Abstract
Transcription factor IIB (TFIIB) recruits RNA polymerase II to promoters and inserts a finger domain into its active site, with unknown consequences. Here we show that that the tip of this finger is important for two transcription initiation functions. First, TFIIB acts as a catalytic cofactor for initial RNA bond formation. It does so via a pair of fingertip aspartates that can bind magnesium, placing TFIIB within a family of proteins that insert finger domains to alter the catalytic functions of RNA polymerase. Second, the TFIIB fingertip mediates the timing of the release of TFIIB that is associated with appropriate promoter escape. These initiation requirements may assist in RNA quality control by minimizing functional synthesis when RNA polymerase becomes inappropriately associated with the genome without having been recruited there by TFIIB.
TFIIB2 is a single-subunit protein that plays a central role in transcription by RNA polymerase II. It consists of three motifs with distinct structures and functions. The C terminus is composed of two cyclin repeats and is responsible for bringing TFIIB to promoters. It does so primarily by recognizing promoter-bound TATA-binding protein but also interacts with DNA sequences that flank the TATA box (1). A second motif is folded by association with a zinc atom (2). This binds a docking domain on RNA polymerase II (3) and recruits the enzyme to promoters to catalyze transcription initiation (4). A third motif is unstructured in solution but folds into a finger-like structure when bound to RNA polymerase (3). A number of functions have been suggested for this finger, mostly involving the escape by the recruited RNA polymerase (discussed below).
TFIIB is unique among the general transcription factors in that it recycles during continuous transcription, i.e. TFIIF travels with the elongating RNA polymerase, and TATA-binding protein, TFIIH, and TFIIE largely remain bound to the promoter (5). Thus there can be a pioneer round of transcription that forms a scaffold upon which reinitiation can occur without requiring the reassembly of all factors from free solution. This allows transcription to occur at a facilitated rate (6). For each round of initiation, a new RNA polymerase must wait for the prior RNA polymerase to escape. This requires the recycling of TFIIB (7) by binding to the scaffold to recruit the next RNA polymerase. These initial steps of transcription initiation, involving initial bond formation, production of small RNAs, and disruption of contacts to the promoter, are collectively termed “escape” (8–10).
The finger domain of TFIIB has been proposed to play multiple roles in promoter escape. Both chemical probing (3) and structural studies (11) have shown that the finger penetrates the main channel of RNA polymerase II to approach the active site of the enzyme. From this location it could influence these early steps by interacting with the template DNA, the newly synthesized RNA, the catalytic center of the RNA polymerase, and perhaps TFIIF or other general transcription factors. Several lines of evidence are consistent with the existence of such interactions and their influence on escape (11–13).
The most direct of these is the known propensity of mutations near the tip of the finger to change the distribution of positions that end up at the 5′-end of the mRNA (14, 15). The ability of the finger domain to influence the start site of transcription is most pronounced in budding yeast. The analogous mutations in human TFIIB do not obviously change the transcription start site but do reduce the number of small abortive RNA transcripts made during initiation (16). The location of the finger deep within the cleft of the budding yeast RNA polymerase suggests that it could contact the RNA roughly when it reaches a length of five nucleotides. Experimental evidence supports the existence of such a contact, which was proposed to both stabilize the RNA within the complex and temporarily block the ability of the RNA to be lengthened by addition of nucleotides (11). Deletions within the potentially similar domains of cofactors for related multi-subunit RNA polymerases have led to somewhat conflicting results but support the general conclusion that a role in catalysis or escape is likely (17–19). These various observations have in common that they suggest a role of the TFIIB finger in these early escape steps, but a cohesive overview of what this role is has not yet emerged.
Whatever the role of TFIIB is in escape, this role must terminate when TFIIB is released. All of the existing proposals have in common that the release of TFIIB is associated with promoter escape and reinitiation. Early experiments suggested that human TFIIB is released before the RNA reaches a length of 10 nucleotides (7). What causes this release is not known, but the finger and zinc-binding domains could be influential, as they both contact the RNA polymerase. Escape in bacterial transcription systems is better characterized and has many properties similar to those just described. In those complexes, the location of the TFIIB finger is assumed by region 3.2 of the major sigma factor, σ70 (20). Mutations in region 3.2 also influence the distributions of short RNAs (20–22). In addition, the production of small RNAs ceases near the same length of 10 (9, 20). As is the case with TFIIB, the sigma factor is also recycled during initiation (23). Region 3.2 of sigma has one property that has not been ascribed to TFIIB; namely, it abets efficient initiation and RNA bond formation by influencing the catalytic center of the RNA polymerase (21).
Overall, the roles of TFIIB in transcription beyond its ability to recruit RNA polymerase are not yet well defined. This is potentially important for understanding the processing of the transcribed RNA. TFIIB release may occur concomitantly with the capping of the 5′-end of the RNA. Its release also appears to coincide with the beginning of the switch in the phosphorylation states of the C-terminal domain of the RNA polymerase (24), which is a critical processing determinant. In addition, yeast TFIIB also associates directly with a 3′-end RNA-processing factor, Ssu72 (25), and can be found at the ends of genes where 3′-processing occurs (26). However, the relationship of the changing status of TFIIB in transcription complexes to the progress of the various initiation and processing events is not known.
