Abstract
Mice were subjected to different dietary manipulations to selectively alter expression of hepatic sterol regulatory element-binding protein 1 (SREBP-1) or SREBP-2. mRNA levels for key target genes were measured and compared with the direct binding of SREBP-1 and -2 to the associated promoters using isoform specific antibodies in chromatin immunoprecipitation studies. A diet supplemented with Zetia (ezetimibe) and lovastatin increased and decreased nuclear SREBP-2 and SREBP-1, respectively, whereas a fasting/refeeding protocol dramatically altered SREBP-1 but had modest effects on SREBP-2 levels. Binding of both SREBP-1 and -2 increased on promoters for 3-hydroxy-3-methylglutaryl-CoA reductase, fatty-acid synthase, and squalene synthase in livers of Zetia/lovastatin-treated mice despite the decline in total SREBP-1 protein. In contrast, only SREBP-2 binding was increased for the low density lipoprotein receptor promoter. Decreased SREBP-1 binding during fasting and a dramatic increase upon refeeding indicates that the lipogenic “overshoot” for fatty-acid synthase gene expression known to occur during high carbohydrate refeeding can be attributed to a similar overshoot in SREBP-1 binding. SREBP co-regulatory protein recruitment was also increased/decreased in parallel with associated changes in SREBP binding, and there were clear distinctions for different promoters in response to the dietary manipulations. Taken together, these studies reveal that there are alternative molecular mechanisms for activating SREBP target genes in response to the different dietary challenges of Zetia/lovastatin versus fasting/refeeding. This underscores the mechanistic flexibility that has evolved at the individual gene/promoter level to maintain metabolic homeostasis in response to shifting nutritional states and environmental fluctuations.
3-Hydroxy-3-methylglutaryl (HMG)3-CoA reductase catalyzes a critical early reaction in the biosynthetic pathway for isoprenoids and cholesterol and is subject to multivalent regulation to ensure optimal pathway activity (1). The molecular events targeted for regulation include primary transcriptional as well as translational and post-translational mechanisms (2). The principal transcriptional regulators for HMG-CoA reductase gene expression are the basic helix-loop-helix (bHLH) leucine zipper sterol regulatory element-binding proteins (SREBPs), which have unique features that distinguish them from other bHLH leucine zipper transcription factors. The first of these is the presence of two closely spaced membrane-spanning helices that bisect the coding sequence and target the SREBPs to the endoplasmic reticulum membrane. These are followed by a carboxyl-terminal domain that interacts with regulatory proteins and controls their trafficking, proteolytic activation, and membrane release (3, 4). SREBPs also have a signature tyrosine residue in the basic DNA-binding domain that is not present in any other bHLH proteins; this one amino acid is key for allowing specific recognition of both the canonical inverted-repeat E-box site, characteristic of most bHLH proteins, and the SREBP-specific direct-repeat-binding element or SRE (5). The dual DNA binding specificity is important to their roles in lipid regulation (6).
There are three major SREBP isoforms in mammals that are encoded by two genes. The Srebf-1 gene produces two overlapping mRNAs that differ only in their specific 5′-terminal exons. The resulting proteins, SREBP-1a and SREBP-1c, are identical except for unique amino-terminal activation domains, which are responsible for their differential co-activator interactions (7). There is a separate Srebf-2 gene and a single SREBP-2 protein with a potent activation domain similar to SREBP-1a. The full-length membrane-bound precursor SREBPs are substrates for regulated intramembrane proteolysis in response to lipid-associated nutritional cues (3). Low cholesterol levels result in membrane release of SREBP-2, whereas low cholesterol and fatty acids trigger release of SREBP-1 (8). The available mouse knock-out studies reveal that there are overlapping but distinct physiological roles for the three SREBPs, but target gene selectivity and the potential roles of SREBP homo- and heterodimers, which have distinct activation properties (9), are not well understood. In the current studies, we fed mice different diets that selectively altered expression of nuclear forms of hepatic SREBP-1 or -2 and performed chromatin immunoprecipitation (ChIP) studies with isoform-specific antibodies to probe target gene specificity and promoter selectivity in SREBP function. The results shown here reveal unique features for SREBP binding and activation of different target promoters along with condition-dependent differential co-regulatory protein recruitment.
