Abstract
In this work, we extend the study of the genes controlling the formation of domes in the rat mammary cell line LA7 under the influence of DMSO. The role of the rat8 gene has already been demonstrated. We have now studied two additional genes. The first, called 133, is the rat ortholog of the human epithelial membrane protein 3 (EMP3), a member of the peripheral myelin protein 22 (PMP22)/EMP/lens-specific membrane protein 20 (MP20) gene family that encodes for tetratransmembrane proteins; it is expressed in the LA7 line in the absence of DMSO but not in its presence. The second gene is the β subunit of the amiloride-sensitive Na+ channel. Studies with antisense oligonucleotides show that the formation of domes is under the control of all three genes: the expression of rat8 is required for both their formation and their persistence; the expression of the Na+ channel β subunit is required for their formation; and the expression of gene 133 blocks the expression of the Na+ channel genes, thus preventing formation of the domes. The formation of these structures is also accompanied by the expression of α6β1 integrin, followed by that of E-cadherin and cytokeratin 8. It appears, therefore, that dome formation requires the activity of the Na+ channel and the rat8-encoded protein and is under the negative control of gene 133. DMSO induces dome formation by blocking this control.
Investigation of complex biological processes such as transepithelial fluid transport has been greatly facilitated by the use of cell culture systems, which are amenable to a genetic analysis of the underlying mechanisms. The cell line LA7, a clonal derivative of the cell line RAMA 25 obtained from a chemically induced mammary tumor in rats (1), has the ability to form hemiblisters or domes in postconfluent cultures. These are discrete, roughly circular areas in which the cell layer is detached from the dish by focal accumulation of fluid. The formation of domes, which is a manifestation of vectorial transepithelial transport of water and solutes, occurs to a certain extent in untreated cultures after they have reached confluence, but is strongly promoted by exposure of the cells, either to the differentiation inducer DMSO or to cAMP analogs, such as 8-Br-cAMP and dibutyryl-cAMP. In a previous study, we demonstrated that the expression of the gene rat8 is required for the formation of domes, as shown by the ability of an antisense oligonucleotide to the rat8 messenger to completely and specifically abolish their production (2). rat8 is homologous to the human gene 9-27, an interferon-inducible gene encoding for the protein Leu-13, that, together with α6β1 integrin and the tetraspan family member CD81 (3), is part of a multimeric membrane complex involved in the transduction of antiproliferative and homotypic adhesion signals in B lymphocytes (4). By in situ hybridization, the cells of the domes express rat8 much more strongly than the surrounding cells of the same culture, and, by immunocytochemistry, they display a strong signal for E-cadherin and cytokeratin 8, while only a few of the surrounding cells display them. The rat8 gene is strongly expressed in tubular structures (in the rat), both in the mammary gland and other organs (2). The formation of domes may, therefore, involve molecular mechanisms that also participate in the formation of tubular structures in the whole animal. In this paper, we describe the identification of additional genes involved in the control of dome formation.
Materials and Methods
Cells.
The cell lines LA7 and 106A10 were cultured as described by Dulbecco et al. (5). To study dome formation, sets of three 35-mm plates seeded with 3 × 105 cells per cm2 were grown to confluence (for 48 hr) and subsequently incubated in the presence of DMSO 1.5% as inducer of dome formation (6).
Isolation of Specific Clones from cDNA Subtraction Libraries.
The strategy employed for the isolation of clones specifically expressed in 106A10 cells parallels the one for obtaining clones specific for LA7 cells, which has been described in detail in our previous paper (2); the only change is that cDNA prepared from mRNA extracted from uninduced 106A10 cells was used as a tester, while the mRNA from DMSO-induced LA7 was used as driver. The PCR-amplified products obtained at the end of the procedure were then cloned by direct ligation into the TA vector (Invitrogen), which was used to transform competent DH5α cells.
For sequencing, we used the universal primer M13 from the TA vector. Nucleic acid sequence homology searches were analyzed by using FastA and Wordsearch programs of the GCG sequence analysis package by searching the complete combined GenBank/EMBL data banks. Amino acid sequence homology searches were also conducted on the complete SwissProt database. Selected amino acid sequences were analyzed by using Blocks (7), while the Scan Prosite program was used to identify potential motifs within translated sequences.
