Abstract
A separation technique employing a microfabricated sieve has been demonstrated by observing the motion of DNA molecules of different size. The sieve consists of a two-dimensional lattice of obstacles whose asymmetric disposition rectifies the Brownian motion of molecules driven through the device, causing them to follow paths that depend on their diffusion coefficient. A nominal 6% resolution by length of DNA molecules in the size range 15–30 kbp may be achieved in a 4-inch (10-cm) silicon wafer. The advantage of this method is that samples can be loaded and sorted continuously, in contrast to the batch mode commonly used in gel electrophoresis.
Keywords: DNA fractionation, Brownian motion, microfabricated array
The separation of DNA fragments from restriction enzyme digests plays a central role in molecular biology (1), and there is a growing realization that new and faster methods are needed to accelerate and simplify genomic analysis. One route to this goal is the exploitation of technologies in the microfabrication industry to create miniature silicon-based devices in which sample preparation, separation, and analysis are integrated in a single microenvironment (2–5). The task is complicated by the fact that, when forced to move by a flow or by the application of an electric field, DNA molecules of different sizes all migrate at the same speed. Traditionally, a sieving medium such as a gel or a solution of polymers has been used to alter the mobility. Recently, however, Duke and Austin (6) and Ertas (7) proposed an alternative approach that takes advantage of the fact that, as the molecules migrate, they also diffuse—and this they do at a size-dependent rate. On theoretical grounds, they showed how a two-dimensional obstacle course could be constructed to sort the swift diffusers from the slow. The basic concept is to use a regular lattice of asymmetric obstacles to rectify the lateral Brownian motion of the molecules so that species of different sizes follow different trajectories through the device. A mixture of molecules injected in a fine stream would be sorted continuously.
We have implemented this idea by using a microfabricated silicon array. That such a device works in principle has been demonstrated by making microscopic observations of the dynamics of fluorescently labeled latex beads and DNA molecules‡‡ and also by the recent work of van Oudenaarden and Boxer (8) with lipids. The experiments are technically demanding, however, and the resolution of different species has not yet been achieved. Progression from a demonstration of principle to practical continuous stream sorting of molecules requires perfection of the technology. In the work reported here, our aim has been to conduct a systematic, quantitative study of the separation process, using molecules of a well-defined size and experimental conditions that are close to optimal. The close correspondence of the results with theoretical predictions permits us to predict, with some confidence, how to construct a device that will work well in practice.
Materials and Methods
Fabrication Procedure.
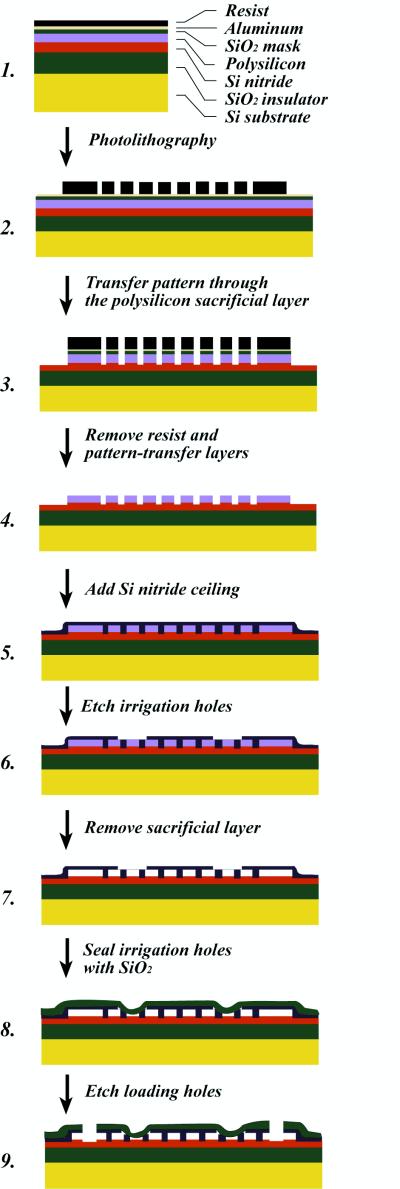
To obtain uniform fluid flows over the entire area of the device, we developed a special technique that integrates the floor and ceiling of the chamber and the array of obstacles in a single monolithic design (9). The fabrication method (Fig. 1) makes use of a sacrificial layer, which is deposited over the silicon nitride floor of the device. This layer is made of polysilicon, chosen because it can easily be etched in alkaline solution. The sacrificial layer represents what will be the open space in the finished device, and so the negative of the desired pattern of obstacles is etched into it. A ceiling layer of silicon nitride is then