MATERIALS AND METHODS
Mouse Studies and RNA Analyses—B6/129 mice (6-week-old males), purchased from Taconic, were fed a normal rodent chow diet and allowed to adapt for 2 weeks to a 12-h light/12-h dark cycle; they were sacrificed at the end of the dark cycle (8 a.m.). The feeding regimens were as follows. Mice were separated into four groups of 4–6 animals/group. One group was maintained on a normal chow diet, and one group was fed normal chow supplemented with a mixture of ezetimibe (Zetia from Merck/Schering-Plough Pharmaceuticals, 0.021%, w/w) and lovastain (Mylan Pharmaceuticals Inc., 0.1%, w/w). The other two groups were subjected to a fasting or fasting/refeeding protocol as described previously (10). All food manipulations were staggered so that animals were all sacrificed at the same time by CO2 asphyxiation at the end of the dark cycle. All of the results reported here were repeated at least twice, and two independent feeding studies were performed with similar results across all experimental measurements.
Where the feeding protocol was combined with adenovirus delivery, mice were administered a total of 1 × 109 plaqueforming units of virus by intravenous injection at the start of the experiment. Replication-defective recombinant adenoviruses, A-CREB and green fluorescent protein (GFP) (11) (gifts from Dr. M. Montminy, Salk Institute), were propagated in 293 cells, purified by CsCl gradient centrifugation, and titered by plaque assay on 293 cells.
After asphyxiation, livers were removed, and ∼20% of them were frozen in liquid nitrogen and stored at –80 °C until RNA had been isolated. Total RNA was isolated using TRIzol (Invitrogen) according to the manufacturer's instructions. cDNA was synthesized and use as template for qPCR as described (10, 12). The remaining 80% was used directly for chromatin isolation as described below for the chromatin immunoprecipitation assays. Primers used for qPCR analysis of mRNAs were taken from the following previous reports (12–14). All qPCR reactions were performed in triplicate.
Chromatin Immunoprecipitation—Equal portions (80–90% total wet weight) of freshly isolated livers from 4 mice/feeding group were pooled and placed in 40 ml of ice-cold phosphate-buffered saline containing a mixture of protease inhibitors (1 μg/ml leupeptin, 1.4 μg/ml pepstatin, 2 μg/ml phenylmethylsulfonyl fluoride, 1 mm EGTA, and 1 mm of EGTA). The tissue was disrupted in a “Tissue-Mizer” at the lowest setting, formaldehyde was added from a 37% stock (v/v) to a final concentration of 1%, and samples were rotated on a shaker for 6 min followed by the addition of glycine to a final concentration of 0.125 m. The samples then were returned to the shaker for an additional 5 min. Cells were collected by centrifugation (2 K in a Sorval RC3B at 4 °C). The cell pellet was washed once with homogenization buffer A (10 mm HEPES, pH 7.6, 25 mm KCl, 1 mm EDTA, 1 mm EGTA, 2 m sucrose, 10% glycerol, 0.15 mm spermine, plus protease inhibitors as listed above). The final pellet was resuspended in buffer A and homogenized in a Dounce homogenizer with a B-pestle to release the nuclei. The solution was layered over buffer A and centrifuged in a Beckman Ultracentrifuge (1 h at 26 K and 4 °C); the nuclear pellet was resuspended in nuclear lysis buffer (1% SDS, 50 mm Tris, pH 7.6, 10 mm EDTA); and nuclei were disrupted using an ultrasonic model W-220F sonicator, five times for 10 s each, to shear chromatin. Chromatin size was checked by agarose electrophoresis to ensure that the average size was between 200 and 500 bp. Aliquots were then used for immunoprecipitation experiments with antibodies described below (under “SDS-PAGE and Immunoblot Analysis”) and processed as described previously (15). Final DNA samples were analyzed by quantitative PCR in triplicate with a standard dilution curve of the input DNA performed in parallel. The data were analyzed by Student's t test, and unless noted otherwise in the individual figure legends, the pairwise comparisons were all significantly different with p values of <0.05.