To address these questions, we have made changes in the human TFIIB finger and studied how these lead to changes in properties associated with the production of RNA. The studies rely on transcription from full, activated transcription complexes. They show that the finger domain plays a critical role in the release of TFIIB during promoter escape. In addition, they define a new role for this domain, in the catalytic function of RNA polymerase. TFIIB is shown to be a member of a family of transcription proteins that can influence the RNA polymerase active catalytic center via finger domains containing amino acids capable of chelating magnesium ions. By comparing the effects of mutations on catalysis and release with those on transcription, roles for TFIIB in transcription initiation and escape can be defined. In addition, the results raise new questions about a potential role of TFIIB in quality control of RNA production.
EXPERIMENTAL PROCEDURES
Mutagenesis of TFIIB—The glutathione S-transferase-tagged expression construct for human TFIIB was generously provided to us by Michael Carey (UCLA). Deletions as well as sitespecific mutations were made using QuikChange site-directed mutagenesis kit (Stratagene).
Purification of Recombinant Protein—The pGEX-2T vector carrying the human coding sequence of wild-type or mutant TFIIB was expressed in BL21(DE3) cells. Cultures containing the plasmid were grown to an A600 of ∼0.6 and induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside. The cultures were grown for another 2 h and then sonicated in PBS buffer supplemented with both DTT and phenylmethylsulfonyl fluoride to 1 mm. The sonicate was centrifuged to separate the insoluble material from the soluble recombinant protein. Glutathione-Sepharose beads were equilibrated with PBS. The supernatant was incubated with beads for 2 h to isolate the expressed protein. The beads were washed extensively with cold PBS supplemented with 1 mm DTT. The beads were resuspended in an equal volume of PBS and 1 mm DTT. Thrombin was added to a total of 0.5 units to separate the bound glutathione S-transferase and TFIIB. The free TFIIB was separated from the beads by gentle centrifugation and dialyzed in Buffer D (20 mm Hepes (pH 7.9), 100 mm KCl, 0.2 mm EDTA, 0.5 mm DTT, and 20% glycerol) at 4 °C.
Immunodepletions—Anti-IIB antibodies (IIB8, Santa Cruz Biotechnology) were bound to protein A-agarose by incubation for 1 h at 4°C. The beads were washed three times in PBS and three additional times in Buffer D. HeLa nuclear extract prepared using the Dignam method (27) was incubated with the agarose beads for 1 h. HeLa nuclear extracts required two incubations over fresh beads to sufficiently remove endogenous TFIIB.
In Vitro Transcription—Transcription was performed using the AdE4 promoter, activated by nine upstream Gal4-binding sites. The transcription reaction contained 200 ng of Gal4-AH (Protein One), 60 μg of depleted HeLa nuclear extract, 5 ng of recombinant TFIIB, and 50 ng of linear DNA template in a total volume of 25 μl. The reaction was preincubated for 30 min at room temperature to allow preinitiation complex formation. Nucleotides were added to concentrations of 400 μm for ATP and CTP and 0.016 μm for cold UTP. Radiolabeled [α-32P]UTP was added concurrently with NTPs to a total activity of 5 μCi for 1 h at 30°C. Transcription was terminated with buffer containing 0.3 m Tris-HCl (pH 7.4), 0.3 m sodium acetate, 0.5% SDS, 2 mm EDTA, and 3 μg/ml tRNA, followed by chloroform/phenol extraction. Isolated transcripts were run on a urea-6% acrylamide gel, dried, and subjected to phosphorimaging (Bio-Rad).
Immobilized Pulldown Assay—Biotinylated primers at the 5′-end were used in PCR reactions to amplify the AdE4 template. Primers were designed further upstream to include the nine Gal4-binding sites. PCR products were gel-purified (Qiagen). Dynabeads M-280 beads (Invitrogen) were washed twice in 2× bind-and-wash buffer (10 mm Tris-HCl (pH 7.5), 1 mm EDTA, and 2 m NaCl). Purified PCR products were incubated with washed Dynabeads in 1× bind-and-wash buffer for 1 h at room temperature. Beads bound to DNA were washed three times with 1× bind-and-wash buffer and two additional times with 10 mm Tris-HCl (pH 7.5). The beads were resuspended to 5 mg/ml. Per reaction, a small aliquot of beads was blocked with Buffer D and bovine serum albumin (1.5 μg/μl) for 30 min at room temperature and washed once with Buffer D. Each reaction of blocked beads was incubated with 200 ng of Gal4-AH, 60 μg of HeLa nuclear extract or depleted HeLa nuclear extract, 30 ng of recombinant TFIIB, 1 unit of hexokinase (Sigma-Aldrich), and 2 mm glucose for 30 min at room temperature. Hexokinase and glucose are used to deplete the extract of any endogenous nucleotides. Each reaction was washed extensively (five times, each time inverting the tube 15 times) to remove nonspecific binding of transcription factors to yield a preinitiation complex. The reactions are used in either the abortive initiation assay or promoter escape assay.