RNA Extraction and Northern Blot Hybridization.
RNA extraction and Northern blot analysis were performed according to standard procedures (8) using 10 μg of total RNA from each cell line. Probes were prepared from PCR-amplified clone inserts and [32P]dCTP-labeled by random priming with Ready Prime (Amersham Pharmacia). Prehybridization, hybridization, and washing conditions were carried out at 45°C according to standard procedures (8). A loading control was performed by using [32P]dCTP-labeled β-actin or 36B4-labeled cDNA probe representing a gene whose level of expression is independent of the action of inducers (9).
Antisense Oligodeoxynucleotide Methodology.
For inhibition studies of the epithelial Na+ channel β subunit mRNA expression, three oligodeoxynucleotides of 20 bases were synthesized: antisense oligomer (5′-GGACGCAAGGAAGGGGACAT-3′) designed as a complementary sequence at the 5′ end of the β subunit coding region, starting at position 921 of the sequence (accession no. U35175); sense oligomer (5′-ATGTCCCCTTCCTTGCGTCC-3′) from the same coding region; and scrambled oligomer (5′-GATGAGAGACGACGAGAGCG-3′), a scrambled sequence with the same nucleotides used for the antisense oligomer. Cell culture conditions were exactly as previously described for rat8 antisense studies (2).
For inhibition studies of gene 133, the following oligonucleotides designed on our cDNA sequence were used: antisense (5′-AGGACAGACAGCAGAGGAT-3′), sense (5′-ATCCTCTGCTGTCTGTCCT-3′), and scrambled (5′-GGAATCAGGGAAACGGAAC-3′). The experiment with 133 antisense was performed with 3 × 105 cells per cm2 according to the procedure already described (2, 10) with minor modifications. Cells were plated, and, after 12 hr, the medium was replaced with a fresh one containing inactivated serum. The flasks were incubated for an additional 24 hr after the addition of 80 μg/ml of one of the three oligomers to each flask, and a control culture flask was left untreated. An additional 40 μg/ml of each oligomer were added to the corresponding flasks twice (every 12 hr). Cells were maintained in culture for 60 hr, inspected for dome formation, photographed, and harvested for RNA extraction.
Immunofluorescence Microscopy.
Detection of α6β1 integrin was performed on cells grown for 3 days on Permanox chamber slides with coverslips (Nunc), as described for cytokeratin 8 and E-cadherin (2). Cells were induced with 1.5% DMSO, fixed in a paraformaldehyde/PBS gradient from 0.5–4% for 20 min, and then incubated with commercial mAb raised against mouse α6β1 integrin (Serotec), according to the manufacturer’s protocol. The secondary Ab used was FITC-labeled anti-mouse IgG (Vector Laboratories). Cells were microscopically examined and photographed with a 4× objective. Detection of cytokeratin 8 and E-cadherin was performed as previously described (2).
Reverse Transcription-PCR.
RT-PCR was performed as previously described on retrotranscribed cDNA from uninduced and induced cells (2). For detection of the three amiloride-sensitive Na+ channel α, β, and γ subunits, the following primers were used: for α subunit, forward primer (5′-GCAACCAGAAACAAATCAGAC-3′) (nucleotides 802–821, accession no. U54700) and reverse primer (5′-ACCATCATCCATAAAGGCAG-3′) (nucleotides 1211–1192); for β subunit, forward primer (5′-ACACCAACACCACCAGTACC-3′) (nucleotides 408–427, accession no. U35175) and reverse primer (5′-GAGACCAAATTCAGTCCCAG-3′) (nucleotides 889–870); for γ subunit, forward primer (5′-TACTGCATGAACACCAACACCC-3′) (nucleotides 146–167, accession no. U37539) and reverse primer (5′-GACCCCATACAAGGACAGCAAG-3′) (nucleotides 445–424). Thirty-five cycles were performed at 94°C (denaturation), 55°C (annealing), and 72°C (elongation). Control samples including not-retrotranscribed RNA from LA7 and 106A10 cells were also included.
Results
Identification of Epithelial Membrane Protein 3 (EMP3) Gene Through Differential Cloning.
The construction of the subtracted cDNA library allowed the isolation of several cDNAs. One of these, clone 133, was completely sequenced, and its sequence was subjected to computer analysis.
