Abstract
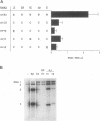
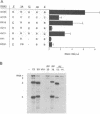
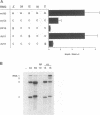
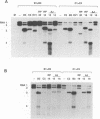
Brome mosaic virus (BMV) and cowpea chlorotic mottle virus (CCMV) are related positive-strand RNA viruses with genomes divided among RNAs 1, 2, and 3. RNAs 1 and 2 encode the viral RNA replication factors, which share extensive conservation with proteins encoded by the animal alphaviruses and diverse plant viruses. In barley protoplasts, CCMV RNAs 1 and 2 support high but distinguishable amplification of either BMV RNA3 (B3) or CCMV RNA3 (C3), while BMV RNAs 1 and 2 show even greater discrimination, amplifying C3 poorly relative to B3. To identify the cis-acting determinants of these template-specific and virus-specific differences in RNA3 accumulation, we constructed and tested a series of B3/C3 hybrids that exchange in turn the 5',3', and intercistronic noncoding regions, which contain all sequences required in cis for efficient B3 and C3 amplification. Despite suggestive prior in vitro results, the 3' noncoding regions were not the major determinant of the differences in amplification of B3 and C3 in vivo. Rather, 3' exchanges had relatively modest effects and did not transfer the distinctive asymmetry of amplification between B3 and C3. Intercistronic exchanges produced larger effects on RNA3 accumulation and transferred some of the polarized characteristics of the wild-type B3 and C3 behaviors. 5' exchanges revealed context-specific effects showing that the contribution of the B3 5' region to RNA3 amplification is dependent on some other B3 segment or segments. Together with previous results implicating the BMV and CCMV 1a genes in trans-acting discrimination between B3 and C3 (P. Traynor and P. Ahlquist, J. Virol. 64:69-77, 1990), these observations should help to guide studies of protein-RNA interactions governing template specificity in bromovirus RNA replication.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahlquist P., Dasgupta R., Kaesberg P. Near identity of 3- RNA secondary structure in bromoviruses and cucumber mosaic virus. Cell. 1981 Jan;23(1):183–189. doi: 10.1016/0092-8674(81)90283-x. [DOI] [PubMed] [Google Scholar]
- Ahlquist P., Dasgupta R., Kaesberg P. Nucleotide sequence of the brome mosaic virus genome and its implications for viral replication. J Mol Biol. 1984 Feb 5;172(4):369–383. doi: 10.1016/s0022-2836(84)80012-1. [DOI] [PubMed] [Google Scholar]
- Ahlquist P., French R., Janda M., Loesch-Fries L. S. Multicomponent RNA plant virus infection derived from cloned viral cDNA. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7066–7070. doi: 10.1073/pnas.81.22.7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlquist P., Janda M. cDNA cloning and in vitro transcription of the complete brome mosaic virus genome. Mol Cell Biol. 1984 Dec;4(12):2876–2882. doi: 10.1128/mcb.4.12.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlquist P., Luckow V., Kaesberg P. Complete nucleotide sequence of brome mosaic virus RNA3. J Mol Biol. 1981 Nov 25;153(1):23–38. doi: 10.1016/0022-2836(81)90524-6. [DOI] [PubMed] [Google Scholar]
- Ahlquist P., Strauss E. G., Rice C. M., Strauss J. H., Haseloff J., Zimmern D. Sindbis virus proteins nsP1 and nsP2 contain homology to nonstructural proteins from several RNA plant viruses. J Virol. 1985 Feb;53(2):536–542. doi: 10.1128/jvi.53.2.536-542.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison R. F., Janda M., Ahlquist P. Infectious in vitro transcripts from cowpea chlorotic mottle virus cDNA clones and exchange of individual RNA components with brome mosaic virus. J Virol. 1988 Oct;62(10):3581–3588. doi: 10.1128/jvi.62.10.3581-3588.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison R. F., Janda M., Ahlquist P. Sequence of cowpea chlorotic mottle virus RNAs 2 and 3 and evidence of a recombination event during bromovirus evolution. Virology. 1989 Sep;172(1):321–330. doi: 10.1016/0042-6822(89)90134-7. [DOI] [PubMed] [Google Scholar]