Abortive Initiation Assay—Preinitiation complexes isolated by the immobilized pulldown assay were resuspended in 25 μl of Buffer D. Beads were incubated with 16 μm dATP, 100 μm UpA, and 5 μCi of [α-32P]CTP for 15 min at 30 °C. Beads were magnetically pulled down, and the supernatant containing the released UpApC product was mixed with formamide/urea loading dye and boiled for 1 min. The products were run on a urea-25% acrylamide gel at 32 watts for 5 h. The gel was then subjected to phosphorimaging.
Promoter Escape Assay—Preinitiation complexes isolated by the immobilized pulldown assay were resuspended in 50 μl of Buffer D. Beads were incubated with either 100 μm ATP, G-stop mixture (100 μm ATP, CTP, and UTP and 20 μm 3′-O-methyl-GTP), or all four nucleotides, the concentration of each being 100 μm. Beads were magnetically pulled down after appropriate time points, and the supernatant was removed. The beads were immediately saturated with SDS loading dye buffer and boiled for 2 min. The beads were pulled down again, and the supernatant was run on a denaturing SDS-acrylamide gel and Western blotted against TFIIB (C-18, Santa Cruz Biotechnology) and polymerase II (8WG16, Covance).
Iron Cleavage Assay (28)—A total reaction mixture of 100 μl containing 100 μm DTT and 100 ng of TFIIB in 8 mm Hepes (pH 7.9) was preincubated at room temperature for 20 min. Iron ammonium sulfate was added to a 20 μm concentration and incubated for various times at 37 °C. An aliquot of 10 μl was taken from each reaction, either containing wild-type or mutant TFIIB. Each was stopped with SDS buffer containing 2 mm EDTA. Samples were run on a denaturing SDS-acrylamide gel and Western blotted against TFIIB (C-18).
RESULTS
The Effect of the Fingertip on Transcription—We made three deletions of TFIIB in which progressively smaller portions of the finger region are missing (Fig. 1A). The largest of these, ΔB, removes the entire motif (residues 44–75). The second largest, D7, removes only seven residues at the very tip of the finger that approaches the active site of the RNA polymerase, 57NDKATKD63. The smallest, D3, removes only three of these same fingertip residues, 59KAT61. The ability of these three proteins to support activated transcription was tested using a HeLa nuclear extract supplemented with artificial activator. First, the extract was immunodepleted of endogenous TFIIB using antibodies bound to agarose beads. The transcription level from this depleted extract is very low (Fig. 1B, lane 1). When recombinant wild-type TFIIB is added back, robust transcription of the adenovirus E4 promoter is observed (lane 2). Addition instead of the larger deletion, ΔB, does not restore transcription (lane 3). These data establish that transcription in this depleted extract depends on added exogenous TFIIB and that transcription also requires the presence of the finger region.
FIGURE 1.
Effect of B-finger deletion proteins on transcription. A, the human TFIIB finger sequence with the D7 and D3 mutations marked. B, transcription of a TFIIB-depleted HeLa extract (lane 1), wild-type (Wt) TFIIB added back (lane 2), and ΔB added back (lane 3). C, transcription as in B: without TFIIB (lane 1), with the wild type (lane 2), with D3 (lane 3), and with D7 (lane 4).
The transcription was repeated using the smaller deletions that are confined to the fingertip region. The three-amino acid deletion, D3, gives transcription levels that were nearly wild type (∼85%) (Fig. 1C, lane 3; compare with the wild type in lane 2). The seven-amino acid deletion, D7, gave much reduced transcription of about one-third of wild-type levels (lane 4 compared with lane 2). The level is nonetheless significantly above the background observed when the extract is not supplemented with TFIIB (lane 4 versus lane 1). A quantitative analysis for D7 and other mutants is presented below. We infer that the three residues at the tip of the finger are mildly important for transcription but that the surrounding four residues are of greater importance.
It is possible that the source of the large transcription defect from the D7 deletion is at the level of protein recruitment, i.e. the loss of transcription might be due to the lack of TFIIB binding to the promoter or its inability to when bound to recruit the RNA polymerase. We assessed these possibilities by using a template pulldown assay (29). In this assay, the template DNA was biotinylated and attached to streptavidin magnetic beads. Immunodepleted nuclear extract and the recombinant TFIIB protein were incubated with the beads, and the presence of proteins was assayed by Western blotting. The recruitment potential of mutant D7 was evaluated in comparison with the wild-type TFIIB and two control situations in which transcription is not observed. The controls are the ΔB protein and the use of wild-type protein but with a template from which the E4 promoter region has been excised. Two different antibodies were used to assess the association of TFIIB and RNA polymerase II with the pulled-down templates.