Clone 133 codes for a 163-aa protein that shows 93.3% (mouse: accession no. AF011750) to 91.4% (human: accession no. X94771 and U87947) sequence identity at the amino acid level with mouse and human EMP3/HNMP1 (hematopoietic neural membrane protein 1) gene, respectively (11–13). Homology with EMP1 and EMP2 and other members of the family ranged from 52% to 89%, clearly suggesting that clone 133 is the rat ortholog of human EMP3 gene, a member of the peripheral myelin protein 22 (PMP22)/EMP/lens-specific membrane protein 20 (MP20) family (14). Members of this family, belonging to the larger tetraspan superfamily, have been shown to be involved in cell–cell interaction and adhesion (12, 14).
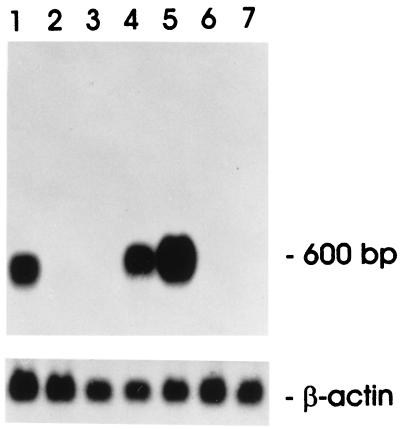
To confirm that clone 133 is specific for 106A10 cells, we performed Northern blot analysis of induced and uninduced LA7 and 106A10 cells. Interestingly, both uninduced 106A10 and uninduced LA7 cells showed expression of this gene (Fig. 1, lanes 1, 4, and 5). However, clone 133 expression was undetectable in LA7 or 106A10 cells incubated with DMSO (Fig. 1, lanes 2, 3, 6, and 7), thus explaining why it was isolated with our subtractive procedure, which used mRNA from DMSO-induced LA7 cells as driver against cDNA obtained from uninduced 106A10. Taken together, these results suggest that EMP3 is a gene that is switched off in mammary carcinoma-derived cells exposed to DMSO, and that this down-regulation could be a necessary but not sufficient step in the differentiation process leading to dome formation by LA7 cells.
Figure 1.
Northern blot analysis of the EMP3 gene expression. Cells were cultured as described in the text and exposed to DMSO at various concentrations. Northern blots were prepared as described in Materials and Methods and hybridized with a radioactive EMP3-specific probe. (Upper) Lane 1, uninduced LA7; lane 2, 1.5% DMSO-induced LA7; lane 3, 3% DMSO-induced LA7; lanes 4 and 5, uninduced 106A10; lane 6, 1.5% DMSO-induced 106A10; and lane 7, 3% DMSO-induced 106A10. (Lower) Hybridization control with β-actin.
Role of Gene 133 in the Formation of Domes.
As the expression of gene 133 is blocked in LA7 cells by DMSO under conditions leading to dome induction, it is possible that the loss of 133 gene expression accompanied differentiation but was not involved in the differentiation itself. To test the importance of the regulation of gene 133 expression in dome formation, we made use of the antisense oligonucleotide methodology. Exposure of confluent LA7 cell cultures to antisense oligonucleotides to the 133 messenger in the absence of DMSO caused the formation of domes, as did the exposure of the cells to DMSO in the absence of antisense (Fig. 3C1). The exposure to 133 mRNA antisense oligonucleotides has, therefore, an effect similar to exposure to DMSO. These results are consistent with the idea that gene 133 is required for down-regulation of dome formation.
