Abstract
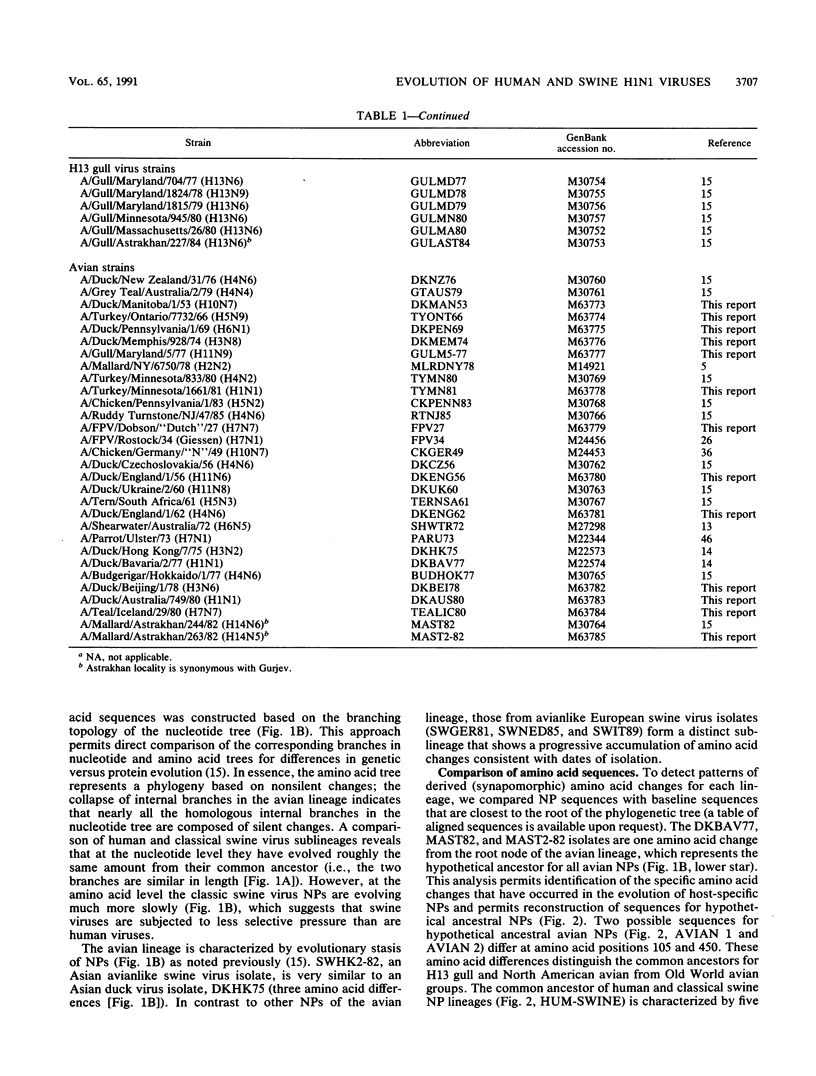
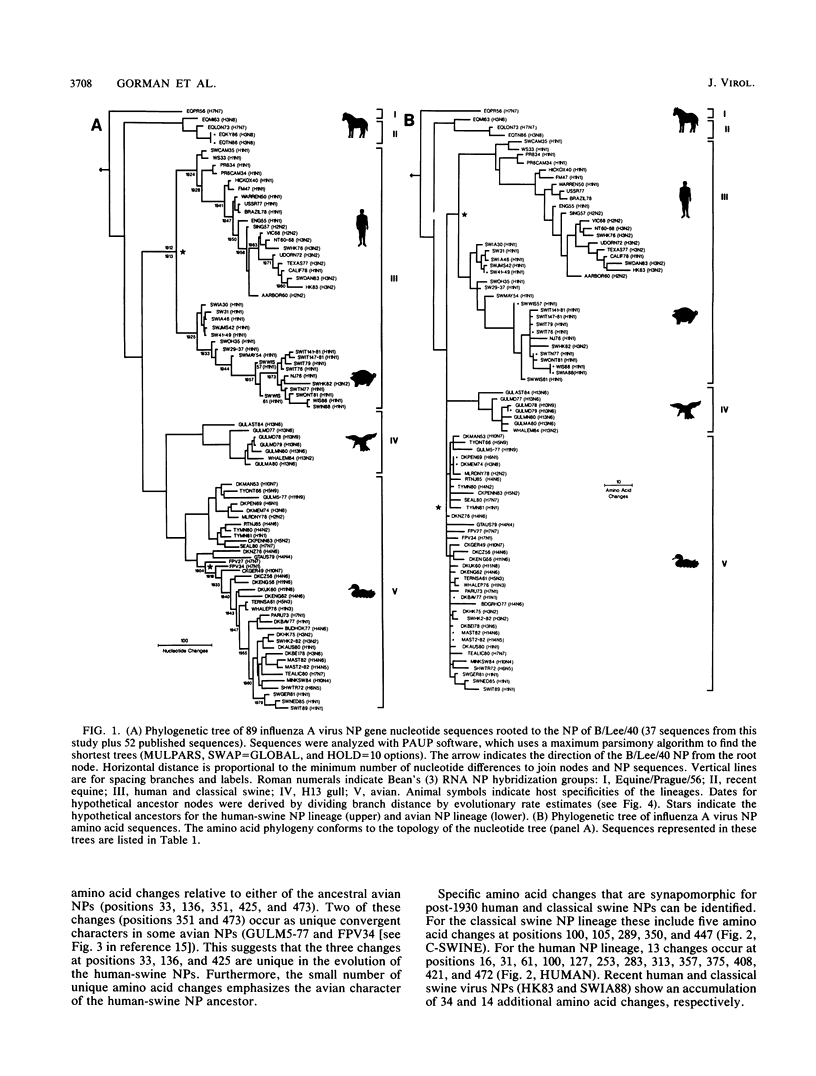
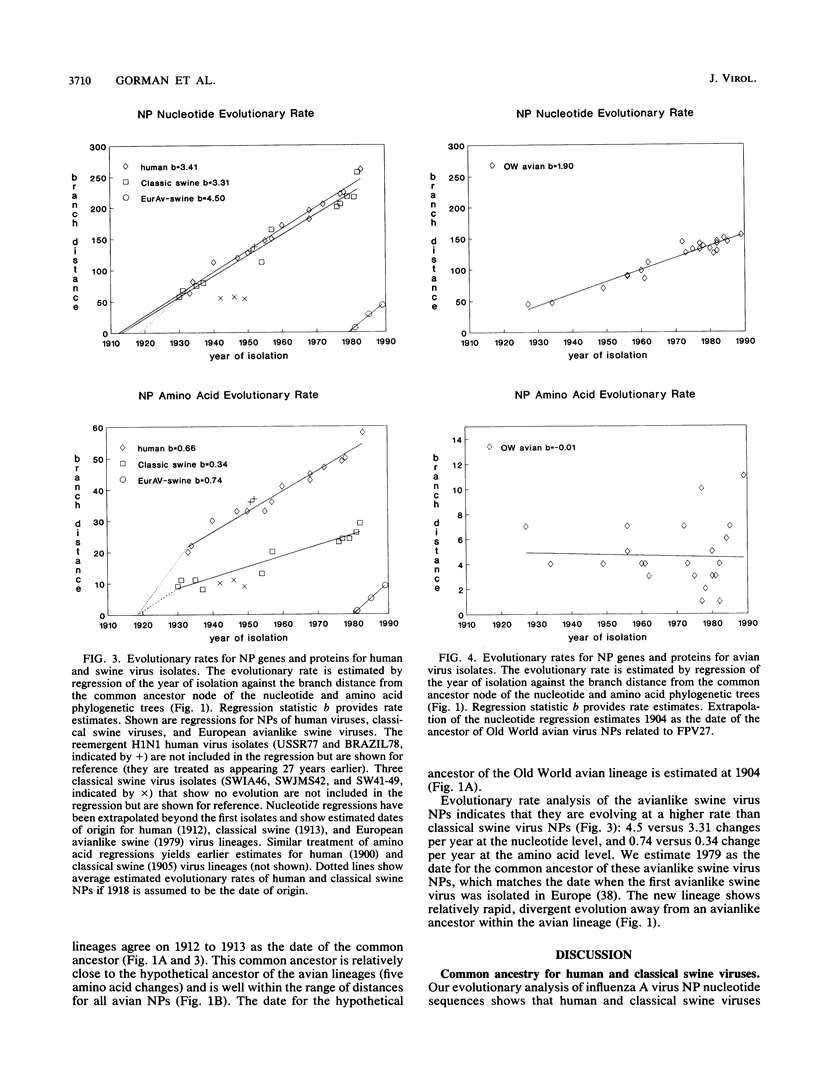
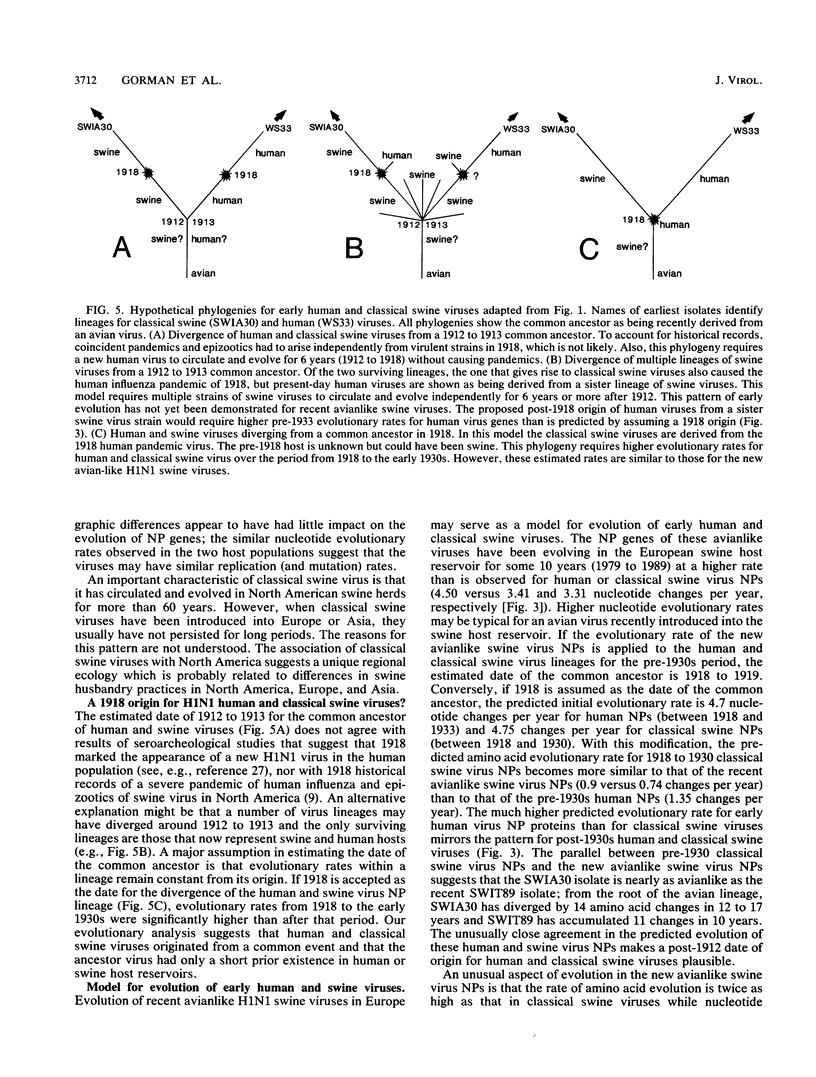
A phylogenetic analysis of 52 published and 37 new nucleoprotein (NP) gene sequences addressed the evolution and origin of human and swine influenza A viruses. H1N1 human and classical swine viruses (i.e., those related to Swine/Iowa/15/30) share a single common ancestor, which was estimated to have occurred in 1912 to 1913. From this common ancestor, human and classical swine virus NP genes have evolved at similar rates that are higher than in avian virus NP genes (3.31 to 3.41 versus 1.90 nucleotide changes per year). At the protein level, human virus NPs have evolved twice as fast as classical swine virus NPs (0.66 versus 0.34 amino acid change per year). Despite evidence of frequent interspecies transmission of human and classical swine viruses, our analysis indicates that these viruses have evolved independently since well before the first isolates in the early 1930s. Although our analysis cannot reveal the original host, the ancestor virus was avianlike, showing only five amino acid differences from the root of the avian virus NP lineage. The common pattern of relationship and origin for the NP and other genes of H1N1 human and classical swine viruses suggests that the common ancestor was an avian virus and not a reassortant derived from previous human or swine influenza A viruses. The new avianlike H1N1 swine viruses in Europe may provide a model for the evolution of newly introduced avian viruses into the swine host reservoir. The NPs of these viruses are evolving more rapidly than those of human or classical swine viruses (4.50 nucleotide changes and 0.74 amino acid change per year), and when these rates are applied to pre-1930s human and classical swine virus NPs, the predicted date of a common ancestor is 1918 rather than 1912 to 1913. Thus, our NP phylogeny is consistent with historical records and the proposal that a short time before 1918, a new H1N1 avianlike virus entered human or swine hosts (O. T. Gorman, R. O. Donis, Y. Kawaoka, and R. G. Webster, J. Virol. 