Expression values for microarray data of QS regulated genes.
(2.22 MB DOC)