Fig. 2 shows that mutant D7 leads to association of a full complement of both RNA polymerase (top row) and TFIIB (bottom row) with the template, i.e. the D7 signals for both TFIIB and polymerase II (lane 4) are comparable with that using wild-type TFIIB (lane 2). Neither protein is recruited when the promoter is excised from the template plasmid (lane 1). The removal of the entire TFIIB finger in ΔB also leads to a failure to significantly recruit RNA polymerase II and stably hold TFIIB at the promoter (lane 3). In experiments involving less stringent washing, ΔB can be found associated with the template, but much less RNA polymerase II is recruited (data not shown). Collectively, these data indicate that D7 efficiently recruits RNA polymerase II but that this RNA polymerase can transcribe at only one-third the normal level.
FIGURE 2.
Recruitment of B-finger deletion proteins to preinitiation complexes. An immobilized template assay was used to determine the relative amounts of polymerase II (Pol II) and TFIIB recruitment for each mutant. Recombinant TFIIB was added to the TFIIB-depleted HeLa extract, and preinitiation complexes were isolated. A negative control for nonspecific binding used the E4 template lacking the TATA box and activator sites, denoted NP (lane 1). TFIIB proteins were wild type (Wt)(lane 2), ΔB(lane 3), D7 (lane 4), and 2-Asp (lane 5).
The Effect of the Fingertip on TFIIB Release—The remodeling of the transcription complex during promoter escape is associated with the release of TFIIB. Thus it is possible that the two-thirds loss of transcription by mutant D7 is due to aberrations in the release of TFIIB. To test this possibility, we established a TFIIB release assay using this same system of templates pulled down from TFIIB-supplemented transcription extracts. In this experiment, the pulled-down template containing the exogenously added TFIIB was incubated with either the NTPs needed for elongation or simply ATP, which restricts open transcription complexes to the promoter without synthesizing RNA. At various times after nucleotide addition, the templates were separated from the supernatants. The amount of TFIIB that was not released to the supernatant and remained associated with the template was assayed by Western blotting.
The results show that the assay can follow the release of TFIIB. The last row of Fig. 3A shows a progressive reduction in retained TFIIB, as transcription elongation proceeds from before NTP addition (time 0) to 40 s after to 80 s after. Essentially all TFIIB has been released by this latter time. It appears that all templates release TFIIB even though all may not be fully active, as discussed for yeast extracts (5). A control with ATP alone that does not support elongation loses little TFIIB during this same time frame (Fig. 3A, top row). We conclude that the assay can follow the release of TFIIB.
FIGURE 3.
TFIIB release from the scaffold and the effect of the B-finger. A, an immobilized template system was used to assay the release of TFIIB by Western blotting as a function of mRNA length. Templates were pulled down before (0 s) or after the addition of nucleotides for 40 and 80 s. Different templates were used to pause the polymerase at specific mRNA lengths of 0, 6, 16, and >16, as indicated. For the 0-length mRNA sample only, ATP was added. For the 6- and 16-nucleotide (nt) template, a mixture of ATP, CTP, UTP, and 3′-O-methyl-GTP was used to pause the polymerase at +6 or +16. To obtain full elongation, ATP, CTP, UTP, and GTP were added. B, quantitative analysis of three independent experiments shows the release of TFIIB using different G-block templates. Diamonds, squares, triangles, and circles refer to transcription blocks at positions 0, 6, and 16 and with no block, respectively. C, the same system was used to assay release using B-finger deletions and the system that halts transcription at +6. The release of TFIIB is indicated by the difference between columns 2 and 1. The wild-type (Wt), D3, and D7 complexes are indicated. Average and S.D. values for TFIIB wild type, D3, and D7 retained are 88 ± 8%, 38 ± 11%, and 14 ± 5%.
The assay was modified to determine at what position along the template TFIIB release occurs. To do this, the templates and the transcription protocols were changed to establish very effective blocks to RNA synthesis at defined points. This procedure was very substantially modified from prior protocols to further increase the effectiveness of read-through, already reported to be low (16, 30). First, double blocks were created by inserting guanines at positions +6 and +7 in the G-less transcription cassettes to create the template G6. Second, even though reactions lacked GTP, a high concentrations of the chain terminator 3′-O-methyl-GTP was added to stall transcription at the double block. Third, extracts were treated with hexokinase and glucose to eliminate the potential of contaminating GTP. Fourth, the templates were pulled down and washed to remove any traces of nucleotide prior to transcription. This drastic treatment completely eliminated read-through transcription on the modified template while allowing it to proceed normally in control experiments where GTP was added (supplemental Fig. 1). A G16 template was also constructed with such double G-blocks at positions +16 and +17, and this exhibited similar behavior.
These templates were used in TFIIB release assays, and the results were compared with the wild-type template (Fig. 3). The effectiveness of the blocks is confirmed in row 2 of Fig. 3B where the G6 template now retains TFIIB (compare this to the release without the block in row 4). By contrast, moving the block to near +16 is associated with the release of TFIIB (row 3). The rates of release in these various contexts are summarized in Fig. 3B. We infer that TFIIB is released between positions +7 and +16, consistent with prior reports in a basal transcription system (7).