64:4893-4902, 1990). This virus provided the ancestors of all known human influenza A virus genes, except for HA, NA, and PB1, which have since been reassorted from avian viruses. We propose that during 1918 a virulent strain of this new avianlike virus caused a severe human influenza pandemic and that the pandemic virus was introduced into North American swine populations, constituting the origin of classical swine virus.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altmüller A., Fitch W. M., Scholtissek C. Biological and genetic evolution of the nucleoprotein gene of human influenza A viruses. J Gen Virol. 1989 Aug;70(Pt 8):2111–2119. doi: 10.1099/0022-1317-70-8-2111. [DOI] [PubMed] [Google Scholar]
- Bean W. J. Correlation of influenza A virus nucleoprotein genes with host species. Virology. 1984 Mar;133(2):438–442. doi: 10.1016/0042-6822(84)90410-0. [DOI] [PubMed] [Google Scholar]
- Briedis D. J., Tobin M. Influenza B virus genome: complete nucleotide sequence of the influenza B/lee/40 virus genome RNA segment 5 encoding the nucleoprotein and comparison with the B/Singapore/222/79 nucleoprotein. Virology. 1984 Mar;133(2):448–455. doi: 10.1016/0042-6822(84)90412-4. [DOI] [PubMed] [Google Scholar]
- Buckler-White A. J., Murphy B. R. Nucleotide sequence analysis of the nucleoprotein gene of an avian and a human influenza virus strain identifies two classes of nucleoproteins. Virology. 1986 Dec;155(2):345–355. doi: 10.1016/0042-6822(86)90198-4. [DOI] [PubMed] [Google Scholar]
- Buonagurio D. A., Nakada S., Parvin J. D., Krystal M., Palese P., Fitch W. M. Evolution of human influenza A viruses over 50 years: rapid, uniform rate of change in NS gene. Science. 1986 May 23;232(4753):980–982. doi: 10.1126/science.2939560. [DOI] [PubMed] [Google Scholar]
