Abstract
Selectin-mediated adhesion of tumor cells to platelets, leukocytes, and endothelial cells may regulate their hematogenous dissemination in the microvasculature. We recently identified CD44 variant isoforms (CD44v) as functional P-, but not E- or L-, selectin ligands on colon carcinoma cells. Moreover, an ∼180-kDa sialofucosylated glycoprotein(s) mediated selectin binding in CD44-knockdown cells. Using immunoaffinity chromatography and tandem mass spectrometry, we identify this glycoprotein as the carcinoembryonic antigen (CEA). Blot rolling assays and flow-based adhesion assays using microbeads coated with CEA immunopurified from LS174T colon carcinoma cells and selectins as substrate reveal that CEA possesses E- and L-, but not P-, selectin ligand activity. CEA on CD44-knockdown LS174T cells exhibits higher HECA-452 immunoreactivity than CEA on wild-type cells, suggesting that CEA functions as an alternative acceptor for selectin-binding glycans. The enhanced expression of HECA-452 reactive epitopes on CEA from CD44-knockdown cells correlates with the increased CEA avidity for E- but not L-selectin. Through the generation of stable knockdown cell lines, we demonstrate that CEA serves as an auxiliary L-selectin ligand, which stabilizes L-selectin-dependent cell rolling against fluid shear. Moreover, CEA and CD44v cooperate to mediate colon carcinoma cell adhesion to E- and L-selectin at elevated shear stresses. The novel finding that CEA is an E- and L-selectin ligand may explain the enhanced metastatic potential associated with tumor cell CEA overexpression and the supportive role of selectins in metastasis.
Several lines of evidence suggest that selectins facilitate cancer metastasis and tumor cell arrest in the microvasculature by mediating specific interactions between selectin-expressing hosts cells and ligands on tumor cells. A variety of tumor cells, including colon carcinoma, express sialylated, fucosylated molecules that could be recognized by selectins (1–6). Enhanced expression of sialylated fucosylated glycans such as sialyl Lex and sialyl Lea on the tumor cell surface correlates with poor prognosis because of tumor progression and metastatic spread (7–10). Earlier studies hypothesized a simple model whereby E-selectin expressed on the surface of activated endothelial cells mediates binding of malignant cells, thereby facilitating their extravasation from the vasculature and the seeding of metastatic foci. This model was corroborated by ample experimental evidence. For instance, the ability of human colon carcinoma cell lines to form lung metastases in nude mice correlates with their adhesion to E-selectin and is markedly diminished by a soluble E-selectin fusion protein (11). Along these lines, the metastatic potential of colon carcinoma cell lines is attenuated by preincubating carcinoma cells with antisialyl Lea antibodies (10). The overexpression of E-selectin in the liver of a transgenic mouse model redirected the metastasis to this organ (12).
During their transit into the circulatory system, tumor cells are exposed to fluid mechanical forces, plasma proteins, and blood cells such as P-selectin-expressing platelets and L-selectin-bearing leukocytes, all of which may affect their survival and escape from the bloodstream. Thus, more complex interactions between tumor cells expressing selectin ligands and host cells (i.e. endothelial cells, platelets, and leukocytes) could potentially occur within the vasculature. Published studies have disclosed a clear role for P-selectin involvement in platelet-tumor cell binding and facilitation of metastasis (13–15). Microscopic observations of tumor cells arrested in the lungs of wild-type mice reveal the presence of a dense coat of platelets surrounding the colon carcinoma cells (13). In contrast, carcinoma cells in P-selectin-deficient mice had a looser and more limited platelet coat (13). The initial seeding and subsequent lodging of metastatic cells in target organs was dramatically attenuated in P-selectin knock-out mice compared with wild-type controls (13–15). Although these studies clearly suggest that platelet P-selectin plays a key role not only in carcinoma cell-platelet adhesion but also in the facilitation of metastasis, they cannot rule out an additional role for endothelial P-selectin that could potentially bind carcinoma cells and mediate their extravasation from the bloodstream. On the other hand, the contribution of L-selectin to cancer metastasis is less developed (14, 16). It is believed that tumor cells can form multicellular complexes with platelets and leukocytes (via an L-selectin-dependent mechanism (17, 18)), which can then arrest in the microvasculature of distant organs and eventually extravasate and establish metastatic colonies. Thus, selectins can act cooperatively to promote tumor cell-host cell interactions and cancer metastasis. To date, the synergistic effects of P- and L-selectin in the facilitation of metastasis have been demonstrated in vivo (14). Interestingly, leukocyte L-selectin can also enhance metastasis by interacting with endothelial L-selectin ligands induced adjacent to established intravascular murine colon carcinoma cell emboli that lack L-selectin ligands on their surfaces (19).
Although molecules that bind E- and L-selectin have previously been identified on tumor cell lines (3, 4, 20), their functional roles and biologic significance have not been substantiated. As has been argued in the literature (21), distinctions must be drawn between molecules that can bind to selectins under certain conditions in vitro and functional ligands that engage selectins under fluid dynamic conditions in vivo. Through the use of the RNA interference technology, we recently discovered that sialofucosylated CD44 variant isoforms (CD44v)2 represent the major functional P-, but not E- or L-, selectin ligands on LS174T colon carcinoma cells (22). During those studies, immunoblot and blot rolling assays using CD44-knockdown cells revealed the presence of an ∼170–180-kDa sialofucosylated glycoprotein capable of supporting selectin-dependent adhesion (22). In this study, we identify this glycoprotein to be the carcinoembryonic antigen (CEA, CD66e), which functions as an E- and L-, but not P-, selectin ligand. CEA expressed by CD44-double knockdown LS174T colon carcinoma cells is more densely decorated with sialofucosylated epitopes than CEA on wild-type LS174T cells, thereby enhancing the avidity of CEA from knockdown relative to wild-type cells for E-selectin. We also disclose that CEA serves as an auxiliary L-selectin ligand, which is engaged in the stabilization of L-selectin-dependent LS174T cell rolling against fluid shear. Moreover, CEA and CD44v synergistically contribute to E- and L-selectin adhesion at elevated shear stresses. Our data imply that CEA overexpression on tumor cells may enhance metastatic potential via a selectin-mediated adhesion process.