Next, we applied these assays to mutant D7, which transcribes at one-third the normal wild-type level, and mutant D3, which transcribes at a nearly wild-type level. Preliminary experiments showed that both of these mutants were released at very early stages compared with the wild-type TFIIB. The amount of TFIIB retained when RNA polymerase stalls near position +6 for these mutants and the wild-type TFIIB is compared in Fig. 3B. For both mutants, TFIIB is nearly completely released from the template at the +6 stage (compare lane 2 with lane 1 for D3 and for D7). This obviously contrasts with the behavior of wild-type TFIIB, which is not significantly released under these same conditions (compare lane 1 with lane 2 for the wild type and also refer to Fig. 3A). We conclude that deletions of either seven or three amino acids at the TFIIB fingertip can lead to early release of TFIIB. However, we note that the early release is not necessarily associated with a defect in transcription. Although both mutants D3 and D7 are released early, D7 is reduced by two-thirds in transcription, whereas D3 transcribes at nearly wild-type levels. This comparison suggests that there may be a different source for the defect in transcription by mutant D7.
A Role for the Fingertip in Catalysis during Initiation—Mutant D7 differs from D3 in that the four amino acids removed include two aspartates. Several proteins that penetrate the active sites of RNA polymerases have regions that contain aspartates, and these strongly influence the catalytic properties of the enzyme. In the case of bacterial σ70, region 3.2 is required for efficient formation of an RNA bond during initiation (21). To assess whether the TFIIB fingertip plays a related catalytic role, we established an assay for initial RNA bond formation.
This assay relies on the pulled-down templates used in the current studies. Immobilized preinitiation complexes are isolated and direct the condensation of the dinucleotide UpA with [α-32P]CTP to create the radioactive RNA product UpApC, which requires catalytic formation of a single RNA bond (lane 4 of Fig. 4A). Several important controls support the relevance of this product. It is not seen when UpA is omitted (lane 2), when the dATP needed for promoter opening is omitted (lane 3), when the RNA polymerase II inhibitor α-amanitin is added (lane 5), or when the promoter is excised from the template (lane 1). Abortive initiation is inhibited by α-amanitin, as observed previously for RNA polymerase II (31). The assay is sensitive because abortive initiation involves repetitive synthesis of the RNA product from a single transcription complex (8, 31).
FIGURE 4.
The B-fingertip is required for abortive initiation. A, formation of 32P-UpApC on the E4 template in the presence of the indicated reagents is shown. NP refers to a template with the E4 promoter removed. B, formation of 32P-UpApC was assayed using the indicated forms of TFIIB, wild type (Wt), D3, and D7.
Levels of abortive product are compared for the D3 and D7 mutants in Fig. 4B. The data show that D3 produces levels comparable with that of wild-type TFIIB (lane 3 versus lane 2), whereas D7 produces approximately one-third of the wild-type level (lane 4 versus lane 2). Because all three forms of TFIIB are recruited in similar amounts, the result implies that the D7 mutation causes the RNA polymerase to be defective in forming an initial RNA bond. We note that the three proteins produced have similar effects on transcription and abortive initiation. It appears that the TFIIB fingertip influences both RNA catalysis and TFIIB release, but it is mainly catalysis that influences the transcription potential of the RNA polymerase. Levels of transcription and abortive products are summarized in Fig. 5C.
FIGURE 5.
The B-fingertip aspartates are needed for transcription and abortive initiation. A, transcription of wild-type (Wt) TFIIB is compared with the D7 and 2-Asp mutants, as indicated. B, abortive initiation is assayed for D7 and 2-Asp, as indicated. C, levels of transcription and abortive initiation averaged from multiple experiments on all mutants are collected in the table.
Roles for Fingertip Aspartic Acids and Magnesium—As noted above, the two aspartates deleted in D7 but not in D3 are candidates for influencing transcription through their potential to influence the ability of RNA polymerase to catalyze initial bond formation. To test this possibility, a double point mutation was constructed in which these aspartates were both changed to alanine (2-Asp). Initially, two assays were applied, one for transcription and another for catalytic abortive initiation.
Both of these assays showed very significant defects. The transcription defect for the 2-Asp mutant is roughly comparable with that for D7, for which deletion covers the two aspartates (Fig. 5A, compare lanes 3 and 4). The defect in the abortive initiation assay was also very significant (Fig. 5B, lane 3 versus lane 1), again much closer to the signal from D7 (lane 2). We conclude that the two aspartate residues of the fingertip play an important role in assisting initiating RNA polymerase to catalyze bond formation. Recruitment of 2-Asp and RNA polymerase is not affected by the loss of the two aspartate residues (Fig. 2, lane 5).
The ability of aspartates to affect catalysis by both RNA and DNA polymerases and accessory transcription factors is thought to rely on their ability to chelate magnesium at the enzyme active site (28, 32–35). In the case of bacterial RNA polymerase, this was confirmed by replacing magnesium with iron and using the iron as a site-specific cleavage reagent (28). The experiment involves the formation of hydroxyl radicals, produced by oxygen, DTT, and the protein-bound Fe2+. The radical is produced by iron bound to a specific site within Escherichia coli RNA polymerase and cleaves polypeptide backbones indiscriminately within ∼15 Å. The idea is to use this established assay to see whether the aspartate residues of the TFIIB fingertip can bind metals and thus cause TFIIB to be degraded by free radicals. Detection relies on the use of an antibody to the TFIIB C terminus.