deposited over the sacrificial layer by chemical vapor deposition. The silicon nitride coats conformally, so that, in addition to coating the top of the polysilicon, it fills the holes that have been etched into it. Small irrigation ducts are then cut through the ceiling to allow a wet etchant (tetramethylammonium hydroxide) to remove the sacrificial layer. Finally, the ducts are resealed with an additional layer of low-temperature silicon dioxide. The resulting structure is a quasi-two-dimensional chamber containing the desired pattern of silicon nitride obstacles. This methodology is an advance over earlier methods, in which the ceiling of the chamber was created by sealing a glass or quartz coverslip to an open micro-machined array (2). Superior uniformity and control of the floor-to-ceiling height is achieved because this dimension is determined by the thickness of the sacrificial layer. The method is also more tolerant of particulate contamination, because the structure is disrupted only in the immediate vicinity of a particle. The silicon nitride ceiling is stable, transparent, and exhibits negligible autofluorescence.
Figure 1.
Production of a sealed monolithic obstacle array.
Samples, Electrophoresis, and Microscopy.
Because it was our intention to test specific theoretical predictions of biased diffusion in two-dimensional obstacle courses (6), we chose a mixture of DNA molecules of well-defined lengths as our sample. Conveniently, the size of large fluorescently labeled DNA molecules can be determined optically by measuring their radius of gyration. Under typical electrophoresis conditions, molecules in the size range 10–50 kbp were predicted to be optimally resolved in a two-dimensional array of micron-sized obstacles (6). Therefore, a mixture of XhoI-cut λ DNA fragments (15 kbp and 33.5 kbp) was stained with fluorescent dye TOTO-1 (1 dye per 10 bp) (Molecular Probes) in 0.5× TBE buffer (45 mM Tris/borate, 1 mM EDTA, pH 8.0), with 0.1% performance-optimized polymer POP-6 (Perkin Elmer Biosystems, Foster City, CA) and 0.1 M DTT. This two-component mixture was moved by electrophoresis through the microfabricated chamber. Individual molecules were imaged in a Kodak Optiphot microscope with an oil immersion objective lens (60×, numerical aperture 1.4), a silicon-intensified target camera (Hamamatsu, Bridgewater, NJ), and excitation at the 488-nm line of an Ar+ laser. A rotating holographic scatter plate was used to reduce speckles.
Array Design.
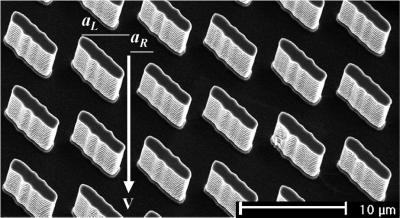
The device consisted of a lattice of rectangular obstacles, each measuring 1.5 × 6 μm and set at an angle of 45° to the direction of the applied electric field. Fig. 2 is a scanning electron micrograph of one such array, which was designed as proposed in ref. 6. A molecule driven through the device must pass through the narrow gaps, or “gates,” between the obstacles. It preferentially follows the channel in the field direction, moving from one gate to the next gate directly below. But there is a small probability, pR, that it diffuses a distance aR ≈1.5 μm to the right, in which case it passes around an obstacle and shifts one channel to the right. The probability pL of shifting to the left is much slimmer, because it requires diffusion over a greater distance (aL ≈5 μm) in a briefer interval of time.
Figure 2.
A scanning electron micrograph of the obstacle course. The obstacles are 0.35 μm high and measure 1.5 × 6.0 μm. The gap between adjacent obstacles is 1.5 μm. An electric field propels the molecules directly through the gaps between the posts with velocity v. Transverse Brownian motion may cause a molecule to skip one channel to the right if it diffuses through displacement aR, or very rarely, one channel to the left if it diffuses through aL.