EXPERIMENTAL PROCEDURES
Adhesion Molecules, Antibodies, and Reagents—The chimeric form of E-selectin-IgG Fc (E-selectin) consisting of the lectin, epidermal growth factor, and consensus repeat domains for human E-selectin linked to each arm of human IgG1 was a generous gift of Wyeth External Research (Cambridge, MA) (23). L-selectin-IgG Fc (L-selectin) and P-selectin-IgG Fc (P-selectin) were purchased from R & D Systems (Minneapolis, MN). Alkaline phosphatase (AP)- and horseradish peroxidase (HRP)-conjugated anti-mouse IgG and AP-conjugated anti-rat IgM were from Southern Biotech (Birmingham, AL). Unlabeled anti-CD66 mAbs GM8G5, 80H3, 9A6, BAP3, and BAC2 were purchased from Abcam (Cambridge, MA). All other unlabeled and phycoerythrin (PE)- or fluorescein isothiocyanate-conjugated antibodies were from BD Biosciences. All other reagents were from Sigma unless otherwise stated.
Cell Culture—The human colorectal carcinoma cell line LS174T was obtained from the American Type Culture Collection (Manassas, VA), and cultured in the recommended medium. Prior to cell lysis, colon carcinoma cells were detached from culture flasks using Enzyme Free Cell Dissociation Media (15 min at 37 °C; Chemicon, Phillipsburg, NJ). For flow cytometric and flow-based adhesion assays, LS174T cells were harvested by mild trypsinization (0.25% trypsin/EDTA for 5 min at 37 °C) and subsequently incubated (107 cells/ml) for 2 h at 37 °C to allow regeneration of surface glycoproteins (5, 18, 24). CHO cells stably transfected with full-length E-selectin (CHO-E) or with phosphatidylinositol glycan-linked extracellular domain of P-selectin (CHO-P) were kindly donated by Affymax (Palo Alto, CA), and processed as described previously (25).
Colon Carcinoma Cell Lysis and Immunoprecipitation Assays—Whole cell lysate was prepared by membrane disruption using 2% Nonidet P-40 followed by differential centrifugation (2, 22). In view of immunoblot assays showing that LS174T cells do not express CD66d (see under “Results”), CEA (CD66e) and CD66c were immunoprecipitated from LS174T colon carcinoma cell lysate with an anti-CD66de mAb, Col-1, and an anti-CD66c mAb, B6.2, respectively, using recombinant protein G-agarose beads (Invitrogen).
Affinity Chromatography and Mass Spectrometry Assays—The putative selectin ligand corresponding to an 180-kDa HECA-452-reactive protein was purified from CD44-knockdown LS174T colon carcinoma cell lysates by affinity chromatography using KappaLock™-agarose supports (Invitrogen) cross-linked using bis(sulfosuccinimidyl)suberate (Pierce) to the HECA-452 mAb. Eluted proteins were separated by SDS-PAGE and immunoblotted with HECA-452. Replica gels were incubated with ProQ Emerald 300 glycoprotein stain (Invitrogen), which only binds to carbohydrate groups at glycosylation sites, thereby leaving the polypeptide core intact. The ProQ Emerald 300-stained band corresponding to the 180-kDa HECA-452-positive protein(s) was excised and digested in-gel with trypsin. Extracted peptides were then analyzed by nanoflow HPLC interfaced to electrospray ionization tandem mass spectrometry (HPLC-MS/MS) using a ThermoFinnigan LTQ mass spectrometer. The MS data were searched against all taxonomies in the NCBI nonredundant protein data base with a 95% significance threshold (p < 0.05) using Mascot (Matrix Science) and with a p < 0.01 confidence using the BioWorks 3.3 software featuring the SEQUEST algorithm (ThermoFinnigan).
SDS-PAGE and Western Blotting—Whole cell lysate or immunopurified CEA or CD66c was diluted with reducing sample buffer and separated using 4–20% SDS-polyacrylamide gels (Bio-Rad) (2, 22). Resolved proteins were transferred to Sequi-blot or Immun-blot polyvinylidene difluoride and blocked with StartingBlock (Pierce) for 15 min. Immunoblots were stained with HECA-452 or anti-CD66de (Col-1) or anti-CD66c (B6.2) mAbs and rinsed with Tris-buffered saline, 0.1% Tween 20. Subsequently, blots were incubated with appropriate AP- or HRP-conjugated secondary antibodies. Western Blue AP substrate (Promega, Madison, WI) and SuperSignal West Pico chemiluminescent substrate (Pierce) were used to develop the AP- and HRP-conjugated antibody-stained immunoblots, respectively.
Blot Rolling Assay—Blots of immunopurified CEA from wild-type or CD44-knockdown LS174T whole cell lysate were stained with anti-CD66de (Col-1) or anti-CD66c (B6.2) or HECA-452 mAbs and rendered translucent by immersion in 90% D-PBS (Dulbecco's phosphate-buffered saline with Ca2+/ Mg2+), 10% glycerol (2, 22). The blots were placed under a parallel plate flow chamber, and human peripheral blood lymphocytes or CHO transfectants expressing P- or E-selectin, resuspended at 5 × 106 cells/ml in 90% D-PBS/10% glycerol, were perfused at the shear stress of 0.5 dynes/cm2 (2, 22). Molecular weight markers were used as guides to aid placement of the flow chamber over stained bands of interest. The number of interacting cells per lane was averaged over five ×10 fields of view (0.55 mm2 each) within each stained region. In select experiments, CHO-E cell suspensions or lymphocytes were pretreated with a function-blocking mAb specific for either E- or L-selectin (20 μg/ml), respectively, for 10 min at room temperature before use in blot rolling assays. Nonspecific adhesion was assessed by perfusing 5 mm EDTA in the flow medium.
Preparation of CEA- or CD66c-coated Microspheres—Immunoprecipitated CEA from wild-type or CD44-knockdown LS174T whole cell lysate was diluted to desired concentrations with binding buffer (0.2 m carbonate/bicarbonate buffer, pH 9.2) and incubated with 10-μm polystyrene microspheres (2.5 × 107 microspheres/ml; Polysciences Inc., Warrington, PA) overnight at 4 °C with constant rotation (2, 22). Microspheres were washed twice with D-PBS and subsequently blocked with D-PBS, 1% BSA for 30 min at room temperature. Microspheres were resuspended (2 × 106 microspheres/ml) in D-PBS, 0.1% BSA for use in flow cytometric and flow chamber assays. Site densities of CEA- or CD66c-coated microspheres were determined by flow cytometry using the B1.1 or B6.2 mAbs, respectively (2, 22).