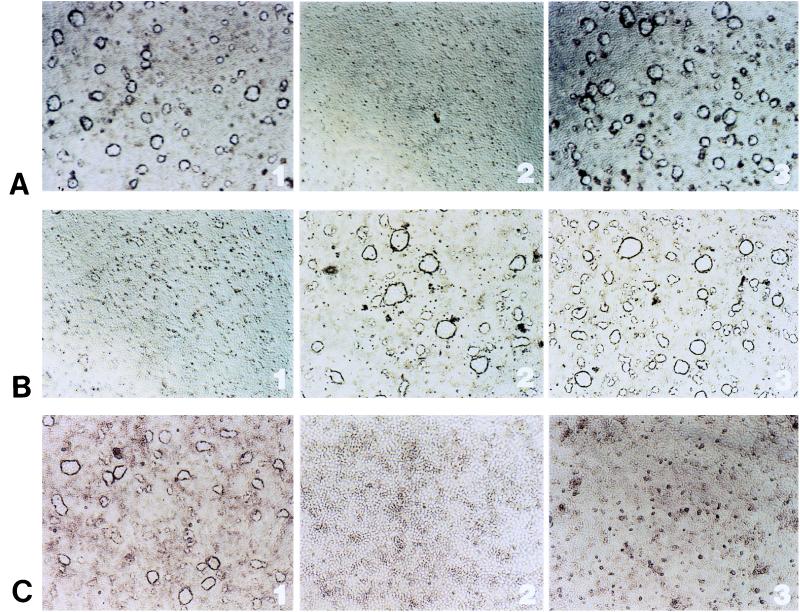
Figure 3.
Effect of antisense oligonucleotides on dome formation and maintenance in LA7 cells. (A1) Domes develop in LA7 cells induced to differentiate with 1.5% DMSO for 36 hr after the addition of the inducer. (A2) Addition of anti-rat8 antisense oligonucleotide causes disappearance of domes in DMSO-induced LA7 cells. (A3) Removal of anti-rat8 antisense oligonucleotide allows for reappearance of domes. (B1) Addition of the epithelial Na+ channel β subunit antisense oligonucleotide causes disappearance of domes in 1.5% DMSO-induced LA7 cells. (B2 and B3) Sense and scrambled oligonucleotides had no effect. (C1) Exposure of LA7 to antisense oligonucleotides to the gene 133 in absence of DMSO causes dome formation. (C2 and C3) The 133 sense and scrambled oligonucleotides had no effect.
The expression of gene 133 also prevents the expression of the β and γ subunits of the amiloride-sensitive epithelial Na+ channel, which is required for the formation of domes (see below). This role of the gene 133 is shown by the effect of its antisense oligonucleotide, which restores the expression of the β and γ subunits of the Na+ channel gene (Fig. 2 C, lanes 2 and 3). Gene 133 may, therefore, be an important regulator in the formation of the domes, by controlling the expression of the channel proteins.
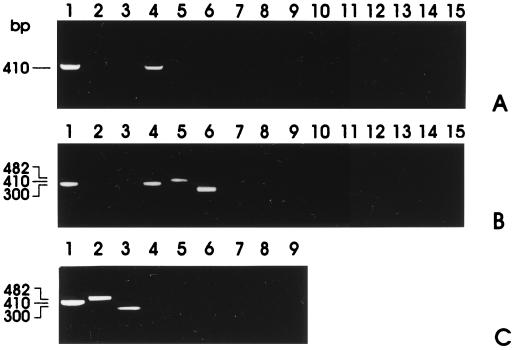
Figure 2.
RT-PCR with primer pairs specific for α, β, and γ subunits of the amiloride-sensitive epithelial Na+ channel performed on uninduced and induced 106A10 and LA7 cells. (A) Lanes 1–3, RT-PCR of uninduced 106A10 cDNA with primers of α (lane 1), β (lane 2), and γ (lane 3) subunits. Lanes 4–6, RT-PCR of DMSO-induced 106A10 cDNA with primers of α (lane 4), β (lane 5), and γ (lane 6) subunits. Lanes 7–12, Negative controls with not-retrotranscribed RNA from samples 1–6. Lanes 13–15, Negative controls without template. (B) Lanes 1–3, RT-PCR of uninduced LA7 cDNA with primers of α (lane 1), β (lane 2), and γ (lane 3) subunits. Lanes 4–6, RT-PCR of DMSO-induced LA7 cDNA with primers of α (lane 4), β (lane 5), and γ (lane 6) subunits. Lanes 7–12, Negative controls with not-retrotranscribed RNA from samples 1–6. Lanes 13–15, Negative controls without template. (C) RT-PCR performed on LA7 cDNA after exposure to antisense oligonucleotides to the EMP3 mRNA. Cells were not exposed to DMSO. Lane 1, α subunits; lane 2, β subunits; and lane 3, γ subunits. Lanes 4–6, Negative controls with not-retrotranscribed RNA from samples 1–3. Lanes 7–9, Negative controls without template.
