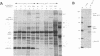Abstract
The human immunodeficiency virus type 1 (HIV-1) Gag-Pol fusion polyprotein is produced via ribosomal frameshifting. Previous studies in vitro and in Saccharomyces cerevisiae have argued against a significant role for RNA secondary structure 3' of the shift site, in contrast with other systems, in which such structure has been shown to be required. Here we show, by expressing the HIV-1 gag-pol domain in cultured vertebrate cells, that a stem-loop structure 3' of the HIV-1 shift site is indeed important for wild-type levels of frameshifting in vivo.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atkins J. F., Weiss R. B., Gesteland R. F. Ribosome gymnastics--degree of difficulty 9.5, style 10.0. Cell. 1990 Aug 10;62(3):413–423. doi: 10.1016/0092-8674(90)90007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Boursnell M. E., Binns M. M., Bilimoria B., Blok V. C., Brown T. D., Inglis S. C. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 1987 Dec 1;6(12):3779–3785. doi: 10.1002/j.1460-2075.1987.tb02713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Digard P., Inglis S. C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989 May 19;57(4):537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Rolley N. J., Jenner A. J., Inglis S. C. Mutational analysis of the RNA pseudoknot component of a coronavirus ribosomal frameshifting signal. J Mol Biol. 1991 Aug 20;220(4):889–902. doi: 10.1016/0022-2836(91)90361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassan M., Berteaux V., Angrand P. O., Rousset J. P. Expression vectors for quantitating in vivo translational ambiguity: their potential use to analyse frameshifting at the HIV gag-pol junction. Res Virol. 1990 Nov-Dec;141(6):597–610. doi: 10.1016/0923-2516(90)90033-F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamorro M., Parkin N., Varmus H. E. An RNA pseudoknot and an optimal heptameric shift site are required for highly efficient ribosomal frameshifting on a retroviral messenger RNA. Proc Natl Acad Sci U S A. 1992 Jan 15;89(2):713–717. doi: 10.1073/pnas.89.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigen W. J., Caskey C. T. Translational frameshifting: where will it stop? Cell. 1987 Jul 3;50(1):1–2. doi: 10.1016/0092-8674(87)90652-0. [DOI] [PubMed] [Google Scholar]
- Dinman J. D., Icho T., Wickner R. B. A -1 ribosomal frameshift in a double-stranded RNA virus of yeast forms a gag-pol fusion protein. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):174–178. doi: 10.1073/pnas.88.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein K. M., Goff S. P. Expression of the gag-pol fusion protein of Moloney murine leukemia virus without gag protein does not induce virion formation or proteolytic processing. J Virol. 1988 Jun;62(6):2179–2182. doi: 10.1128/jvi.62.6.2179-2182.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flower A. M., McHenry C. S. The gamma subunit of DNA polymerase III holoenzyme of Escherichia coli is produced by ribosomal frameshifting. Proc Natl Acad Sci U S A. 1990 May;87(10):3713–3717. doi: 10.1073/pnas.87.10.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura T., Esteban R., Esteban L. M., Wickner R. B. Portable encapsidation signal of the L-A double-stranded RNA virus of S. cerevisiae. Cell. 1990 Aug 24;62(4):819–828. doi: 10.1016/0092-8674(90)90125-x. [DOI] [PubMed] [Google Scholar]
- Geballe A. P., Spaete R. R., Mocarski E. S. A cis-acting element within the 5' leader of a cytomegalovirus beta transcript determines kinetic class. Cell. 1986 Sep 12;46(6):865–872. doi: 10.1016/0092-8674(86)90068-1. [DOI] [PubMed] [Google Scholar]
- Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981 Jan;23(1):175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Hatfield D., Oroszlan S. The where, what and how of ribosomal frameshifting in retroviral protein synthesis. Trends Biochem Sci. 1990 May;15(5):186–190. doi: 10.1016/0968-0004(90)90159-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Madhani H. D., Masiarz F. R., Varmus H. E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988 Nov 4;55(3):447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Power M. D., Masiarz F. R., Luciw P. A., Barr P. J., Varmus H. E. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988 Jan 21;331(6153):280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- Jacks T., Townsley K., Varmus H. E., Majors J. Two efficient ribosomal frameshifting events are required for synthesis of mouse mammary tumor virus gag-related polyproteins. Proc Natl Acad Sci U S A. 1987 Jun;84(12):4298–4302. doi: 10.1073/pnas.84.12.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T. Translational suppression in gene expression in retroviruses and retrotransposons. Curr Top Microbiol Immunol. 1990;157:93–124. doi: 10.1007/978-3-642-75218-6_4. [DOI] [PubMed] [Google Scholar]
- Jacks T., Varmus H. E. Expression of the Rous sarcoma virus pol gene by ribosomal frameshifting. Science. 1985 Dec 13;230(4731):1237–1242. doi: 10.1126/science.2416054. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Loeb D. D., Swanstrom R., Everitt L., Manchester M., Stamper S. E., Hutchison C. A., 3rd Complete mutagenesis of the HIV-1 protease. Nature. 1989 Aug 3;340(6232):397–400. doi: 10.1038/340397a0. [DOI] [PubMed] [Google Scholar]