Flow Cytometry—CEA, CD66c, and HECA-452 site densities on microspheres were quantified by single color immunofluorescence and flow cytometry (FACSCalibur, BD Biosciences) using PE-conjugated anti-CD66 (B1.1), anti-CD66c (B6.2), or HECA-452 antibodies. Background levels were determined by incubating cell or microsphere suspensions with properly matched PE-conjugated isotype control antibodies (2, 22). CEACAM expression on colon carcinoma cells was studied by using primary anti-CD66 mAbs (CD66a, GM8G5; CD66b, 80H3; CD66c, 9A6; CD66de, Col-1; CD66f, BAP3; and CEACAM7, BAC2) with appropriate PE-conjugated secondary and isotype control antibodies.
Flow-based Adhesion Assays—To simulate the physiological shear environment of the vasculature, colon carcinoma cells or CEA-coated microspheres suspended in D-PBS, 0.1% BSA were perfused over immobilized IgG- or E-, P-, or L-selectin-coated dishes at prescribed wall shear stresses using a parallel plate flow chamber (250-μm channel depth, 5.0-mm channel width) (2, 22, 26). The extent of adhesion was quantified by perfusing cells/microspheres at 1 × 106/ml and enumerating the total number of tethering events in a single ×10 field of view during a 2- or 5-min period. Average rolling velocities were computed as the distance traveled by the centroid of the translating cell/microsphere divided by the time interval at the given wall shear stress (2, 22). In select experiments, wild-type and CD44-knockdown LS174T cells or CEA-coated microspheres were perfused over substrates with 5 mm EDTA in the flow medium.
Preparation of CEA siRNA Oligonucleotides—Short interfering (si) RNA oligonucleotides targeting CEA were generated using the siRNA design program from Whitehead Institute (Massachusetts Institute of Technology) as described previously (22). The siRNA sequences were used to construct 60-mer short hairpin RNA oligonucleotides, which were then synthesized (Operon, Inc., Huntsville, AL) and ligated into the pSUPER.puro.gfp expression vector (Oligoengine, Inc., Seattle) under the control of the H1 promoter. The following oligonucleotide was used (underlined, sense and antisense sequences; boldface, restriction enzyme sites; italicized, polymerase III termination signal; boldface italicized, loop with linker): 5′-GATCCCCGGACCCTCACTCTATTCAATTCAAGAGATTGAATAGAGTGAGGGTCCTTTTTC-3′. The ligated product was transformed into competent DH5α Escherichia coli cells, amplified in the presence of ampicillin, and the plasmid was purified using the EndoFree maxi kit (Qiagen, Valencia, CA). Sequence insertion was verified by restriction and confirmed by direct sequencing. An empty vector was used as a negative control in all short hairpin RNA experiments.
Generation of Stable CEA-knockdown and CEA/CD44-double Knockdown Colon Carcinoma Cell Lines—8 × 106 wild-type or CD44-knockdown LS174T cells were plated in 100-mm dishes and grown overnight reaching an ∼50% confluency. The cells were then transfected with 32 μg of pSUPER.puro.gfp. CEA using Lipofectamine 2000 for 24 h. Upon reaching confluency, transfected cells were passed and 5 × 106 cells seeded per Petri dish in growth medium in triplicate. After 24 h, the medium was replaced by a fresh aliquot containing 2.5 μg/ml puromycin. Cells were then grown continually without passaging for 15 days, replenishing the puromycin-containing medium every 2–3 days. Single cell colonies were isolated and cultured using standard techniques.
Statistical Analysis—Data are expressed as the mean ± S.E. for at least three independent experiments. Statistical significance of differences between means was determined by analysis of variance. If means were shown to be significantly different (p < 0.05), multiple comparisons were performed by the Tukey test.
RESULTS
Identification of CEA as an E- and L-selectin Ligand Expressed by Wild-type and CD44-knockdown LS174T Colon Carcinoma Cells—We recently reported the presence of an ∼170–180-kDa sialofucosylated glycoprotein(s) in CD44-knockdown LS174T colon carcinoma cells that was capable of mediating selectin binding under flow (22). To delineate the identity of the selectin ligand(s), the putative target was purified from CD44-knockdown LS174T colon carcinoma cell lysates by affinity chromatography using KappaLock™-agarose beads coated with a HECA-452 mAb (Fig. 1), which detects sialofucosylated epitopes. Eluted samples were separated by SDS-PAGE and immunoblotted with HECA-452 to confirm the retention of the putative target throughout purification. Replica gels were incubated with ProQ Emerald 300 glycoprotein stain, which fluorescently labels periodate-oxidized glycans while leaving the polypeptide backbone intact. The ProQ Emerald 300-stained gel band corresponding to the ∼180-kDa HECA-452-reactive protein(s) was excised and digested in-gel with trypsin (Fig. 1). Extracted peptides were then analyzed by nano-flow HPLC interfaced to electrospray ionization MS/MS (Fig. 1). Bioinformatics analysis of the MS data revealed peptide fragment matches for carcinoembryonic antigen (CEA; CD66e) in two separate sample submissions.
FIGURE 1.
Schematic diagram of the process of HECA-452 immunoaffinity chromatography, in-gel glycoprotein staining of eluted samples, tandem mass spectrometry, and bioinformatics analysis of trypsin-digested samples used to identify carcinoembryonic antigen as the 180-kDa sialofucosylated glycoprotein selectin ligand band in whole cell lysates of CD44-knockdown LS174T cells. Panel 1, HECA-452-reactive molecules were purified from the whole cell lysates of CD44-knockdown LS174T cells by immunoaffinity chromatography using KappaLock™-agarose supports cross-linked with bis(sulfosuccinimidyl)suberate to the HECA-452 mAb. HECA-452-reactive molecules were eluted using a low pH elution buffer. Panel 2, samples were then separated by SDS-PAGE and stained in-gel using ProQ Emerald 300 glycoprotein stain. The fluorescently labeled band at 180 kDa was subsequently excised from the gel. Panel 3, proteins were extracted and trypsin-digested and then subjected to tandem mass spectrometry. Bioinformatics analysis of the mass spectrometry data revealed the presence of CEA in the sample.