- Cox N. J., Kitame F., Kendal A. P., Maassab H. F., Naeve C. Identification of sequence changes in the cold-adapted, live attenuated influenza vaccine strain, A/Ann Arbor/6/60 (H2N2). Virology. 1988 Dec;167(2):554–567. [PubMed] [Google Scholar]
- DAVENPORT F. M., HENNESSY A. V., DRESCHER J., MULDER J., FRANCIS T., Jr FURTHER OBSERVATIONS ON THE RELEVANCE OF SEROLOGIC RECAPITULATIONS OF HUMAN INFECTION WITH INFLUENZA VIRUSES. J Exp Med. 1964 Dec 1;120:1087–1097. doi: 10.1084/jem.120.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAVENPORT F. M., HENNESSY A. V., FRANCIS T., Jr Epidemiologic and immunologic significance of age distribution of antibody to antigenic variants of influenza virus. J Exp Med. 1953 Dec;98(6):641–656. doi: 10.1084/jem.98.6.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacso C. C., Couch R. B., Six H. R., Young J. F., Quarles J. M., Kasel J. A. Sporadic occurrence of zoonotic swine influenza virus infections. J Clin Microbiol. 1984 Oct;20(4):833–835. doi: 10.1128/jcm.20.4.833-835.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammelin M., Altmüller A., Reinhardt U., Mandler J., Harley V. R., Hudson P. J., Fitch W. M., Scholtissek C. Phylogenetic analysis of nucleoproteins suggests that human influenza A viruses emerged from a 19th-century avian ancestor. Mol Biol Evol. 1990 Mar;7(2):194–200. doi: 10.1093/oxfordjournals.molbev.a040594. [DOI] [PubMed] [Google Scholar]
- Gammelin M., Mandler J., Scholtissek C. Two subtypes of nucleoproteins (NP) of influenza A viruses. Virology. 1989 May;170(1):71–80. doi: 10.1016/0042-6822(89)90353-x. [DOI] [PubMed] [Google Scholar]