The qPCR oligonucleotide pairs for the mouse promoters were as follows: HMG-CoA reductase, forward (–274) 5′-GCTCGGAGACCAATAGGA-3′ and reverse (–64) 5′-CCGCCAATAAGGAAGGAT-3′; LDL receptor, forward (–166) 5′-GAACTTCCCACTGCTGC-3′ and reverse (+4) 5′-CACGCCCAGAGTCATTC-3′; squalene synthase, forward (–245) 5′-ATCGCGCCAGGCTCCTCCGGCTTC-3′ and reverse (–27) 5′-CTCCCGCTCCCACCTGTGTTTAGA-3′; fatty-acid synthase promoter, forward (–115) 5′-GCGCAGCCCCGACGCTCATT-3′ and reverse (–20) 5′-CGGCGCTATTTAAACCGCGG-3′.
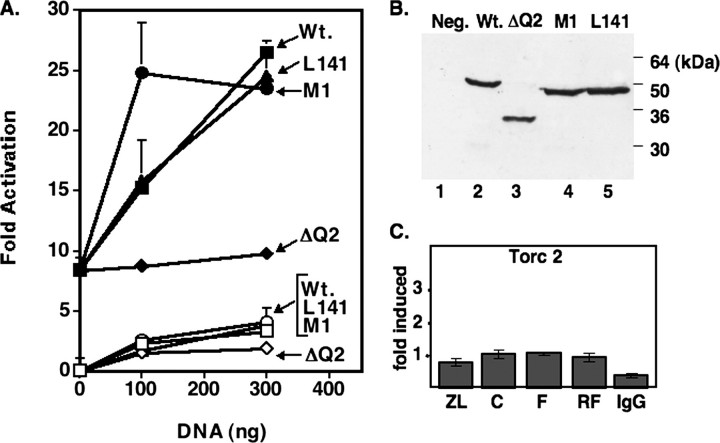
Transient Transfection Assay in Drosophila SL2 Cells—Drosophila SL2 cells (16) were cultured in Shields and Sang insect medium (Sigma) containing 10% heat-inactivated fetal bovine serum and were seeded at 480,000 cells/well in 6-well dishes on day 0. On day 1, cells were transfected by the calcium phosphate co-precipitation method with each dish receiving 2 μg of each test plasmid, 10.75 μg of salmon sperm DNA, and 1 μgofthe control plasmid, pPAC-β-gal, containing the coding region of the Escherichia coli β-galactosidase gene driven by the Drosophila actin 5C promoter. The pPAC constructs used for activation studies in SL2 cells contained the coding regions of the Sp1 or SREBP-1a (residues 1–490) or SREBP-2 (residues 1–486) gene under the control of the Drosophila actin 5C promoter and have been described previously (17). The pPAC NF-Y constructs containing the coding regions for the three individual CBF/NF-Y subunits (A, B, and C) were also described previously (18). Cells were harvested on day 3, and luciferase and β-galactosidase activity were measured in cell extracts as described previously (18). pPAC CREB was described previously (19); the coding sequences for the CREB mutants containing point mutations that inactivate the kinase-inducible domain (KID) (M1 and L141) or ΔQ2 (which deletes the constitutive glutamine domain (20, 21)) were inserted into the pPAC vector for expression in SL2 cells as well. The expression levels for CREB and CREB mutant proteins were compared using an antibody raised against human CREB (gift from M. Montminy). Briefly, transfection experiments were performed as described above with differing amounts of expression vectors as described in the legend to Fig. 5 and a constant amount of the pPAC-β-gal control plasmid. Protein extracts from the transfected cells were first normalized for transfection efficiency by measuring the β-galactosidase activity of individual extracts, and normalized amounts were analyzed by immunoblotting as described below. All transfections were repeated at least twice with similar results.
FIGURE 5.
Activation of HMG-CoA reductase promoter by SREBP, NF-Y, and CREB in SL2 cells. A, transient DNA transfection. All samples received HMG-CoA reductase reporter plus an SREBP-2 expression vector. Open symbols indicate no NF-Y, and closed symbols indicate plus all three NF-Y subunits. The amount of DNA for each CREB or mutant CREB expression vector included in the transfection is noted on the x axis. Mutations M1 and L141 are in the KID; ΔQ2 denotes the deletion of the “constitutive” glutamine-rich domain. B, protein extracts from SL2 cells transfected with the wild type (Wt.) or the indicated CREB mutant were analyzed for protein expression by immunoblotting with an antibody to CREB. C, TORC2 binding to HMG-CoA reductase promoter under the different feeding conditions was analyzed by ChIP. There was no statistical difference between the different feeding groups, but the difference between all samples and the IgG control was significant at p < 0.05. Refer to the legend for Fig. 2 for symbols and notations.