We next performed a series of experiments to confirm the identity of the selectin ligand. Immunoblot analysis using an anti-CD66de mAb, Col-1, revealed the presence of CEA with an apparent molecular mass of ∼180 kDa in CD44-knockdown LS174T cell lysate and the lack of CD66d immunoreactivity at ∼35 kDa (Fig. 2A, lane 1). CEA is enriched in HECA-452-immunoprecipitated specimens relative to whole cell lysates (Fig. 2A, lane 3). The presence of HECA-452 reactivity on CEA was also disclosed by staining immunopurified CEA with a HECA-452 mAb (Fig. 2A, lanes 5 and 6).
FIGURE 2.
A, Western blots of whole cell lysate or immunopurified HECA-452-reactive epitopes or immunopurified CEA from CD44-knockdown LS174T colon carcinoma cells. Anti-CD66de (Col-1) (lanes 1, 3, and 5) or HECA-452 (lanes 2,4, and 6) mAbs were used to stain Western blots of CD44-knockdown LS174T whole cell lysate (lanes 1 and 2), immunoprecipitated HECA-452-reactive epitopes (lanes 3 and 4), and immunoprecipitated (IP) CEA (lanes 5 and 6) from CD44-knockdown LS174T cells. B, selectin-dependent adhesion to SDS-PAGE resolved and blotted CEA immunoprecipitated from CD44-knockdown LS174T whole cell lysate. CHO-E cells, lymphocytes, or CHO-P cells were perfused at the wall shear stress level of 0.5 dynes/cm2 over SDS-PAGE immunoblots of immunopurified CEA from whole cell lysates of CD44-knockdown LS174T cells. In select experiments, CHO-E cells and lymphocytes were pretreated with an anti-E-selectin or an anti-L-selectin function-blocking mAb (20 μg/ml), respectively, before use in blot rolling assays. The saturating concentration of the mAb (20 μg/ml) was maintained in the perfusion assays. Data represent the mean ± S.E. of n ≥ 3 experiments. ND, not done.
Although previous studies have reported that CEA (27) and CEA family members such as CD66c (28) bind E-selectin under static/no-flow conditions, the capacity of CEA to interact with E-selectin under physiologically relevant flow conditions as well as its potential cross-reactivity with L- and P-selectin have yet to be examined. To address these issues, E-selectin- and P-selectin-transfected CHO cells as well as L-selectin-expressing human peripheral blood lymphocytes were perfused over the SDS-PAGE-resolved immunopurified CEA protein band. Our data reveal that E- and L-, but not P-, selectin-expressing cells bind avidly and extensively to immunopurified CEA from CD44-knockdown colon carcinoma cells (Fig. 2B), suggesting that CEA possesses E- and L-, but not P-, selectin ligand activity. The specificity of these adhesive interactions was assessed by incubating CHO-E cell suspensions or lymphocytes with an anti-E-selectin or an anti-L-selectin function-blocking mAb, respectively (Fig. 2B). Moreover, CHO-E cells or lymphocytes suspended in flow medium containing 5 mm EDTA failed to adhere to any region of the blot (data not shown).
Using mAbs specific for the various members of the CEA family of immunoglobulins (CD66a–c, -e, and -f and CEACAM7) along with indirect single color immunofluorescence flow cytometry, we determined that CEA (CD66e) and CD66c, but not CD66a, -b, and -f or CEACAM7, are expressed on the surface of wild-type LS174T colon carcinoma cells (Fig. 3A). Similarly, only CEA and CD66c are present on the surface of CD44-knockdown LS174T cells at levels equivalent to those on wild-type controls (data not shown). The presence of CEA and CD66c in wild-type LS174T cell lysate was confirmed by immunoblot analysis (Fig. 3, B and C). In accord with our data using CD44-knockdown colon carcinoma cells, CEA is HECA-452-positive with a molecular mass of ∼180 kDa (Fig. 3B). In contrast, CD66c is HECA-452-negative (Fig. 3C).
FIGURE 3.

A, representative flow cytometric histograms of CEACAM expression by wild-type LS174T cells. CEACAM expression on colon carcinoma cells was investigated by using primary anti-CD66 mAbs (CD66a, GM8G5; CD66b, 80H3; CD66c, 9A6; CD66de, Col-1; CD66f, BAP3; and CEACAM7, BAC2) in conjunction with appropriate PE-conjugated secondary and isotype control antibodies. B, Western blots of whole cell lysate or immunoprecipitated (IP) CEA from wild-type LS174T colon carcinoma cells. Anti-CD66de (Col-1) (lanes 1 and 3) or HECA-452 (lanes 2 and 4) mAbs were used to stain Western blots of whole cell lysate (lanes 1 and 2) and immunoprecipitated CEA (lanes 3 and 4) from wild-type LS174T colon carcinoma cells. C, Western blots of whole cell lysate or immunoprecipitated CD66c from wild-type LS174T colon carcinoma cells. Anti-CD66c (B6.2) (lanes 1 and 3) or HECA-452 (lanes 2 and 4) mAbs were used to stain Western blots of whole cell lysate (lanes 1 and 2) and immunoprecipitated CD66c (lanes 3 and 4) from wild-type LS174T cells.
CEA, but Not CD66c, Is a Sialofucosylated Selectin Ligand on LS174T Colon Carcinoma Cells—We next used a cell-free flowbased adhesion assay (2, 22) to compare the adhesion of microspheres coated with CEA immunopurified from wild-type versus CD44-knockdown LS174T colon carcinoma cells to selectin substrates in shear flow. This technique allows quantitative comparisons of CEA-mediated adhesion to selectin substrates at prescribed CEA and selectin site densities under physiological flow conditions. By coating microspheres with equivalent levels of CEA from wild-type and CD44-knockdown LS174T cells (Fig. 4A), we determined that CEA from CD44-knockdown cells is much more densely decorated with HECA-452-reactive epitopes relative to wild-type LS174T CEA (Fig. 4B). Taken together, these data suggest that CEA serves as an alternative glycosylation acceptor on colon carcinoma cells.
FIGURE 4.