Results and Discussion
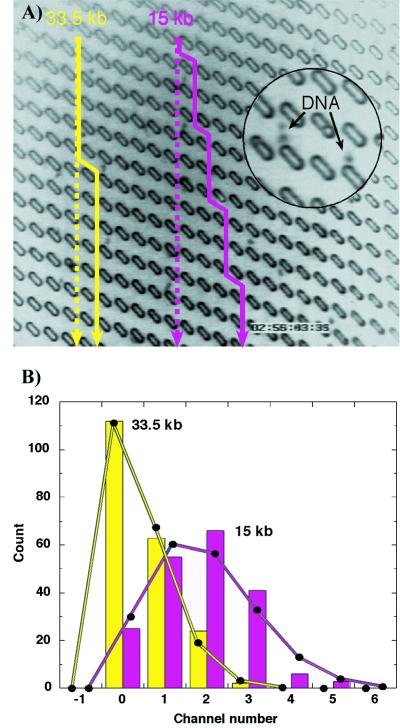
Fig. 3A illustrates the trajectory of individual 15-kbp and 33.5-kbp DNA molecules as they are transported through 14 gates in the same field of view of the array by a vertical electric field of 1.4 V/cm. The distribution of the lateral displacement during many such trajectories is shown in Fig. 3B. If the molecular motion is independent from one cell to the next, we expect the distribution to be described by binomial statistics (assuming that pL is negligible; measured pL < 0.001 for ≈200 molecules of each length traveling through 14 gates). Good agreement between the binomial distribution and our measurements indicates that Brownian motion is strong enough to negate any correlations in the way molecules pass through successive gates. The probabilities of lateral deflection per gate can then be calculated from the data: pR(15 kbp) = 0.127 ± 0.006, and pR(33.5 kbp) = 0.042 ± 0.003. Because there is a significant difference between these figures (Δp = 0.085 ± 0.007), we conclude that the 15-kbp and 33.5-kbp DNA fragments travel at different angles through the array.
Figure 3.
(A) The trajectories of XhoI-digested bacteriophage λ DNA fragments. The straight lines show the average electric field direction. The solid yellow line traces the path of a 33.5-kbp fragment; the purple line traces a 15-kbp fragment. The frame size is 130 × 114 μm2. Total dimension of the separation device is 3 × 4 cm, and electric field strength is 1.4 V/cm. (A video clip illustrating the process is available at http://suiling.princeton.edu.) (B) Histogram of the deflection of approximately 200 DNA fragments of each size after they have passed through 14 gates (the vertical distance in A). Scale at bottom indicates the total number of channels that the molecule has shifted to the right. The solid lines are binomial distributions with the same mean and area as the experimental data.
A simplified theory (6) suggests that the trajectory of molecules within our device is governed by the dimensionless parameter D/va, where D is the diffusion coefficient of the molecule, v is its electrophoretic velocity, and a is the short side of the obstacle. We have verified the linear relation between v and applied field E, which yields an electrophoretic mobility of 1.47 ± 0.02 and 1.29 ± 0.02 (μm/s)/(V/cm) for 15-kbp and 33.5-kbp molecules, respectively. Because the 15-kbp molecules interact less frequently with the obstacles, we take v (15 kbp) as the free-flow electrophoretic velocity for the following analysis. Molecules that diffuse very slowly are likely to travel straight through the sieve, without being deflected by the obstacles. At the other extreme, molecules that diffuse very rapidly are turned to the left or the right with equal probability; they also, on average, travel straight down. In an intermediate range, 0.02 < D/va < 0.3, the probability of rightward deflection increases monotonically with D/va, whereas the probability of leftward deflection remains negligible (6); this variance permits more swiftly diffusing molecules to be sorted from less mobile ones. Thus, the fractionation of a given mixture of molecules in a particular sieve may by optimized by choosing an appropriate drift velocity.