Role of the Genes for the Subunits of the Amiloride-Sensitive Epithelial Na+ Channel in the Formation of Domes.
The formation of domes requires the expression of many genes, including those involved in the transcellular transport of water and ions, as described for the kidney cell line MDCK, which also forms domes (15). To investigate the role of the amiloride-sensitive epithelial Na+ channel in the process of dome formation, we designed specific primers for each of its three subunits (16, 17) and performed expression analysis by RT-PCR with LA7 and 106A10 cell lines in the presence of various inducers of differentiation or antisense oligonucleotide mRNAs.
The data show that, whereas in the untreated 106A10 and LA7 cultures only the α subunit is appreciably expressed (Fig. 2 A and B, lane 1), exposure to DMSO causes strong expression of the β and γ subunits in LA7 (Fig. 2B, lanes 5 and 6) but not in 106A10 (Fig. 2A, lanes 5 and 6).
To determine the role of these genes in dome formation, LA7 cultures were exposed to the antisense oligonucleotides specific for the RNAs for the αβ and γ subunits; the presence of the antisense oligonucleotides to the β subunit blocked the formation of the domes (Fig. 3B1), while antisense oligonucleotides to the α and γ subunits had no effect. The presence of the β subunit antisense oligonucleotides did not reduce the expression of rat8 (as determined by Northern blot analysis), showing that the expression of the complete Na+ channel is required for the development of domes, independently from the action of rat8. A similar effect is caused by antisense oligonucleotides to the 133 messenger showing that both DMSO and gene 133 control the expression of genes for the β and γ subunits of the Na+ channel (Fig. 2 B and C, and Fig. 3C1).
Role of rat8 in Dome Formation.
The data reported previously show that the expression of rat8 is required, but by itself is not sufficient to cause dome formation; the genes for the subunits of the Na+ channel must also be expressed. It was not clear, however, whether rat8 activity is required only to initiate the formation of domes, or is also continuously required for the persistence of the domes once they are formed. For this purpose, rat8 antisense was added to dome-rich cultures induced by DMSO (Fig. 3A1). This resulted in a rapid dome collapse within 5–6 hr (Fig. 3A2). The subsequent removal of rat8 antisense caused the reappearance of the domes, showing that their disappearance after the addition of the antisense was not the result of nonspecific damage to the cells (Fig. 3A3). The expression of rat8 is therefore continuously required for the persistence of the domes.
Epithelial Marker Expression.
In our previous work (2), it was shown that E-cadherin and cytokeratin 8, detected by immunocytochemistry, are strongly expressed in the domes but are not recognizable in nondoming cells. In an attempt to identify additional proteins selectively localized in domes, we examined the expression of α6β1 integrin. The choice of α6β1 integrin was dictated on one hand by the notion that this integrin is involved in epithelial cell–matrix and cell–cell adhesion, and on the other hand by the finding that in B lymphocytes the tetra-spanning CD81 and the protein Leu-13 (homologous to rat8) form a complex with α6β1 integrin. We found that α6β1 integrin mRNA is expressed in both induced and uninduced cells (3). However, as shown in Fig. 4 A, α6β1 integrin immunoreactivity is high in the cells forming domes, whereas it is present only in scattered cells of the uninduced cultures (Fig. 4B).
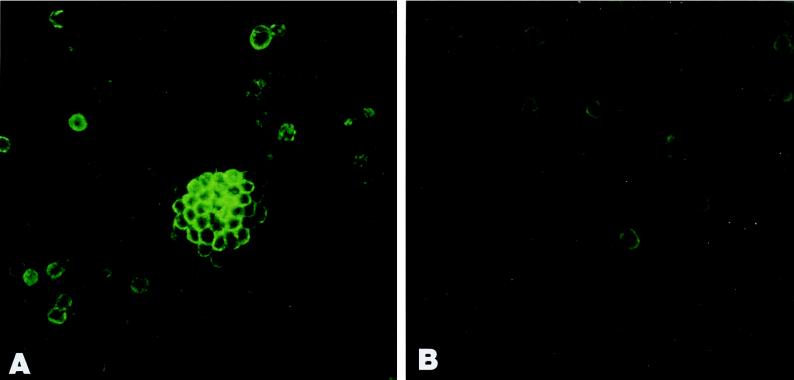
Figure 4.