Site densities of CEA (A) and HECA-452-reactive epitopes (B) on polystyrene microspheres coated with CEA immunopurified from either wild-type (bold line) or CD44-knockdown (thin line) LS174T cells. Microspheres were stained with PE-conjugated anti-CD66 B1.1 (A), fluorescein isothiocyanate-conjugated HECA-452 (B), or PE- or fluorescein isothiocyanate-conjugated isotype control antibodies (dashed lines). C, extent of adhesion of microspheres (106/ml) coated with CEA immunopurified from wild-type (black bars) or CD44-knockdown (white bars) LS174T colon carcinoma cells to 10 μg/ml E-, L-, or P-selectin at a wall shear stress level of 1 dyne/cm2 for 2 min. Data represent the mean ± S.E. *, p < 0.05 with respect to microspheres coated with CEA immunopurified from wild-type LS174T cells. ND, not done. Average rolling velocities of microspheres (106/ml) coated with CEA immunopurified from wild-type or CD44-knockdown LS174T cells on 10 μg/ml E-selectin (D) or L-selectin (E) at prescribed wall shear stresses. Data represent the mean ± S.E. *, p < 0.05 with respect to microspheres coated with CEA immunopurified from wild-type LS174T cells.
We hypothesized that the difference in HECA-452 immunoreactivity of CEA in wild-type and CD44-knockdown cells could impact the biophysics of CEA-selectin interactions in shear flow. To test this hypothesis, we perfused the CEA-coated microspheres over purified selectin substrates under prescribed wall shear stress levels. As expected from our blot rolling assays, microspheres coated with CEA from either cell type were capable of tethering and rolling over E- and L-, but not P-, selectin substrates, albeit with varying efficiencies (Fig. 4, C–E). Most importantly, the extent of tethering of CD44-knockdown LS174T CEA-coated microspheres to E-selectin was lower than that of microspheres decorated with CEA from wild-type LS174T cells (Fig. 4C). This difference is attributed to the slower average rolling velocities of the former microspheres (and thus lower number of beads entering the field of observation (29)) relative to wild-type LS174T CEA-coated beads over E-selectin over a wide range of wall shear stresses varying from 0.5 to 1.5 dynes/cm2 (Fig. 4D). In contrast, no difference was detected in either the extent of tethering (Fig. 4C) or the average rolling velocities (Fig. 4E) of microspheres coated with CEA from either cell type over L-selectin. The specificity of CEA-selectin interactions in these assays was evaluated through the use of nonspecific IgG-coated microspheres and by preincubating the selectin-functionalized dishes with the respective function-blocking anti-selectin mAb prior to the perfusion of CEA-coated microspheres. In both cases, no microsphere tethered to selectin substrates during the entire length of the flow experiment (Fig. 4C). As an additional control experiment, CEA-bearing microspheres, perfused over selectin substrates in the presence of 5 mm EDTA in the perfusion buffer, failed to tether to either E- or L-selectin under flow (Fig. 4C).
In view of an earlier report suggesting that CD66c on human neutrophils possesses E-selectin ligand activity (28), we determined whether CD66c on LS174T cells serves as a selectin ligand. To this end, we immunopurified CD66c from the whole cell lysate of wild-type and CD44-knockdown LS174T colon carcinoma cells using the anti-CD66c mAb B6.2. Microspheres coated with CD66c from either cell type failed to tether to E-selectin substrates beyond background levels under flow (data not shown). Cumulatively, these data suggest that the HECA-452-negative CD66c (Fig. 3C) does not possess E-selectin-ligand activity in LS174T colon carcinoma cells.
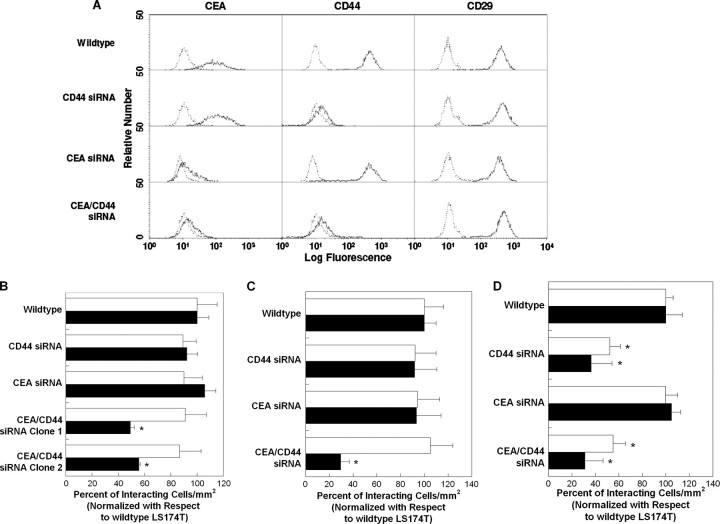
CEA and CD44 Cooperate to Mediate Colon Carcinoma Cell Adhesion to E- and L-selectin at Elevated Shear Stresses—To assess the functional role of CEA in the adhesion of colon carcinoma cells to selectins under flow, we generated stable CEA-knockdown and CEA/CD44-double knockdown LS174T cell lines by transfecting wild-type and CD44-knockdown cells, respectively, with a CEA shRNA plasmid, isolating single cell clones and propagating these clones in puromycin-containing media. As shown in Fig. 5A, this procedure resulted in the generation of CEA-knockdown and CEA/CD44-double knockdown LS174T cells with markedly reduced CEA surface expression (>95% decrease in mean fluorescence intensity) relative to wild-type and CD44-knockdown LS174T cells transfected with a control plasmid, as evidenced by flow cytometry using the anti-CD66de mAb Col-1. Evidence for the specificity of this genetic intervention was provided by the flow cytometric analysis of other LS174T cell surface adhesion molecules such as CD29 (Fig. 5A).
FIGURE 5.