SDS-PAGE and Immunoblot Analysis—SL2 nuclear extracts and liver chromatin extracts were analyzed for immunoblotting as described (22, 23). The antibodies used were as follows: a polyclonal raised against human CREB (a gift from M. Montminy (24)) pCREB (Cell Signaling, catalog No. 9191), polyclonal antibodies raised against mouse SREBP-1 and SREBP-2 (gifts from J. Horton (13)), polyclonal antibody against TORC2 (gift from Paul Brindle (25)), CBP (Santa Cruz Biotechnology, sc-369), FXR (Santa Cruz Biotechnology, sc-H130), NF-Y A subunit (Rockland, catalog No. 100-401-100), Ac-H3 (Upstate, catalog No. 06-599), and β-actin (Sigma, catalog No. A1978). The blots were developed with the ECL kit from Pierce.
RESULTS
We were interested in exploring the overlapping and unique functional properties of SREBP-1 and SREBP-2. Because they are co-expressed in many of the same tissues, we conducted this evaluation by combining different feeding protocols designed specifically to alter the levels of either the SREBP-1 or -2 protein in liver cell nuclei, using gene expression and promoter-specific ChIP studies with antibodies that react specifically with each SREBP isoform.
Mice were fed a standard laboratory chow diet and separated into a control group and three treatment groups. In one treatment group, the chow was supplemented with a mixture of lovastatin and Zetia (Z/L) for 7 days. The remaining two groups were subjected to a 24-h fast and either sacrificed at the end of the fasting period or refed a high carbohydrate chow for an additional 12 h after fasting prior to sacrifice. The time of the dietary modifications were staggered so that all animals were sacrificed at the same time, which was at the end of the 12-h dark cycle.
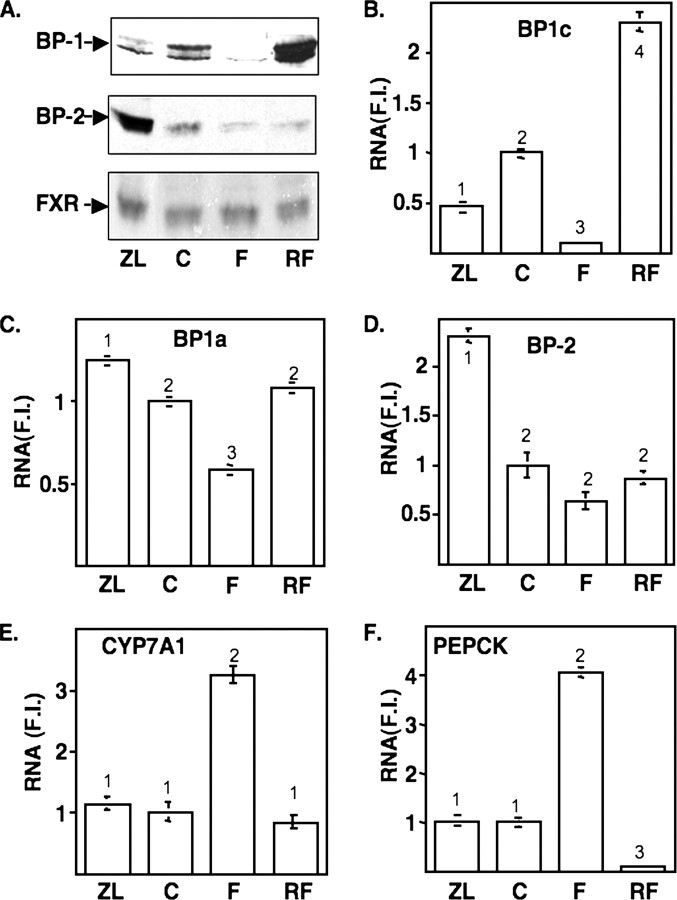
Lovastatin and Zetia were added to limit cholesterol synthesis and uptake, respectively. The statin inhibits HMG-CoA reductase activity to limit endogenous cholesterol synthesis (2), and the Zetia inhibits the NPC1L1-mediated absorption of dietary sterol (26). Under these conditions, SREBP-2 nuclear protein levels were induced 10-fold, whereas the levels of nuclear SREBP-1 actually declined 60–70% (Fig. 1A).
FIGURE 1.