- Malim M. H., Hauber J., Fenrick R., Cullen B. R. Immunodeficiency virus rev trans-activator modulates the expression of the viral regulatory genes. Nature. 1988 Sep 8;335(6186):181–183. doi: 10.1038/335181a0. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergener K., Fäcke M., Welker R., Brinkmann V., Gelderblom H. R., Kräusslich H. G. Analysis of HIV particle formation using transient expression of subviral constructs in mammalian cells. Virology. 1992 Jan;186(1):25–39. doi: 10.1016/0042-6822(92)90058-w. [DOI] [PubMed] [Google Scholar]
- Moosmayer D., Reil H., Ausmeier M., Scharf J. G., Hauser H., Jentsch K. D., Hunsmann G. Expression and frameshifting but extremely inefficient proteolytic processing of the HIV-1 gag and pol gene products in stably transfected rodent cell lines. Virology. 1991 Jul;183(1):215–224. doi: 10.1016/0042-6822(91)90134-W. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa S., Bishop D. H. Identification and analysis of the gag-pol ribosomal frameshift site of feline immunodeficiency virus. Virology. 1992 Feb;186(2):389–397. doi: 10.1016/0042-6822(92)90004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscovici C., Moscovici M. G., Jimenez H., Lai M. M., Hayman M. J., Vogt P. K. Continuous tissue culture cell lines derived from chemically induced tumors of Japanese quail. Cell. 1977 May;11(1):95–103. doi: 10.1016/0092-8674(77)90320-8. [DOI] [PubMed] [Google Scholar]
- Park J., Morrow C. D. Overexpression of the gag-pol precursor from human immunodeficiency virus type 1 proviral genomes results in efficient proteolytic processing in the absence of virion production. J Virol. 1991 Sep;65(9):5111–5117. doi: 10.1128/jvi.65.9.5111-5117.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C., Chang N. T., Chang T. W. Identification and characterization of human immunodeficiency virus type 1 gag-pol fusion protein in transfected mammalian cells. J Virol. 1991 May;65(5):2751–2756. doi: 10.1128/jvi.65.5.2751-2756.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reil H., Hauser H. Test system for determination of HIV-1 frameshifting efficiency in animal cells. Biochim Biophys Acta. 1990 Aug 27;1050(1-3):288–292. doi: 10.1016/0167-4781(90)90183-3. [DOI] [PubMed] [Google Scholar]
- Rosen C. A., Pavlakis G. N. Tat and Rev: positive regulators of HIV gene expression. AIDS. 1990 Jun;4(6):499–509. [PubMed] [Google Scholar]
- Ross E. K., Fuerst T. R., Orenstein J. M., O'Neill T., Martin M. A., Venkatesan S. Maturation of human immunodeficiency virus particles assembled from the gag precursor protein requires in situ processing by gag-pol protease. AIDS Res Hum Retroviruses. 1991 May;7(5):475–483. doi: 10.1089/aid.1991.7.475. [DOI] [PubMed] [Google Scholar]
- Smith A. J., Cho M. I., Hammarskjöld M. L., Rekosh D. Human immunodeficiency virus type 1 Pr55gag and Pr160gag-pol expressed from a simian virus 40 late replacement vector are efficiently processed and assembled into viruslike particles. J Virol. 1990 Jun;64(6):2743–2750. doi: 10.1128/jvi.64.6.2743-2750.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steimer K. S., Puma J. P., Power M. D., Powers M. A., George-Nascimento C., Stephans J. C., Levy J. A., Sanchez-Pescador R., Luciw P. A., Barr P. J. Differential antibody responses of individuals infected with AIDS-associated retroviruses surveyed using the viral core antigen p25gag expressed in bacteria. Virology. 1986 Apr 15;150(1):283–290. doi: 10.1016/0042-6822(86)90289-8. [DOI] [PubMed] [Google Scholar]
- Tsuchihashi Z., Kornberg A. Translational frameshifting generates the gamma subunit of DNA polymerase III holoenzyme. Proc Natl Acad Sci U S A. 1990 Apr;87(7):2516–2520. doi: 10.1073/pnas.87.7.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchihashi Z. Translational frameshifting in the Escherichia coli dnaX gene in vitro. Nucleic Acids Res. 1991 May 11;19(9):2457–2462. doi: 10.1093/nar/19.9.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D. H., Sugimoto N., Jaeger J. A., Longfellow C. E., Freier S. M., Kierzek R. Improved parameters for prediction of RNA structure. Cold Spring Harb Symp Quant Biol. 1987;52:123–133. doi: 10.1101/sqb.1987.052.01.017. [DOI] [PubMed] [Google Scholar]
- Wilson W., Braddock M., Adams S. E., Rathjen P. D., Kingsman S. M., Kingsman A. J. HIV expression strategies: ribosomal frameshifting is directed by a short sequence in both mammalian and yeast systems. Cell. 1988 Dec 23;55(6):1159–1169. doi: 10.1016/0092-8674(88)90260-7. [DOI] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis using M13-derived vectors: an efficient and general procedure for the production of point mutations in any fragment of DNA. Nucleic Acids Res. 1982 Oct 25;10(20):6487–6500. doi: 10.1093/nar/10.20.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Dam E. B., Pleij C. W., Bosch L. RNA pseudoknots: translational frameshifting and readthrough on viral RNAs. Virus Genes. 1990 Jul;4(2):121–136. doi: 10.1007/BF00678404. [DOI] [PMC free article] [PubMed] [Google Scholar]