Immunofluorescence microscopy of DMSO-induced (A) and uninduced (B) LA7 cultures incubated with anti α6β1 integrin mAb. Cells reacting with the Ab are those forming domes (A). LA7 cells not involved in dome formation show scattered immunoreactivity (B). The secondary Ab used was fluorescein-conjugated anti-mouse IgG.
To investigate the possible relationship among E-cadherin, cytokeratin 8, and α6β1 integrin, we compared the time sequence of their appearance in DMSO-induced LA7 cells. Because small domes start appearing approximately 27 hr after DMSO addition, immunohistochemistry was performed every 3 hr from this time. α6β1 integrin was first detectable at 30 hr after DMSO addition, while both E-cadherin and cytokeratin 8 were detected after only 3 additional hr. This result suggests that the expression of the α6β1 integrin influences the expression of the other two proteins.
Discussion
The formation of domes in the presence of DMSO depends on a complex genetic regulation that involves gene rat8, gene 133 (the rat ortholog of human EMP3 gene), and the amiloride-sensitive epithelial Na+ channel. As previously pointed out (4), cAMP-response elements are also involved in the control of dome formation, but probably through a different pathway, which is not considered here. The 133 gene, which encodes for an integral membrane glycoprotein involved in the control of cell growth and cell–cell interactions (12), may play a central role in DMSO-induced dome formation. Indeed, we have shown that the addition of antisense oligonucleotides to mRNA 133 activates the expression of the β subunit of the Na+ channel, thereby allowing, by the reconstruction of a complete channel, the formation of domes. This suggests that an important function of gene 133 is to inhibit the expression of Na+ channel β subunit. Interestingly, a similar effect, i.e., block of 133 expression, is elicited by the DMSO. Thus, the main role of DMSO in the process of dome formation seems to be the suppression of gene 133 expression. DMSO cannot act by activating the expression of rat8 because its expression in LA7 cells is similar in the presence and absence of DMSO.
Presently, we can recognize three main players in the formation of domes: the Na+ channel, which appears to be directly responsible for dome formation; gene 133, which regulates the expression of the Na+ channel β and γ subunits; and rat8, which is likely to act independently of these genes, but whose expression is continuously needed for the persistence of the domes.
A possible function of the rat8 protein may be inferred from its role in B human lymphocytes, where its homologous Leu-13 forms a complex in the cell membrane with several proteins, among which are CD19 and CD21. However, rat8 protein cannot form the same complex in LA7 cells as in B lymphocytes because C19 is not expressed in these cells, by either Western blot or identification of mRNA, and C21 is not detectable by Western blot although it is weakly expressed as a mRNA as detected by RT-PCR (data not shown). rat8-encoded protein may, however, form complexes with proteins, such as the subunits of the Na+ channel and possibly proteins involved in cell adhesion or junction formation. In agreement with our finding that α6β1 integrin expression is strongest in domes, the association of 9-27 (Leu-13)-containing complexes with integrins has recently been demonstrated in B lymphocytes (3). In this regard, the fact that α6β1 integrin is detected in our system before the appearance of E-cadherin and cytokeratin 8 could suggest that integrin expression is required for the induction of the later molecules. It is also noteworthy that the product of gene 133 is a member of the tetraspan superfamily, like Tapa-1 (CD81) and other components of the 9-27 (Leu-13)-containing complex that is known to be involved in signal transduction (3). It is therefore conceivable that in LA7 cells the product of gene 133 directly or indirectly transmits inhibitory signals that repress the expression of the Na+ channel β and γ subunits.
The important function of the β subunit of the Na+ channel in the transepithelial transport of ions and fluids is underscored by its prominent role in the Liddle syndrome, a dominantly inherited disease of the kidney caused by increased Na+ channel current (18). The disease is thought to result from mutations in the conserved C-terminal motif of the β subunit of the Na+ channel, which disrupt the normal interaction with Nedd4, a ubiquitin-protein ligase that down-regulates Na+ channel activity (18). It will be interesting to investigate whether the 133 gene regulates Na+ reabsorption in kidney tubules.
An as-yet-unanswered question is how the coordinated activity of the genes described here is specifically localized to regions of the monolayer involved in dome formation, leaving the surrounding cells apparently unchanged. The availability of a simple in vitro system, such as the one described here, may help to shed light on these questions.