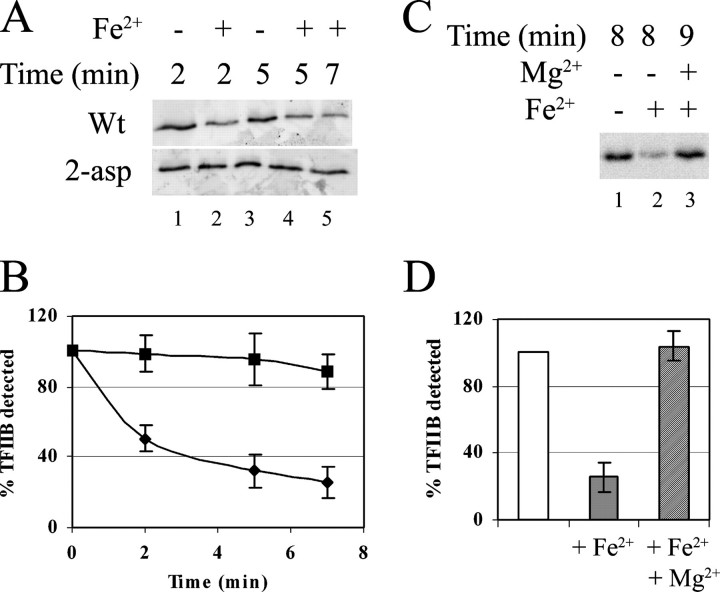
The degradation of full-length TFIIB by iron-induced free radical formation is shown in Fig. 6A. The reactions were initiated and quenched at various times before assaying for residual full-length TFIIB on a Western blot. Discrete shortened intermediate products were not observed, possibly because of free radical cleavage or modification of the C terminus, which is in close proximity to the finger region in isolated TFIIB (36–38). For wild-type TFIIB (top row), there is loss of signal by 2 min (lane 2 compared with lane 1) with a slight further loss later (lane 5). When the protein is not loaded with iron, there is no obvious loss of signal (lane 3 compared with lane 1). Thus the assay successfully detects the cleavage of wild-type TFIIB by radicals generated by site-bound iron.
FIGURE 6.
Isolated TFIIB has the ability to bind metals. A, isolated TFIIB was incubated with or without iron (as indicated), and free radical reactions were allowed to proceed for the indicated times. Full-length TFIIB was assayed by Western blotting. Wt, wild-type. B, averages of iron-treated samples of wild type (diamonds) and 2-Asp (squares) derived from three independent experiments are shown. C, reactions were as in A, but for 8–9 min. In lane 3, magnesium was added prior to iron. D, averages of iron-treated versus iron- and magnesium-treated samples from three independent experiments are shown.
The lower panel of Fig. 6A shows a parallel experiment using the 2-Asp mutant. In this case, there is no detectable degradation at 2 min (lane 2 versus lane 1) and only a slight degradation by 7 min (lane 5). These results, summarized quantitatively in Fig. 5B, show far less degradation than occurs with the wild-type protein, suggesting that the protein with deleted aspartates has a much reduced ability to bind metal. In the application of this assay to bacterial RNA polymerase, the preference of the aspartates for magnesium was demonstrated by observing the protection from degradation when added magnesium displaces the iron. Fig. 6C (summarized in the legend to Fig. 6D) shows that this also occurs with wild-type TFIIB, i.e. an 8-min treatment significantly degrades the protein (lane 2 versus the lane 1 control lacking iron), but adding magnesium protects against this degradation (lane 3), implying that magnesium and iron compete for the same site, the aspartates on TFIIB. Taken together with the above data, the results show that the TFIIB finger has multiple roles in overall transcription, in catalytic formation of the initial RNA bond, in metal binding, in factor recruitment, and in TFIIB release during escape.
DISCUSSION
Of the multiple roles uncovered here for the fingertip region of TFIIB, two were the least expected. First, the data show that two aspartates in this region are required to power catalysis during initiation of RNA synthesis, i.e. the initiating RNA polymerase is not capable of efficient RNA bond formation in the absence of this assistance. Chemical probing data suggest that these aspartates can bind magnesium, which may confer the potential to reconfigure the active site of the enzyme. The second unexpected result was the role for the TFIIB fingertip in determining the timing of the release of TFIIB during promoter escape. The data showed that normal release occurs when the RNA is 6–16 nucleotides long. However, even a three-amino acid deletion at the tip of the finger triggers efficient release prior to position +6. It is noteworthy that this very early release has no obvious consequence for transcription. Because promoter escape and the accompanying release of TFIIB are thought to be central events in coupling transcription to RNA processing (39), it is possible that the TFIIB fingertip may have a role in this processing.