A, representative flow cytometric histograms of CEA, CD44, and CD29 expression by wild-type, CD44-knockdown, CEA-knockdown, and CEA/CD44-double knockdown LS174T cells. Cells were stained by indirect single color immunofluorescence using the anti-CD66de mAb Col-1 (solid line) or an isotype control antibody (dashed line). Alternatively, cells were stained with the PE-conjugated anti-CD44 mAb 515 (solid line) or PE-conjugated isotype control antibody (dashed line). In other experiments, cells were stained with the PE-conjugated anti-CD29 mAb MAR4 (solid line) or PE-conjugated isotype control antibody (dashed line). B, extent of adhesion of wild-type, CD44-knockdown, CEA-knockdown, and two distinct CEA/CD44-double knockdown LS174T cell lines (106/ml) to E-selectin (0.75 μg/ml) under physiological flow conditions. The average number of wild-type LS174T cells per mm2 that tethered and rolled on E-selectin at 1.0 and 2.0 dynes/cm2 was 380 ± 60 and 290 ± 30, respectively. Data represent the mean ± S.E. of n = 3 experiments. Black bars represent data acquired at the wall shear stress level of 1.0 dyne/cm2, whereas white bars represent data at 2.0 dynes/cm2. *, p < 0.05 with respect to wild-type, CD44-knockdown, and CEA-knockdown LS174T cells. C, extent of adhesion of wild-type, CD44-knockdown, CEA-knockdown, and CEA/CD44-double knockdown LS174T cells (106/ml) to L-selectin (1.5 μg/ml) under physiological flow conditions. The average number of wild-type LS174T cells per mm2 that tethered and rolled on L-selectin at 1.0 and 2.0 dynes/cm2 was 700 ± 100 and 600 ± 100, respectively. Data are normalized with respect to wild-type LS174T cells and represent the mean ± S.E. of n = 3–4 experiments. Black bars represent data acquired at the wall shear stress level of 1.0 dyne/cm2, and white bars represent data at 2.0 dynes/cm2. *, p < 0.05 with respect to wild-type, CD44-knockdown, and CEA-knockdown LS174T cells. D, extent of adhesion of wild-type, CD44-knockdown, CEA-knockdown, and CEA/CD44-double knockdown LS174T cells (106/ml) to P-selectin (1.5μg/ml) under physiological flow conditions. The average number of wild-type LS174T cells per mm2 that tethered and rolled on P-selectin at 1.0 and 2.0 dynes/cm2 was 1200 ± 200 and 610 ± 90, respectively. Data are normalized with respect to wild-type LS174T cells and represent the mean ± S.E. of n = 3 experiments. Black bars represent data acquired at the wall shear stress level of 1.0 dynes/cm2, and white bars represent data at 2.0 dynes/cm2. *, p < 0.05 with respect to wild-type and CEA-knockdown LS174T cells.
In flow-based adhesion assays, CEA-knockdown LS174T colon carcinoma cells tethered to E-, L-, and P-selectin substrates under flow at levels comparable with those of wild-type controls (Fig. 5, B–D). However, CEA knockdown significantly increased the average rolling velocity of colon carcinoma cells on L-selectin (Table 1), but not on E-selectin (Table 2) or P-selectin (data not shown), relative to wild-type cells. On the other hand, CEA/CD44-double knockdown LS174T colon carcinoma cells displayed a markedly reduced capacity to tether and roll on purified E-selectin (∼50% of control) and L-selectin (∼70% of control), but not P-selectin, substrates at a wall shear stress of 2.0 dynes/cm2, whereas no difference was observed at 1.0 dyne/cm2 (Fig. 5, B–D). These results were reproducible using two distinct CEA/CD44-double knockdown cell lines (Fig. 5B). Moreover, CEA/CD44-double knockdown LS174T colon carcinoma cells rolled with higher rolling velocities over E-selectin relative to wild-type, CD44-knockdown, and CEA-knockdown LS174T cells at 2 dynes/cm2, although no difference was evident at 1.0 dyne/cm2 (Table 2). Although CEA/CD44-double knockdown rolled faster than wild-type controls on L-selectin, no significant difference was detected between double, CEA-, or CD44-knockdown cells. Altogether, these data indicate that CEA serves as an auxiliary L-selectin ligand, which is engaged in the stabilization of LS174T cell rolling on L-selectin against fluid shear. Moreover, CEA and CD44 cooperate to mediate colon carcinoma cell adhesion to E- and L-selectin at elevated shear stresses.
TABLE 1.
Average rolling velocities (μm/s) of wild type, CD44-knockdown, CEA-knockdown, and CEA/CD44-double knockdown LS174T cells (106/ml) perfused over a surface coated with 1.5 μg/ml L-selectin at the physiological shear stress levels of 1.0 or 2.0 dynes/cm2
Data represent the means ± S.E.
|
Average rolling velocity |
||
|---|---|---|
| 1.0 dyne/cm2 | 2.0 dynes/cm2 | |
| μm/s | ||
| Wild-type LS174T | 182 ± 9 | 320 ± 10 |
| CD44-knockdown LS174T | 236 ± 8a | 440 ± 10a |
| CEA-knockdown LS174T | 245 ± 7a | 400 ± 20a |
| CEA/CD44-knockdown LS174T | 238 ± 8a | 440 ± 20a |
p < 0.05 with respect to wild-type LS174T cells.
TABLE 2.
Average rolling velocities (μm/s) of wild-type, CD44-knockdown, CEA-knockdown, and CEA/CD44-double knockdown LS174T cells (106/ml) perfused over a surface coated with 0.75 μg/ml E-selectin at the physiological shear stress levels of 1.0 or 2.0 dynes/cm2
Data represent the mean ± S.E.
|
Average rolling velocity |
||
|---|---|---|
| 1.0 dyne/cm2 | 2.0 dynes/cm2 | |
| μm/s | ||
| Wild-type LS174T | 5.3 ± 0.3 | 7.2 ± 0.4 |
| CD44-knockdown LS174T | 5.1 ± 0.3 | 6.9 ± 0.5 |
| CEA-knockdown LS174T | 5.2 ± 0.4 | 7.1 ± 0.5 |
| CEA/CD44-knockdown LS174T | 5.2 ± 0.4 | 10 ± 1a,b,c |
Values are p < 0.05 with respect to wild-type LS174T cells.
Values are p < 0.05 with respect to CD44-knockdown LS174T cells.
Values are p < 0.05 with respect to CEA-knockdown LS174T cells.
DISCUSSION
CEA is one of the most extensively used clinical tumor markers due in part to its limited expression in adult normal tissue and high expression levels in positive tumors. It is expressed in a number of tumors of epithelial origin, including colorectal carcinoma, lung adenocarcinoma, and mucinous ovarian carcinoma (30). CEA has been reported to promote the metastatic potential of colon cancer cells (31, 32). The capacity of different colorectal cell lines to grow in nude mouse spleen and liver models correlates positively with CEA production (33). CEA has emerged as a suitable target antigen for the detection of primary and metastatic colorectal and some other carcinomas (30), and it is presently being evaluated as a possible target for antibody-mediated therapy (34).