- Allison R., Thompson C., Ahlquist P. Regeneration of a functional RNA virus genome by recombination between deletion mutants and requirement for cowpea chlorotic mottle virus 3a and coat genes for systemic infection. Proc Natl Acad Sci U S A. 1990 Mar;87(5):1820–1824. doi: 10.1073/pnas.87.5.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastin M., Kaesberg P. A possible replicative form of brome mosaic virus RNA 4. Virology. 1976 Jul 15;72(2):536–539. doi: 10.1016/0042-6822(76)90185-9. [DOI] [PubMed] [Google Scholar]
- Bujarski J. J., Ahlquist P., Hall T. C., Dreher T. W., Kaesberg P. Modulation of replication, aminoacylation and adenylation in vitro and infectivity in vivo of BMV RNAs containing deletions within the multifunctional 3' end. EMBO J. 1986 Aug;5(8):1769–1774. doi: 10.1002/j.1460-2075.1986.tb04425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujarski J. J., Kaesberg P. Genetic recombination between RNA components of a multipartite plant virus. 1986 May 29-Jun 4Nature. 321(6069):528–531. doi: 10.1038/321528a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael G. G., McMaster G. K. The analysis of nucleic acids in gels using glyoxal and acridine orange. Methods Enzymol. 1980;65(1):380–391. doi: 10.1016/s0076-6879(80)65049-6. [DOI] [PubMed] [Google Scholar]
- Dasgupta R., Harada F., Kaesberg P. Blocked 5' termini in brome mosaic virus RNA. J Virol. 1976 Apr;18(1):260–267. doi: 10.1128/jvi.18.1.260-267.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher T. W., Hall T. C. Mutational analysis of the sequence and structural requirements in brome mosaic virus RNA for minus strand promoter activity. J Mol Biol. 1988 May 5;201(1):31–40. doi: 10.1016/0022-2836(88)90436-6. [DOI] [PubMed] [Google Scholar]
- French R., Ahlquist P. Characterization and engineering of sequences controlling in vivo synthesis of brome mosaic virus subgenomic RNA. J Virol. 1988 Jul;62(7):2411–2420. doi: 10.1128/jvi.62.7.2411-2420.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French R., Ahlquist P. Intercistronic as well as terminal sequences are required for efficient amplification of brome mosaic virus RNA3. J Virol. 1987 May;61(5):1457–1465. doi: 10.1128/jvi.61.5.1457-1465.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French R., Janda M., Ahlquist P. Bacterial gene inserted in an engineered RNA virus: efficient expression in monocotyledonous plant cells. Science. 1986 Mar 14;231(4743):1294–1297. doi: 10.1126/science.231.4743.1294. [DOI] [PubMed] [Google Scholar]
- Goldbach R. Genome similarities between plant and animal RNA viruses. Microbiol Sci. 1987 Jul;4(7):197–202. [PubMed] [Google Scholar]
- Haseloff J., Goelet P., Zimmern D., Ahlquist P., Dasgupta R., Kaesberg P. Striking similarities in amino acid sequence among nonstructural proteins encoded by RNA viruses that have dissimilar genomic organization. Proc Natl Acad Sci U S A. 1984 Jul;81(14):4358–4362. doi: 10.1073/pnas.81.14.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa M., Meshi T., Watanabe Y., Okada Y. Replication of chimeric tobacco mosaic viruses which carry heterologous combinations of replicase genes and 3' noncoding regions. Virology. 1988 May;164(1):290–293. doi: 10.1016/0042-6822(88)90648-4. [DOI] [PubMed] [Google Scholar]
- Janda M., French R., Ahlquist P. High efficiency T7 polymerase synthesis of infectious RNA from cloned brome mosaic virus cdna and effects of 5' extensions on transcript infectivity. Virology. 1987 May;158(1):259–262. doi: 10.1016/0042-6822(87)90265-0. [DOI] [PubMed] [Google Scholar]