These data place TFIIB within the small family of proteins that penetrate the active site of RNA polymerases to alter the catalytic properties of the enzyme, likely via magnesium ion chelation. The TFIIB fingertip properties are reminiscent of those of region 3.2 of σ70. Both regions penetrate the main channel of RNA polymerase to deliver multiple aspartates, and both regions are required to power efficient catalysis of RNA bond formation during initiation (21). The family also includes the elongation factors TFIIS (33), GreA (40), and GreB (34). These proteins have long extensions with acidic amino acids that penetrate the secondary channel of stalled RNA polymerases and modify the enzyme so that it acts as a nuclease rather than a polymerase. It is not known how these various systems function except in the general sense that they must drastically reorganize the catalytic center of RNA polymerases. This reorganization may be required when RNA polymerases are not in elongation mode, i.e. during initiation at promoters and when elongation stalls within a gene. Under these conditions, apparently the enzyme active site is not optimally configured and thus optimal catalysis requires the assistance of exogenous factors. There have been conflicting results concerning the roles of upstream binding factor or TFIIB-like factors such as transcription factor B that assist in transcription by other RNA polymerases and apparently can act between recruitment and elongation (17–19).
Are there functional advantages associated with the dependence of RNA polymerase II initiation and escape on the fingertip region of TFIIB? One speculative possibility is that this reliance ensures that improperly bound RNA polymerases will have poor catalytic function. RNA polymerase II and other RNA polymerases have a strong propensity to bind DNA nonspecifically (41), and RNA polymerase II has been observed within inactive genes (42). If such interactions lack TFIIB, as might be expected, then the inappropriately bound RNA polymerases would have difficulty producing RNA. Thus the reliance on TFIIB could contribute to minimizing improper, unregulated transcription (43).
These data confirm that TFIIB is released after the RNA reaches a length of 6 but before a length of 16 and now show that the fingertip is needed to delay the release until this latter length is approached. There have not been prior studies of the determinants of TFIIB release, but several studies have implicated the fingertip region in the general process of promoter escape (see Introduction). When the RNA reaches a length of five nucleotides, it can readily approach the fingertip (11) and make a contact that helps retain the RNA within the transcription initiation complex. This stabilizing contact is either no longer required or lost as the RNA lengthens (11), which could contribute to the TFIIB release beyond +6 that is observed here. When the fingertip is mutated, RNAs of lengths +7 and +9 are observed to no longer accumulate (16). This may reflect the removal of blocks associated with the finger, as our data suggest that the mutant TFIIB may be released prematurely. Thus all of these data are consistent with the view that the initiating RNA polymerase holds TFIIB by its fingertip until the RNA length exceeds approximately five nucleotides and then releases TFIIB shortly afterward. At this point, the RNA polymerase begins to remodel its active site into the catalytically efficient elongation mode. The retention of TFIIB prior to this point allows for efficient RNA synthesis during the abortive phase of initiation, which might be required for appropriate regulation of transcription.
This remodeling of the transcription complex during promoter escape is thought to be linked closely to RNA-processing events. The data here imply that the release of TFIIB will occur after Ser-5 RNA polymerase C-terminal domain phosphorylation and precede 5′ mRNA capping and Ser-2 CTD phosphorylation. Any error in the timing of these events could conceivably lead to the improper reconfiguration of associated factors and thus induce subsequent degradation of the mRNA by the quality control systems. Thus the fingertip may coordinate this timing so that transcription and processing are properly coupled. This too may minimize the production of RNA from inappropriate chromosomal sites, as the lack of TFIIB at such sites may lead to the lack of appropriate processing and hence to degradation of wrongly initiated RNAs.
Supplementary Material
Acknowledgments
We thank Yin C. Lin for development of assays; Vivian Y. Shi for technical advice; Michael Carey for the glutathione S-transferase-TFIIB clone; and Guillaume Chanfreau, Albert J. Courey, Hao A. Duong, and laboratory members for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM49048. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
Footnotes
The abbreviations used are: TF, transcription factor; PBS, phosphate-buffered saline; DTT, dithiothreitol.