CEA functions as a chemoattractant and as a cell adhesion molecule (35). CEA is involved in both homophilic binding between the N domains of anti-parallel CEA molecules on apposing cell surfaces and heterophilic binding to non-CEA molecules (30). However, the possibility of CEA-dependent binding to selectins has been largely overlooked. Interestingly, selectins play critical roles in the hematogenous dissemination of tumor cells, including colon carcinomas (13–15). We recently reported that CD44v represents the major functional P-, but not E- or L-, selectin ligand on colon carcinoma cells (22). Moreover, we observed the expression of sialofucosylated glycoproteins with molecular masses of ∼180 and ∼130 kDa that were capable of supporting selectin-dependent adhesion of CD44-knockdown LS174T colon carcinoma cells in shear flow (22). Through the use of immunoaffinity chromatography in conjunction with tandem mass spectrometry, we identify CEA as the 180-kDa selectin ligand expressed by both wild-type and CD44-knockdown LS174T colon carcinoma cells. By employing a biochemical/bioengineering approach, involving SDS-PAGE analysis of whole cell lysates, blot rolling, and cell-free flow-based adhesion assays, we demonstrate that CEA is an E- and L-, but not P-, selectin ligand on LS174T colon carcinoma cells. Although E-selectin binding activity by commercially available CEA has been reported under static/no-flow conditions (27), the biological significance and biochemical characterization of this interaction were not previously investigated. As has been appropriately argued by Varki (21), the challenge is to “... tell the difference between what can bind... in vitro, and what does bind under fluid dynamic conditions in vivo.” To our knowledge, this is the first study to document the capacity of CEA to interact with E- and L-selectin in a dynamic flow environment.
The level of adsorbed CEA immunopurified from either wild-type or CD44-knockdown LS174T cells as well as the degree of HECA-452 immunoreactivity on the polypeptide backbone was directly quantified by flow cytometry. Using this technique, we determined that CEA from CD44-knockdown LS174T cells exhibits a significantly higher number of HECA-452-reactive epitopes relative to that of CEA expressed by wild-type LS174T. Our data also reveal that this difference in extent of sialofucosylation of CEA directly affects the avidity of this glycoprotein for E-, but not L-, selectin. The emergence of CEA as an alternate glycosylation acceptor explains the elevated E-selectin-dependent adhesion to the ∼180-kDa region previously observed in blot rolling assays using cell lysates from CD44-knockdown cells relative to wild-type controls (22). In contrast to a previously published report suggesting CD66c expressed on human neutrophils possesses E-selectin ligand activity (28), our data indicate that CD66c on LS174T colon carcinoma cells is not an E-selectin ligand. The difference might be reconciled by the fact that LS174T CD66c is devoid of any HECA-452 reactivity, whereas the CD66c functions as a presenter molecule of the sialyl Lex oligosaccharide structures on neutrophils (28).
By generating stable CEA-knockdown and CEA/CD44-double knockdown LS174T colon carcinoma cell lines, we disclose that CEA functions as an auxiliary L-selectin ligand, which participates in stabilizing LS174T colon carcinoma cell rolling on L-selectin against fluid shear. Moreover, CEA and CD44 cooperate to mediate tethering of LS174T cells to E- and L-selectin substrates at a wall shear stress level of 2.0 dynes/cm2 but not at lower stresses. These data point to the potential presence of an additional yet unidentified glycoprotein(s) capable of mediating E- and L-selectin-dependent adhesion under flow. In view of our recently published data (22), a sialofucosylated glycoprotein(s) with an apparent molecular mass of ∼130 kDa emerges as a primary potential E- and L-selectin ligand candidate, without excluding the possibility for the involvement of another yet unidentified ∼180-kDa glycoprotein in this process. Glycolipids (25) may also contribute to E-, but not L-, selectin-dependent rolling of LS174T colon carcinoma cells (36). It is noteworthy that the individual removal of either CEA or CD44 (22) is not sufficient to affect the avidity of LS174T colon carcinoma cells for E-selectin in shear flow, underscoring their synergistic contribution to E-selectin-dependent binding. Although the individual knockdown of CD44 or CEA increases the average rolling velocity of LS174T cells without affecting the extent of their tethering onto L-selectin substrates in shear flow, CEA and CD44 act in a cooperative mode in supporting colon carcinoma cell tethering to L-selectin at elevated shear stresses.
In conclusion, we have demonstrated that CEA possesses E- and L-selectin ligand activity. Moreover, CEA and CD44v act synergistically to mediate the tethering and rolling of LS174T colon carcinoma cells on E- and L-selectin substrates. Our findings offer a unifying perspective on the apparent enhanced metastatic potential associated with CEA overexpression on many types of tumor cells, including colon carcinoma and the critical role of selectins in metastatic spread. Our data support further research to investigate CEA as a potential therapeutic target to combat metastasis and contribute to the complexity of the possible functions from this ubiquitous adhesion molecule.
Acknowledgments
We thank Drs. Robert Cole and Robert N. O'Meally who performed the peptide analysis and protein identification in the Mass Spectrometry and Proteomics Facility at Johns Hopkins School of Medicine with support from the Institute for Cell Engineering. We also thank Wyeth External Research for the generous gifts of E-selectin Fc IgG chimera protein and Affymax for the E- and P-selectin-transfected CHO cells.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 CA101135 (NCI) (to K. K.). This work was also supported by a National Science Foundation graduate research fellowship (to S. N. T.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: CD44v, variant isoforms of CD44; CEA, CD66e carcinoembryonic antigen; CHO, Chinese hamster ovary; BSA, bovine serum albumin; mAb, monoclonal antibody; PE, phycoerythrin; AP, alkaline phosphatase; HRP, horseradish peroxidase; siRNA, short interfering RNA; HPLC, high pressure liquid chromatography; MS, mass spectrometry; MS/MS, tandem mass spectrometry.