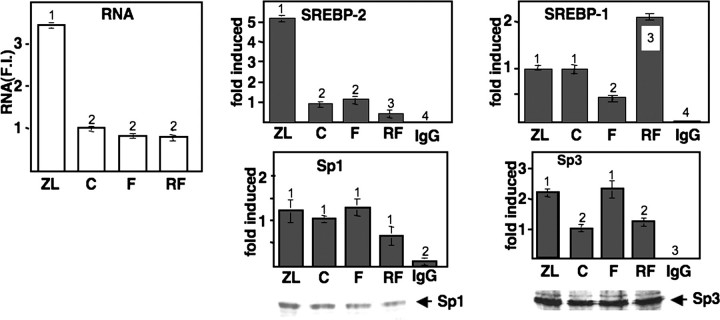
Dietary regulation expression of SREBPs in mouse liver. A, immunoblot showing nuclear expression of SREBP-1 (BP-1), SREBP-2 (BP-2), or FXR (NR1H4) across different treatment groups as described under “Materials and Methods.” Chromatin were prepared from freshly isolated pooled liver nuclei (from 4 animals/treatment group) and processed for immunoblotting as described under “Materials and Methods.” ZL, Zetia plus lovastatin; C, chow control; F, fasted; RF, fasted and refed. The immunoblot shown here is representative of the results obtained form several different gels analyzed with samples from at least four different feeding experiments. These patterns of expression are consistent with those reported by others for similar feeding protocols (13, 30, 31). B–F, total RNA from mice from the same pools from A were analyzed for expression of SREBP-1c (B), SREBP-1a (C), SREBP-2 (D), cholesterol 7 α-hydroxylase (CYP7A1)(E), or PEPCK (F) by qPCR as described under “Materials and Methods.” Expression of each mRNA indicated was normalized to the expression of ribosomal protein L32 in each sample, and the ratio in the chow sample was set at 1.0. All values are plotted relative to this value (F.I. = -fold induced). Student's t test was used to evaluate statistical significance, and values that are statistically significant (p < 0.05) are indicated by different numbers at the top of each bar. Comparisons between individual samples that resulted in p > 0.05 are labeled with the same number.
In response to fasting, SREBP-1 protein declined to almost undetectable levels, whereas 12 h of refeeding following the fast resulted in a superinduction to a value severalfold higher than that observed in control animals. In contrast, nuclear SREBP-2 protein declined slightly by the fasting protocol and did not return to control levels over the acute high carbohydrate refeeding protocol. The SREBP-1 antibody used here reacts with both the SREBP-1a and -1c isoforms (13), which differ only by unique amino-terminal activation domains (27). Our immunoblotting and ChIP experiments likely reflect changes in nuclear SREBP-1c protein. This is because the ratio of SREBP-1c to -1a mRNA is ∼10:1 in the liver (28), the residual SREBP-1a is difficult to detect when SREBP-1c is specifically deleted (13), and the protein levels follow fluctuations similar to those of SREBP-1c mRNA (see below).
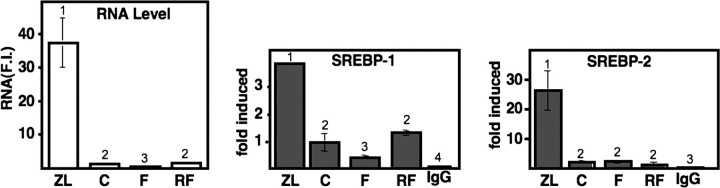
SREBP-1c mRNA levels paralleled the changes in SREBP-1 protein, declining significantly as a result of both Z/L feeding and fasting and rising well above the control value in response to the refeeding regimen (Fig. 1B). SREBP-1a mRNA levels declined with fasting but did not exhibit the dramatic shifts displayed by SREBP-1c, and there was only a minor change in response to Z/L (Fig. 1C). SREBP-2 mRNA was increased by Z/L, as would be predicted because it contains a SREBP site in its promoter (29); however, there were only minor changes in response to the fasting/refeeding (Fig. 1D). Nuclear SREBP-2 protein levels are increased more robustly than the mRNA because in addition to the SREBP-2 gene being SREBP-responsive, the processing of the extranuclear membrane-bound precursor is also increased by sterol deprivation (30). Phosphoenolpyruvate carboxykinase (PEPCK) and cholesterol 7 α-hydroxylase RNA levels were analyzed as controls. Both were induced by fasting, and PEPCK was dramatically decreased by the refeeding protocol as predicted (Fig. 1, E and F (10)).