- Gorman O. T., Bean W. J., Kawaoka Y., Webster R. G. Evolution of the nucleoprotein gene of influenza A virus. J Virol. 1990 Apr;64(4):1487–1497. doi: 10.1128/jvi.64.4.1487-1497.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman O. T., Donis R. O., Kawaoka Y., Webster R. G. Evolution of influenza A virus PB2 genes: implications for evolution of the ribonucleoprotein complex and origin of human influenza A virus. J Virol. 1990 Oct;64(10):4893–4902. doi: 10.1128/jvi.64.10.4893-4902.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HENNESSY A. V., DAVENPORT F. M. Epidemiologic implications of the distribution by age of antibody response to experimental influenza virus vaccines. J Immunol. 1958 Feb;80(2):114–121. [PubMed] [Google Scholar]
- HENNESSY A. V., DAVENPORT F. M., FRANCIS T., Jr Studies of antibodies to strains of influenza virus in persons of different ages in sera collected in a postepidemic period. J Immunol. 1955 Nov;75(5):401–409. [PubMed] [Google Scholar]
- Hinshaw V. S., Bean W. J., Jr, Webster R. G., Easterday B. C. The prevalence of influenza viruses in swine and the antigenic and genetic relatedness of influenza viruses from man and swine. Virology. 1978 Jan;84(1):51–62. doi: 10.1016/0042-6822(78)90217-9. [DOI] [PubMed] [Google Scholar]
- Huddleston J. A., Brownlee G. G. The sequence of the nucleoprotein gene of human influenza A virus, strain A/NT/60/68. Nucleic Acids Res. 1982 Feb 11;10(3):1029–1038. doi: 10.1093/nar/10.3.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka Y., Krauss S., Webster R. G. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J Virol. 1989 Nov;63(11):4603–4608. doi: 10.1128/jvi.63.11.4603-4608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kida H., Shortridge K. F., Webster R. G. Origin of the hemagglutinin gene of H3N2 influenza viruses from pigs in China. Virology. 1988 Jan;162(1):160–166. doi: 10.1016/0042-6822(88)90405-9. [DOI] [PubMed] [Google Scholar]
- Kundin W. D. Hong Kong A-2 influenza virus infection among swine during a human epidemic in Taiwan. Nature. 1970 Nov 28;228(5274):857–857. doi: 10.1038/228857a0. [DOI] [PubMed] [Google Scholar]