References
- 1.Deng, W., and Roberts, S. G. (2005) Genes Dev. 19 2418–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buratowski, S., and Zhou, H. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 5633–5637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, H. T., and Hahn, S. (2004) Cell 119 169–180 [DOI] [PubMed] [Google Scholar]
- 4.Tubon, T. C., Tansey, W. P., and Herr, W. (2004) Mol. Cell. Biol. 24 2863–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yudkovsky, N., Ranish, J. A., and Hahn, S. (2000) Nature 408 225–229 [DOI] [PubMed] [Google Scholar]
- 6.Jiang, Y., and Gralla, J. D. (1993) Mol. Cell. Biol. 13 4572–4577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zawel, L., Kumar, K. P., and Reinberg, D. (1995) Genes Dev. 9 1479–1490 [DOI] [PubMed] [Google Scholar]
- 8.Carpousis, A. J., and Gralla, J. D. (1980) Biochemistry 19 3245–3253 [DOI] [PubMed] [Google Scholar]
- 9.Carpousis, A. J., and Gralla, J. D. (1985) J. Mol. Biol. 183 165–177 [DOI] [PubMed] [Google Scholar]
- 10.Dvir, A. (2002) Biochim. Biophys. Acta 1577 208–223 [DOI] [PubMed] [Google Scholar]
- 11.Bushnell, D. A., Westover, K. D., Davis, R. E., and Kornberg, R. D. (2004) Science 303 983–988 [DOI] [PubMed] [Google Scholar]
- 12.Freire-Picos, M. A., Krishnamurthy, S., Sun, Z. W., and Hampsey, M. (2005) Nucleic Acids Res. 33 5045–5052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, H. T., Warfield, L., and Hahn, S. (2007) Nat. Struct. Mol. Biol. 14 696–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pinto, I., Ware, D. E., and Hampsey, M. (1992) Cell 68 977–988 [DOI] [PubMed] [Google Scholar]
- 15.Zhang, D. Y., Carson, D. J., and Ma, J. (2002) Nucleic Acids Res. 30 3078–3085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pal, M., Ponticelli, A. S., and Luse, D. S. (2005) Mol. Cell 19 101–110 [DOI] [PubMed] [Google Scholar]
- 17.Santangelo, T. J., Cubonova, L., James, C. L., and Reeve, J. N. (2007) J. Mol. Biol. 367 344–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Panov, K. I., Friedrich, J. K., Russell, J., and Zomerdijk, J. C. (2006) EMBO J. 25 3310–3322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Werner, F., and Weinzierl, R. O. (2005) Mol. Cell. Biol. 25 8344–8355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murakami, K. S., Masuda, S., and Darst, S. A. (2002) Science 296 1280–1284 [DOI] [PubMed] [Google Scholar]
- 21.Kulbachinskiy, A., and Mustaev, A. (2006) J. Biol. Chem. 281 18273–18276 [DOI] [PubMed] [Google Scholar]
- 22.Cashel, M., Hsu, L. M., and Hernandez, V. J. (2003) J. Biol. Chem. 278 5539–5547 [DOI] [PubMed] [Google Scholar]
- 23.Raffaelle, M., Kanin, E. I., Vogt, J., Burgess, R. R., and Ansari, A. Z. (2005) Mol. Cell 20 357–366 [DOI] [PubMed] [Google Scholar]
- 24.Komarnitsky, P., Cho, E. J., and Buratowski, S. (2000) Genes Dev. 14 2452–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu, W. H., Pinto, I., Chen, B. S., and Hampsey, M. (1999) Genetics 153 643–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh, B. N., and Hampsey, M. (2007) Mol. Cell 27 806–816 [DOI] [PubMed] [Google Scholar]
- 27.Dignam, J. D., Lebovitz, R. M., and Roeder, R. G. (1983) Nucleic Acids Res. 11 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zaychikov, E., Martin, E., Denissova, L., Kozlov, M., Markovtsov, V., Kashlev, M., Heumann, H., Nikiforov, V., Goldfarb, A., and Mustaev, A. (1996) Science 273 107–109 [DOI] [PubMed] [Google Scholar]
- 29.Johnson, K. M., Wang, J., Smallwood, A., and Carey, M. (2004) Methods Enzymol. 380 207–219 [DOI] [PubMed] [Google Scholar]
- 30.Holstege, F. C., Fiedler, U., and Timmers, H. T. (1997) EMBO J. 16 7468–7480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang, Y., Yan, M., and Gralla, J. D. (1995) J. Biol. Chem. 270 27332–27338 [DOI] [PubMed] [Google Scholar]
- 32.Beese, L. S., and Steitz, T. A. (1991) EMBO J. 10 25–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeon, C., Yoon, H., and Agarwal, K. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 9106–9110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Opalka, N., Chlenov, M., Chacon, P., Rice, W. J., Wriggers, W., and Darst, S. A. (2003) Cell 114 335–345 [DOI] [PubMed] [Google Scholar]
- 35.Sosunova, E., Sosunov, V., Kozlov, M., Nikiforov, V., Goldfarb, A., and Mustaev, A. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 15469–15474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grossmann, J. G., Sharff, A. J., O'Hare, P., and Luisi, B. (2001) Biochemistry 40 6267–6274 [DOI] [PubMed] [Google Scholar]
- 37.Zheng, L., Hoeflich, K. P., Elsby, L. M., Ghosh, M., Roberts, S. G., and Ikura, M. (2004) Eur. J. Biochem. 271 792–800 [DOI] [PubMed] [Google Scholar]
- 38.Roberts, S. G., and Green, M. R. (1994) Nature 371 717–720 [DOI] [PubMed] [Google Scholar]
- 39.Cho, E. J., Kobor, M. S., Kim, M., Greenblatt, J., and Buratowski, S. (2001) Genes Dev. 15 3319–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laptenko, O., Lee, J., Lomakin, I., and Borukhov, S. (2003) EMBO J. 22 6322–6334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chandler, D. W., and Gralla, J. (1981) Nucleic Acids Res. 9 6031–6046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steinmetz, E. J., Warren, C. L., Kuehner, J. N., Panbehi, B., Ansari, A. Z., and Brow, D. A. (2006) Mol. Cell 24 735–746 [DOI] [PubMed] [Google Scholar]
- 43.Struhl, K. (2007) Nat. Struct. Mol. Biol. 14 103–105 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.