Acknowledgments
We thank Drs. E. Cattaneo, L. Conti, and S. Morara for their help and contribution to immunofluorescence microscopy. We also thank Victoria Starnes for typing the manuscript. This work was partially funded by grants from Associazione Italiana per la Ricerca sul Cancro (to R.D.) and P. F. Biotecnologie (to P.V.). This is manuscript no. 31 of the Genoma 2000/Istituto di Tecnologie Biomediche Avanzate Project funded by Cariplo.
Abbreviations
- EMP3
epithelial membrane protein 3
- PMP22
peripheral myelin protein 22
- MP20
lens-specific membrane protein 20
- RT
reverse transcription
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. Y10889).
References
- 1.Bennett D C, Peachey L A, Durbin H, Rudland P S. Cell. 1978;15:283–298. doi: 10.1016/0092-8674(78)90104-6. [DOI] [PubMed] [Google Scholar]
- 2.Zucchi I, Montagna C, Susani L, Vezzoni P, Dulbecco R. Proc Natl Acad Sci USA. 1998;95:1079–1084. doi: 10.1073/pnas.95.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horvath G, Serru V, Clay D, Billard M, Boucheix C, Rubinstein E. J Biol Chem. 1998;273:30537–30543. doi: 10.1074/jbc.273.46.30537. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi S, Dos C, Levy S, Levy R. J Immunol. 1990;145:2207–2213. [PubMed] [Google Scholar]
- 5.Dulbecco R. Proc Natl Acad Sci USA. 1979;76:1256–1260. doi: 10.1073/pnas.76.3.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dulbecco R, Okada S. Proc R Soc London Ser B. 1980;208:399–408. doi: 10.1098/rspb.1980.0058. [DOI] [PubMed] [Google Scholar]
- 7.Henikoff J G, Pietrokovski S, Henikoff S. Nucleic Acids Res. 1997;25:222–225. doi: 10.1093/nar/25.1.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 9.Rio M C, Bellocq J P, Gairard B, Rasmussen U B, Krust A, Koehl C, Calderoli H, Schiff V, Renaud R, Chambon P. Proc Natl Acad Sci USA. 1987;84:9243–9247. doi: 10.1073/pnas.84.24.9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szczylik C, Skorski T, Nicolaides N C, Manzella L, Malaguarnera L, Venturelli D, Gewirtz A M, Calabretta B. Science. 1991;253:562–565. doi: 10.1126/science.1857987. [DOI] [PubMed] [Google Scholar]
- 11.Taylor V, Suter U. Gene. 1996;175:115–120. doi: 10.1016/0378-1119(96)00134-5. [DOI] [PubMed] [Google Scholar]
- 12.Ben-Porath I, Benvenisty N. Gene. 1996;183:69–75. doi: 10.1016/s0378-1119(96)00475-1. [DOI] [PubMed] [Google Scholar]
- 13.Bolin L M, McNeil T, Lucian L A, Devaux B, Franz-Bacon K, Gorman M D, Zurawski S, Murray R, McClanahan T K. J Neurosci. 1997;17:5493–5502. doi: 10.1523/JNEUROSCI.17-14-05493.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor V, Welcher A A, Program A E, Suter U. J Biol Chem. 1995;270:28824–28833. doi: 10.1074/jbc.270.48.28824. [DOI] [PubMed] [Google Scholar]
- 15.Misfeldt D S, Hamamoto S T, Pitelka D R. Proc Natl Acad Sci USA. 1976;73:1212–1216. doi: 10.1073/pnas.73.4.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Canessa C M, Horisberger J D, Rossier B C. Nature (London) 1993;361:467–470. doi: 10.1038/361467a0. [DOI] [PubMed] [Google Scholar]
- 17.Canessa C M, Schild L, Buell G, Thorens B, Gautschi I, Horisberger J D, Rossier B C. Nature (London) 1994;367:463–467. doi: 10.1038/367463a0. [DOI] [PubMed] [Google Scholar]
- 18.Snyder P M, Price M, McDonald F, Adams C M, Volk K A, Zeiher B G, Stokes J B, Welsh M J. Cell. 1995;83:969–978. doi: 10.1016/0092-8674(95)90212-0. [DOI] [PubMed] [Google Scholar]