- MULDER J., MASUREL N. Pre-epidemic antibody against 1957 strain of Asiatic influenza in serum of older people living in the Netherlands. Lancet. 1958 Apr 19;1(7025):810–814. doi: 10.1016/s0140-6736(58)91738-0. [DOI] [PubMed] [Google Scholar]
- Mandler J., Gorman O. T., Ludwig S., Schroeder E., Fitch W. M., Webster R. G., Scholtissek C. Derivation of the nucleoproteins (NP) of influenza A viruses isolated from marine mammals. Virology. 1990 May;176(1):255–261. doi: 10.1016/0042-6822(90)90250-u. [DOI] [PubMed] [Google Scholar]
- Mandler J., Scholtissek C. Localisation of the temperature-sensitive defect in the nucleoprotein of an influenza A/FPV/Rostock/34 virus. Virus Res. 1989 Feb;12(2):113–121. doi: 10.1016/0168-1702(89)90058-0. [DOI] [PubMed] [Google Scholar]
- Nakajima K., Desselberger U., Palese P. Recent human influenza A (H1N1) viruses are closely related genetically to strains isolated in 1950. Nature. 1978 Jul 27;274(5669):334–339. doi: 10.1038/274334a0. [DOI] [PubMed] [Google Scholar]
- Nakajima K., Nakajima S., Shortridge K. F., Kendal A. P. Further genetic evidence for maintenance of early Hong Kong-like influenza A(H3N2) strains in swine until 1976. Virology. 1982 Jan 30;116(2):562–572. doi: 10.1016/0042-6822(82)90148-9. [DOI] [PubMed] [Google Scholar]
- Nakajima K., Nobusawa E., Nakajima S. Genetic relatedness between A/Swine/Iowa/15/30(H1N1) and human influenza viruses. Virology. 1984 Nov;139(1):194–198. doi: 10.1016/0042-6822(84)90341-6. [DOI] [PubMed] [Google Scholar]
- Nardelli L., Pascucci S., Gualandi G. L., Loda P. Outbreaks of classical swine influenza in Italy in 1976. Zentralbl Veterinarmed B. 1978 Dec;25(10):853–857. doi: 10.1111/j.1439-0450.1978.tb01062.x. [DOI] [PubMed] [Google Scholar]
- Nerome K., Ishida M., Oya A., Kanai C., Suwicha K. Isolation of an influenza H1N1 virus from a pig. Virology. 1982 Mar;117(2):485–489. doi: 10.1016/0042-6822(82)90486-x. [DOI] [PubMed] [Google Scholar]