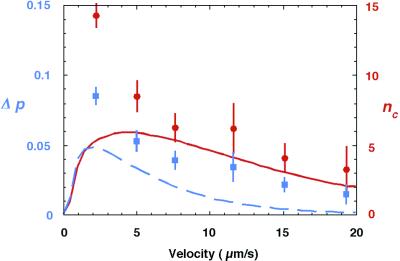
To test these predictions, we carried out a series of measurements in which we varied the electric field. Fig. 4 shows that Δp decreases monotonically with the drift velocity over the range of values investigated. Because visualization by fluorescence microscopy requires large molecules, which diffuse slowly, we were unable to reduce the drift velocity sufficiently to observe the predicted decline in resolution at high values of D/va. The best performance is observed at 1.4 V/cm (v = 2.1 μm/s) and would amount to a band separation Δx of ≈6.4 mm at the bottom of a 10-cm sieve, with a bandwidth σ of about 0.2 mm (15 kbp) and 0.1 mm (33.5 kbp). Here, we have used the central limit theorem to derive the extrapolated bandwidth from the measured variance of the distributions. Given the narrowness of the bands, high performance requires that the molecules be injected into the array in a very fine jet. In this experiment, we used a wide (diameter 500 μm) loading nozzle which introduced initial broadening, perturbed the electric field, and diminished the resolution. To give a clearer idea of the resolution that could be achieved with a better loading procedure, we have calculated the extrapolated band capacity nC, defined as the number of bands that can be resolved between 15 kbp and 33.5 kbp in a 10-cm sieve (corresponding to N = 12,500 gates). Using nC = Δx/1.5[σ(15 kbp) + σ(33.5 kbp)] (10), we find that up to 14 bands could be distinguished, which corresponds to a resolution of 6% in molecular length (resolution = ln(L1/L2)/nC, where L1 = 33.5 kbp and L2 = 15 kbp, assuming a uniform resolution over the range). Note that although the lateral separation is proportional to the total number of gates N, band broadening is proportional to N1/2. Thus, in principle, the resolution could be enhanced indefinitely by increasing the length of the sieve.
Figure 4.
Difference Δp in the probability per gate that 15-kbp and 33.5-kbp molecules skip to the right (blue). Extrapolated number of bands nC that could be detected between 15 kbp and 33.5 kbp in a 10-cm sieve (red). Lines are theoretical predictions for Δp (blue) and nC (red). The theoretical curve of Δp is taken from Eq. 1 of ref. 6, substituting in the two values of D/va, and calculating pR and pL for both fragments: Δp = [pR(15 kbp) − pL(15 kbp)] − [pR(33.5 kbp) − pL(33.5 kbp)]. Theoretical curve for band capacity nC uses variance σ2 = N [pR(1 − pR) + pL(1 − pL) + pRpL)], given in ref. 6, where N = 12,500 gates.
Theoretical predictions of Δp and nC may be obtained from an estimate of the molecular diffusion coefficients. The 15-kbp and 33.5-kbp DNA fragments have respective radii of gyration RG = 0.31 μm and 0.43 μm. We have previously found that, when such molecules are confined in a 0.35-μm-deep chamber (as here), the partial screening of hydrodynamic interactions makes their Brownian dynamics approximately half as fast as it is in free solution (11). Therefore, taking half the Zimm diffusion coefficient (12) as the appropriate value, we obtain D(15 kbp) = 0.6 μm2/s and D(33.5 kbp) = 0.4 μm2/s. The predicted regime of good separation, 0.02 < D/va < 0.3, then corresponds to drift velocities in the range v = 1–15 μm/s. The performance of the device is compared with the theoretical predictions in Fig. 4. Although the experimental data agree with the general trend of the theory, the device actually performs better than expected, especially in the region of optimal resolution. We attribute this discrepancy to several simplifying assumptions in the theory. It is supposed that the field is uniform, that the molecules are point-like particles, and that the motion is independent from one cell to the next. In reality, the field lines are deformed by the insulating obstacles, the different geometric sizes of the molecules cause them to interact with obstacles in slightly different ways, and correlations are apparent in the passage through successive gates at high velocities (the measured probability distributions differ somewhat from the binomial distribution for E > 5 V/cm). It is gratifying that these features do not impair the operation of the device.