References
- 1.Hanley, W. D., Burdick, M. M., Konstantopoulos, K., and Sackstein, R. (2005) Cancer Res. 65 5812–5817 [DOI] [PubMed] [Google Scholar]
- 2.Hanley, W. D., Napier, S. L., Burdick, M. M., Schnaar, R. L., Sackstein, R., and Konstantopoulos, K. (2006) FASEB J. 20 337–339 [DOI] [PubMed] [Google Scholar]
- 3.Kaytes, P. S., and Geng, J. G. (1998) Biochemistry 37 10514–10521 [DOI] [PubMed] [Google Scholar]
- 4.Kim, Y. J., Borsig, L., Han, H. L., Varki, N. M., and Varki, A. (1999) Am. J. Pathol. 155 461–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mannori, G., Crottet, P., Cecconi, O., Hanasaki, K., Aruffo, A., Nelson, R. M., Varki, A., and Bevilacqua, M. P. (1995) Cancer Res. 55 4425–4431 [PubMed] [Google Scholar]
- 6.Stone, J. P., and Wagner, D. D. (1993) J. Clin. Investig. 92 804–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kannagi, R., Izawa, M., Koike, T., Miyazaki, K., and Kimura, N. (2004) Cancer Sci. 95 377–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakamori, S., Kameyama, M., Imaoka, S., Furukawa, H., Ishikawa, O., Sasaki, Y., Kabuto, T., Iwanaga, T., Matsushita, Y., and Irimura, T. (1993) Cancer Res. 53 3632–3637 [PubMed] [Google Scholar]
- 9.Nakayama, T., Watanabe, M., Katsumata, T., Teramoto, T., and Kitajima, M. (1995) Cancer 75 2051–2056 [DOI] [PubMed] [Google Scholar]
- 10.Sato, M., Narita, T., Kimura, N., Zenita, K., Hashimoto, T., Manabe, T., and Kannagi, R. (1997) Anticancer Res. 17 3505–3511 [PubMed] [Google Scholar]
- 11.Mannori, G., Santoro, D., Carter, L., Corless, C., Nelson, R. M., and Bevilacqua, M. P. (1997) Am. J. Pathol. 151 233–243 [PMC free article] [PubMed] [Google Scholar]
- 12.Biancone, L., Araki, M., Araki, K., Vassalli, P., and Stamenkovic, I. (1996) J. Exp. Med. 183 581–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borsig, L., Wong, R., Feramisco, J., Nadeau, D. R., Varki, N. M., and Varki, A. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 3352–3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borsig, L., Wong, R., Hynes, R. O., Varki, N. M., and Varki, A. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 2193–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, Y. J., Borsig, L., Varki, N. M., and Varki, A. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 9325–9330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borsig, L. (2004) News Physiol. Sci. 19 16–21 [DOI] [PubMed] [Google Scholar]
- 17.Jadhav, S., Bochner, B. S., and Konstantopoulos, K. (2001) J. Immunol. 167 5986–5993 [DOI] [PubMed] [Google Scholar]
- 18.Jadhav, S., and Konstantopoulos, K. (2002) Am. J. Physiol. 283 C1133–C1143 [DOI] [PubMed] [Google Scholar]
- 19.Laubli, H., Stevenson, J. L., Varki, A., Varki, N. M., and Borsig, L. (2006) Cancer Res. 66 1536–1542 [DOI] [PubMed] [Google Scholar]
- 20.Li, L., Short, H. J., Qian, K. X., Elhammer, A. P., and Geng, J. G. (2001) Biochem. Biophys. Res. Commun. 288 637–644 [DOI] [PubMed] [Google Scholar]
- 21.Varki, A. (1997) J. Clin. Investig. 100 Suppl. 11, 31–35 [Google Scholar]
- 22.Napier, S. L., Healy, Z. R., Schnaar, R. L., and Konstantopoulos, K. (2007) J. Biol. Chem. 282 3433–3441 [DOI] [PubMed] [Google Scholar]
- 23.Somers, W. S., Tang, J., Shaw, G. D., and Camphausen, R. T. (2000) Cell 103 467–479 [DOI] [PubMed] [Google Scholar]
- 24.McCarty, O. J. T., Jadhav, S., Burdick, M. M., Bell, W. R., and Konstantopoulos, K. (2002) Biophys. J. 83 836–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burdick, M. B., Schnaar, R. L., Collins, B. E., Bochner, B. S., and Konstantopoulos, K. (2001) Biochem. Biophys. Res. Commun. 284 42–49 [DOI] [PubMed] [Google Scholar]
- 26.Burdick, M. M., and Konstantopoulos, K. (2004) Am. J. Physiol. 287 C539–C547 [DOI] [PubMed] [Google Scholar]
- 27.Anostario, M., Jr., Li, S. H., and Huang, K. S. (1994) Anal. Biochem. 221 317–322 [DOI] [PubMed] [Google Scholar]
- 28.Kuijpers, T. W., Hoogerwerf, M., van der Laan, L. J., Nagel, G., van der Schoot, C. E., Grunert, F., and Roos, D. (1992) J. Cell Biol. 118 457–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCarty, O. J. T., Mousa, S. A., Bray, P. F., and Konstantopoulos, K. (2000) Blood 96 1789–1797 [PubMed] [Google Scholar]
- 30.Hammarstrom, S. (1999) Semin. Cancer Biol. 9 67–81 [DOI] [PubMed] [Google Scholar]
- 31.Hashino, J., Fukuda, Y., Oikawa, S., Nakazato, H., and Nakanishi, T. (1994) Clin. Exp. Metastasis 12 324–328 [DOI] [PubMed] [Google Scholar]
- 32.Minami, S., Furui, J., and Kanematsu, T. (2001) Cancer Res. 61 2732–2735 [PubMed] [Google Scholar]
- 33.Tibbetts, L. M., Doremus, C. M., Tzanakakis, G. N., and Vezeridis, M. P. (1993) Cancer 71 315–321 [DOI] [PubMed] [Google Scholar]
- 34.Blumenthal, R. D., Hansen, H. J., and Goldenberg, D. M. (2005) Cancer Res. 65 8809–8817 [DOI] [PubMed] [Google Scholar]
- 35.Kim, J. C., Koo, K. H., Kim, B. S., Park, K. C., Bicknell, D. C., and Bodmer, W. F. (1999) Int. J. Cancer 82 880–885 [DOI] [PubMed] [Google Scholar]
- 36.Burdick, M. M., McCaffery, J. M., Kim, Y. S., Bochner, B. S., and Konstantopoulos, K. (2003) Am. J. Physiol. 284 C977–C987 [DOI] [PubMed] [Google Scholar]