- Nerome K., Ishida M., Oya A., Oda K. The possible origin H1N1 (Hsw1N1) virus in the swine population of Japan and antigenic analysis of the isolates. J Gen Virol. 1982 Sep;62(Pt 1):171–175. doi: 10.1099/0022-1317-62-1-171. [DOI] [PubMed] [Google Scholar]
- Okazaki K., Kawaoka Y., Webster R. G. Evolutionary pathways of the PA genes of influenza A viruses. Virology. 1989 Oct;172(2):601–608. doi: 10.1016/0042-6822(89)90202-x. [DOI] [PubMed] [Google Scholar]
- Reinhardt U., Scholtissek C. Comparison of the nucleoprotein genes of a chicken and a mink influenza A H 10 virus. Arch Virol. 1988;103(1-2):139–145. doi: 10.1007/BF01319816. [DOI] [PubMed] [Google Scholar]
- Schnurrenberger P. R., Woods G. T., Martin R. J. Serologic evidence of human infection with swine influenza virus. Am Rev Respir Dis. 1970 Sep;102(3):356–361. doi: 10.1164/arrd.1970.102.3.356. [DOI] [PubMed] [Google Scholar]
- Scholtissek C., Bürger H., Bachmann P. A., Hannoun C. Genetic relatedness of hemagglutinins of the H1 subtype of influenza A viruses isolated from swine and birds. Virology. 1983 Sep;129(2):521–523. doi: 10.1016/0042-6822(83)90194-0. [DOI] [PubMed] [Google Scholar]
- Scholtissek C., Bürger H., Kistner O., Shortridge K. F. The nucleoprotein as a possible major factor in determining host specificity of influenza H3N2 viruses. Virology. 1985 Dec;147(2):287–294. doi: 10.1016/0042-6822(85)90131-x. [DOI] [PubMed] [Google Scholar]
- Scholtissek C., Naylor E. Fish farming and influenza pandemics. Nature. 1988 Jan 21;331(6153):215–215. doi: 10.1038/331215a0. [DOI] [PubMed] [Google Scholar]
- Scholtissek C., von Hoyningen V., Rott R. Genetic relatedness between the new 1977 epidemic strains (H1N1) of influenza and human influenza strains isolated between 1947 and 1957 (H1N1). Virology. 1978 Sep;89(2):613–617. doi: 10.1016/0042-6822(78)90203-9. [DOI] [PubMed] [Google Scholar]