Next, we analyzed both the expression of HMG-CoA reductase mRNA and SREBP binding to its promoter using ChIP (Fig. 2). The mRNA was induced over 30-fold by the Z/L feeding protocol, and as reported previously, the fasting level declined and returned to the control level following high carbohydrate refeeding (31). The dramatic induction of the mRNA by Z/L was paralleled by a similar dramatic increase in SREBP-2 protein binding to the promoter. However, the levels of promoter-associated SREBP-2 during the fasting and fasting/refeeding treatments were not significantly different from those found in the chow-fed control group.
FIGURE 2.
mRNA expression for and SREBP binding to the HMG-CoA reductase gene. mRNA expression (left panel) and binding of SREBP-1 or SREBP-2 by ChIP. RNA was analyzed as described in the legend for Fig. 1. ChIP analyses were performed with antibodies to either SREBP-1 or SREBP-2 as described under “Materials and Methods.” The level of binding was assessed by qPCR following immunoprecipitation, and the value obtained for chow (C) was set at 1.0; all values are plotted relative to this value (F.I. = -fold induced). qPCR reactions were performed in triplicate, and the resulting error bars are displayed. The relative amount of PCR product generated when a control IgG fraction was used in the immunoprecipitation is also provided (IgG). Significance was evaluated as described for Fig. 1. ZL, Zetia plus lovastatin; C, chow control; F, fasted; RF, fasted and refed.
As mentioned above, total nuclear SREBP-1 protein levels declined following the Z/L treatment, but interestingly, the binding of SREBP-1 to the HMG-CoA reductase promoter was actually increased by more than 3-fold. In contrast, promoter association of SREBP-1 declined significantly following fasting and returned to control levels following the refeeding, which paralleled changes in HMG-CoA reductase mRNA.
In a previous study, we showed that cholesterol depletion in cultured cells results in an increase in binding of the SREBP co-regulatory transactivators NF-Y and CREB, which is accompanied by a substantial increase in acetylation of promoter proximal histone H3 (32). Therefore, we also analyzed the binding of NF-Y and CREB as well as H3 acetylation in our animal feeding studies (Fig. 3). The binding of both NF-Y and CREB was increased by the 7-day Z/L feeding regimen, and histone H3 acetylation also increased significantly.
FIGURE 3.
SREBP co-regulatory protein and histone H3 acetylation at HMG-CoA reductase promoter. The binding of NF-Y (A) and CREB (B), acetylation level of histone H3 (C), and binding of CBP (D) to the HMG-CoA reductase promoter were analyzed by ChIP as detailed under “Materials and Methods.” All symbols and notations are as described in the legend for Fig. 2. Immunoblots measuring expression of the A-subunit of NF-Y and CREB are also displayed. Significance was evaluated as described for Fig. 1. There was no significant difference in any of the GFP or PEPCK samples.
NF-Y association declined and rebounded following the fasting and fasting/refeeding protocols, respectively, whereas CREB binding was not altered and actually declined during the refeeding. Additionally, there were no significant changes in histone H3 acetylation by the relatively acute fasting/refeeding treatment. Immunoblotting results showed that there were no changes in the total nuclear concentration of the A-subunit of NF-Y or CREB across any of the samples. Because CREB and SREBP both interact with the transcriptional co-activator CBP (33), we also measured its association with the HMG-CoA reductase promoter. CBP binding increased with Z/L treatment and was increased by the refeeding protocol as well, although at a lower level. The increase in HMG-CoA reductase mRNA from the fasting nadir back to the base-line levels following the refeeding protocol corresponded to a large increase in HMG-CoA reductase mRNA (10-fold). This was accompanied by an increase in CBP binding between the two groups; the binding level was also significantly higher than in the control group. However, the level of RNA in the refeeding group was about the same as the control sample. This apparent paradox likely reflects the fact that the acute refeeding phase was not a steady homeostatic state as seen in the control sample. The acute increase in CBP caused by refeeding ensured a rapid increase in HMG-CoA reductase gene expression, and when homeostasis was reestablished the level of CBP binding would likely return to the control level.