Conclusions
We have demonstrated the separation of macromolecules by using the principle of rectified Brownian motion. Previously, a pulsed, one-dimensional asymmetric potential has been used to propel colloidal particles at different speeds (13–15). Compared with this technique and with traditional methods such as gel electrophoresis, our approach has the advantage of accomplishing sorting continuously. Different components in a mixture of molecules, injected in a fine stream at the top corner of the device, are carried to different locations at the bottom edge, making it ideally suited for integrating separation with sample preparation and subsequent analytical steps. Moreover, by obviating the need for a viscous sieving medium, the method facilitates automation and offers new levels of convenience. It is not limited to DNA analysis, but can also be applied to the separation of proteins, colloidal particles, and cells by suitable adjustment of the obstacle dimensions, flow velocity, and supporting solvents.
Acknowledgments
We thank D. Ertas, J. Han, and J. Tegenfeldt for helpful discussions, and N. Yao for technical assistance with scanning electron microscopy. Microfabrication of the silicon wafers was done at the Cornell Nanofabrication Facility. This work was supported by National Institutes of Health grants (HG01506 and GM55453), the Royal Society, and a Biotechnology and Biological Sciences Research Council grant (E08580). The Niels Bohr Institute supported T.D. with a visiting professorship.
Footnotes
Chou, C. F., Duke, T. A. J., Chan, S. S., Bakajin, O., Austin, R. H. & Cox, E. C. (1998) Bull. Am. Phys. Soc. K14.08; and Chou, C. F., Bakajin, O., Turner, S. W., Duke, T. A. J., Chan, S. S., Cox, E. C., Craighead, H. G. & Austin, R. H. (1999) Bull. Am. Phys. Soc. WC31.05.
References
- 1.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual (2nd Ed.) Plainview, New York: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 2.Volkmuth W D, Austin R H. Nature (London) 1992;358:600–602. doi: 10.1038/358600a0. [DOI] [PubMed] [Google Scholar]
- 3.Hoch H C, Jelinski L W, Craighead H G, editors. Nanofabrication and Biosystems: Integrating Materials Science, Engineering, and Biology. Cambridge, U.K.: Cambridge Univ. Press; 1996. [Google Scholar]
- 4.Harrison D J, van den Berg A, editors. Micro Total Analysis Systems. Boston: Kluwer Academic; 1998. [Google Scholar]
- 5.Burns M A, Johnson B N, Brahmasandra S N, Handique K, Webster J R, Krishnan M, Sammarco T S, Man P M, Jones D, Heldsinger D, et al. Science. 1998;282:484–487. doi: 10.1126/science.282.5388.484. [DOI] [PubMed] [Google Scholar]
- 6.Duke T A J, Austin R H. Phys Rev Lett. 1998;80:1552–1555. [Google Scholar]
- 7.Ertas D. Phys Rev Lett. 1998;80:1548–1551. [Google Scholar]
- 8.van Oudenaarden A, Boxer S G. Science. 1999;285:1046–1048. doi: 10.1126/science.285.5430.1046. [DOI] [PubMed] [Google Scholar]
- 9.Turner S W, Perez A M, Lopez A, Craighead H G. J Vac Sci Technol B. 1998;38:3835–3840. [Google Scholar]
- 10.Giddings J C. Unified Separation Science. New York: Wiley; 1991. pp. 101–106. [Google Scholar]
- 11.Bakajin O B, Duke T A J, Chou C F, Chan S S, Austin R H, Cox E C. Phys Rev Lett. 1998;80:2737–2740. doi: 10.1103/PhysRevLett.86.1378. [DOI] [PubMed] [Google Scholar]
- 12.Zimm B H. J Chem Phys. 1956;24:269–278. [Google Scholar]
- 13.Rousselet J, Salome L, Ajdari L, Prost J. Nature (London) 1994;370:446–448. doi: 10.1038/370446a0. [DOI] [PubMed] [Google Scholar]
- 14.Faucheaux L P, Libchaber A. J Chem Soc Faraday Trans. 1995;91:3163–3166. [Google Scholar]
- 15.Gorre-Talini L, Jeanjean S, Silberzan P. Phys Rev E. 1997;56:2025–2034. [Google Scholar]