- Shortridge K. F., Cherry A., Kendal A. P. Further studies of the antigenic properties of H3N2 strains of influenza A isolated from swine in South East Asia. J Gen Virol. 1979 Jul;44(1):251–254. doi: 10.1099/0022-1317-44-1-251. [DOI] [PubMed] [Google Scholar]
- Shortridge K. F., Webster R. G., Butterfield W. K., Campbell C. H. Persistence of Hong Kong influenza virus variants in pigs. Science. 1977 Jun 24;196(4297):1454–1455. doi: 10.1126/science.867041. [DOI] [PubMed] [Google Scholar]
- Shortridge K. F., Webster R. G. Geographical distribution of swine (Hsw1N1) and Hong Kong (H3N2) influenza virus variants in pigs in Southeast Asia. Intervirology. 1979;11(1):9–15. doi: 10.1159/000149006. [DOI] [PubMed] [Google Scholar]
- Snyder M. H., Buckler-White A. J., London W. T., Tierney E. L., Murphy B. R. The avian influenza virus nucleoprotein gene and a specific constellation of avian and human virus polymerase genes each specify attenuation of avian-human influenza A/Pintail/79 reassortant viruses for monkeys. J Virol. 1987 Sep;61(9):2857–2863. doi: 10.1128/jvi.61.9.2857-2863.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuler H., Schröder B., Bürger H., Scholtissek C. Sequence of the nucleoprotein gene of influenza A/parrot/Ulster/73. Virus Res. 1985 Jul;3(1):35–40. doi: 10.1016/0168-1702(85)90039-5. [DOI] [PubMed] [Google Scholar]
- Webster R. G., Hinshaw V. S., Laver W. G. Selection and analysis of antigenic variants of the neuraminidase of N2 influenza viruses with monoclonal antibodies. Virology. 1982 Feb;117(1):93–104. doi: 10.1016/0042-6822(82)90510-4. [DOI] [PubMed] [Google Scholar]
- Winter G., Fields S. The structure of the gene encoding the nucleoprotein of human influenza virus A/PR/8/34. Virology. 1981 Oct 30;114(2):423–428. doi: 10.1016/0042-6822(81)90223-3. [DOI] [PubMed] [Google Scholar]
- Yamane N., Arikawa J., Odagiri T., Ishida N. Annual examination of influenza virus infection among pigs in Miyagi prefecture, Japan: the appearance of Hsw1N1 virus. Acta Virol. 1979 May;23(3):240–248. [PubMed] [Google Scholar]