The KID of CREB contains a consensus phosphorylation site for cAMP-dependent protein kinase (PKA), and upon phosphorylation KID interacts with the CBP co-activator to stimulate target gene expression (33). Our current results show that the increased expression of SREBP-2 due to Z/L feeding results in an increase in the recruitment of CREB to the HMG-CoA reductase promoter without a change in total CREB levels (Fig. 3) or phosphorylation status (data not shown). To evaluate whether the increase in CREB binding plays a direct role in activation of HMG-CoA reductase expression by Z/L, we repeated the feeding study; but along with the dietary changes, we injected additional groups of mice with an adenovirus expressing GFP and a dominant negative version of CREB (11) or a control virus expressing only GFP (Fig. 4).
FIGURE 4.
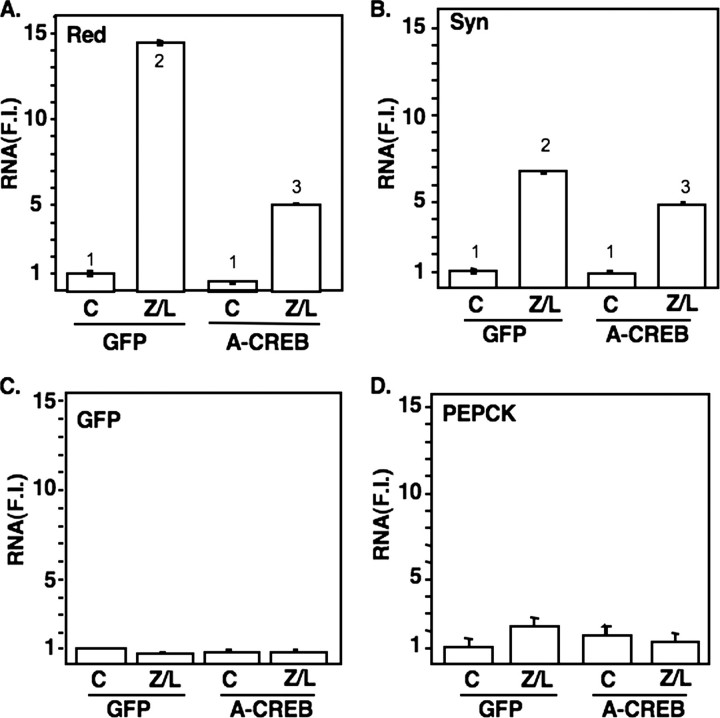
CREB is required for induction of HMG-CoA reductase in response to Zetia and lovastatin. Groups of mice (3/group) were fed a chow diet or a diet supplemented with Zetia and lovastatin as described for Figs. 1, 2, 3. Animals on each diet were infected with a control adenovirus construct expressing GFP or an adenovirus expressing A-CREB, a dominant negative version of CREB. RNA was harvested and pooled for analysis by qPCR as described for Figs. 1, 2, 3 and under “Materials and Methods.” Genes analyzed were: A, HMG-CoA reductase (Red); B, cytosolic HMG-CoA synthase (Syn); C, adenovirally encoded GFP (GFP); and D, PEPCK. The significance was evaluated as described for Fig. 1. There were no significant differences in the GFP or PEPCK control groups across all samples. F.I. = -fold induced.
We observed a dramatic increase in HMG-CoA reductase mRNA expression in the control animals; this was significantly attenuated in the group expressing the dominant negative CREB protein (Fig. 4A). Expression of another gene of the early cholesterol synthetic pathway, HMG-CoA synthase, was also significantly elevated by the Z/L treatment; however, its expression was not decreased by the dominant negative CREB virus (Fig. 4B). Additionally, there were minimal effects of the dominant negative CREB virus on PEPCK gene expression following Z/L treatment, and both viruses showed equal infectivity, as indicated by the similar levels of GFP expression across all treatment groups (Fig. 4, C and D).
In addition to the KID, CREB contains another activation domain that is reported to function constitutively (20, 34). During fasting, the KID is phosphorylated through glucagon and cAMP signaling, and TORC proteins interact with the b-zip region of CREB to stimulate gene expression (24). However, HMG-CoA reductase expression is repressed during fasting, which suggests that CREB phosphorylation and TORC recruitment might not contribute to HMG-CoA reductase activation. Additionally, because SREBPs also interact with the b-zip domain of CREB4 (19), we reasoned that concerted activation by SREBPs and CREB would not require TORC because of steric limitations and that synergy might instead occur through the constitutive activation domain instead of KID.