- Kamer G., Argos P. Primary structural comparison of RNA-dependent polymerases from plant, animal and bacterial viruses. Nucleic Acids Res. 1984 Sep 25;12(18):7269–7282. doi: 10.1093/nar/12.18.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiberstis P. A., Loesch-Fries L. S., Hall T. C. Viral protein synthesis in barley protoplasts inoculated with native and fractionated brome mosaic virus RNA. Virology. 1981 Jul 30;112(2):804–808. doi: 10.1016/0042-6822(81)90331-7. [DOI] [PubMed] [Google Scholar]
- Kroner P. A., Young B. M., Ahlquist P. Analysis of the role of brome mosaic virus 1a protein domains in RNA replication, using linker insertion mutagenesis. J Virol. 1990 Dec;64(12):6110–6120. doi: 10.1128/jvi.64.12.6110-6120.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroner P., Richards D., Traynor P., Ahlquist P. Defined mutations in a small region of the brome mosaic virus 2 gene cause diverse temperature-sensitive RNA replication phenotypes. J Virol. 1989 Dec;63(12):5302–5309. doi: 10.1128/jvi.63.12.5302-5309.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh L. E., Dreher T. W., Hall T. C. Mutational analysis of the core and modulator sequences of the BMV RNA3 subgenomic promoter. Nucleic Acids Res. 1988 Feb 11;16(3):981–995. doi: 10.1093/nar/16.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller W. A., Bujarski J. J., Dreher T. W., Hall T. C. Minus-strand initiation by brome mosaic virus replicase within the 3' tRNA-like structure of native and modified RNA templates. J Mol Biol. 1986 Feb 20;187(4):537–546. doi: 10.1016/0022-2836(86)90332-3. [DOI] [PubMed] [Google Scholar]
- Miller W. A., Dreher T. W., Hall T. C. Synthesis of brome mosaic virus subgenomic RNA in vitro by internal initiation on (-)-sense genomic RNA. Nature. 1985 Jan 3;313(5997):68–70. doi: 10.1038/313068a0. [DOI] [PubMed] [Google Scholar]
- Miller W. A., Hall T. C. RNA-dependent RNA polymerase isolated from cowpea chlorotic mottle virus-infected cowpeas is specific for bromoviral RNA. Virology. 1984 Jan 15;132(1):53–60. doi: 10.1016/0042-6822(84)90090-4. [DOI] [PubMed] [Google Scholar]
- Miller W. A., Hall T. C. Use of micrococcal nuclease in the purification of highly template dependent RNA-dependent RNA polymerase from brome mosaic virus-infected barley. Virology. 1983 Feb;125(1):236–241. doi: 10.1016/0042-6822(83)90077-6. [DOI] [PubMed] [Google Scholar]
- Pacha R. F., Allison R. F., Ahlquist P. cis-acting sequences required for in vivo amplification of genomic RNA3 are organized differently in related bromoviruses. Virology. 1990 Feb;174(2):436–443. doi: 10.1016/0042-6822(90)90097-b. [DOI] [PubMed] [Google Scholar]
- Sacher R., French R., Ahlquist P. Hybrid brome mosaic virus RNAs express and are packaged in tobacco mosaic virus coat protein in vivo. Virology. 1988 Nov;167(1):15–24. doi: 10.1016/0042-6822(88)90049-9. [DOI] [PubMed] [Google Scholar]
- Schenborn E. T., Mierendorf R. C., Jr A novel transcription property of SP6 and T7 RNA polymerases: dependence on template structure. Nucleic Acids Res. 1985 Sep 11;13(17):6223–6236. doi: 10.1093/nar/13.17.6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynor P., Ahlquist P. Use of bromovirus RNA2 hybrids to map cis- and trans-acting functions in a conserved RNA replication gene. J Virol. 1990 Jan;64(1):69–77. doi: 10.1128/jvi.64.1.69-77.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynor P., Young B. M., Ahlquist P. Deletion analysis of brome mosaic virus 2a protein: effects on RNA replication and systemic spread. J Virol. 1991 Jun;65(6):2807–2815. doi: 10.1128/jvi.65.6.2807-2815.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Belkum A., Abrahams J. P., Pleij C. W., Bosch L. Five pseudoknots are present at the 204 nucleotides long 3' noncoding region of tobacco mosaic virus RNA. Nucleic Acids Res. 1985 Nov 11;13(21):7673–7686. doi: 10.1093/nar/13.21.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]