To test these ideas, we investigated the requirements for SREBP-CREB activation using an SL2-based transfection assay (Fig. 5) where high levels of expression from the HMG-CoA reductase promoter requires the addition of exogenously supplied expression vectors for SREBP and co-regulatory proteins CREB and NF-Y (22). The activity of the HMG-CoA reductase promoter transfected alone was set at 1, and the addition of the expression vector for SREBP did not change this low value, consistent with our previous observations (22). When an expression vector for full-length wild type CREB was included at two different concentrations, a low level of activation was observed. The addition of expression vectors for all three NF-Y subunits stimulated the promoter ∼8-fold, and the additional inclusion of an expression vector for wild type CREB further stimulated promoter activity significantly.
Similar results were observed when two mutant CREB expression constructs that inactivate the KID were substituted (M1 and L141 (35)), whereas a mutant with a deletion of the constitutive activation domain (ΔQ2 (20)) failed to stimulate HMG-CoA reductase promoter activity beyond that achieved by NF-Y. When NF-Y subunits were omitted, stimulation by the CREB constructs was very low, but the same trends were observed. Because all of the tested mutants were expressed at levels similar to those of wild type CREB, these results indicate that the constitutive activation domain of CREB is essential for stimulation of the HMG-CoA reductase promoter in collaboration with SREBP, but the KID is not required.
The data so far presented indicate that the increase in HMG-CoA reductase in response to a low cholesterol signal in animals requires SREBPs working with CREB through its constitutive activation domain. As mentioned above, we did not think that the CREB-interacting TORC proteins would be involved in SREBP activation. However, to investigate a putative role of TORCs in SREBP-CREB synergy, we analyzed TORC recruitment to the HMG-CoA reductase promoter using an antibody to the major hepatic form of TORC, TORC2, in a ChIP study (Fig. 5C (25)). The results show that there was no change in TORC recruitment to the HMG-CoA reductase promoter following Z/L feeding or during fasting/refeeding, indicating that the increased recruitment of CREB following Zetia/lovastatin treatment is not likely to be accompanied by the co-recruitment of TORC proteins.
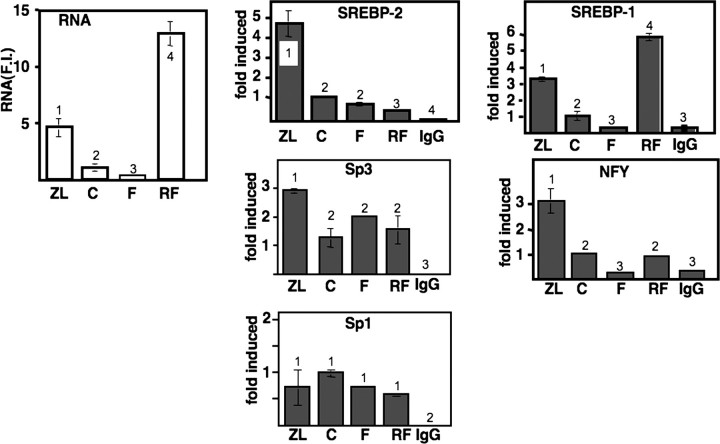
To extend our investigation of differential SREBP binding to other key target gene promoters, we analyzed the LDL receptor (Fig. 6). Induction of LDL receptor RNA was much more modest, 3.3-fold, following Z/L feeding, and the fasting/refeeding regimen did not result in any significant change from the chow fed control. The more modest increase in LDL receptor mRNA was also paralleled by a more modest 5-fold increase in binding of SREBP-2 to the promoter relative to the more than 25-fold increase observed for HMG-CoA reductase. There was no significant change in SREBP-2 association by fasting; however, this association was reduced in the refeeding treatment group. There was no increase in SREBP-1 binding to the LDL receptor promoter following Z/L feeding, highlighting a significant difference with HMG-CoA reductase. SREBP-1 binding decreased following fasting and was increased in the refeeding group to a level higher than observed in the chow control group.
FIGURE 6.
LDL receptor promoter and mRNA analysis. mRNA expression (left panel) and binding of SREBP-1 or SREBP-2 by ChIP for the LDL receptor gene was performed essentially as described in the legend for Fig. 2 but with primers specific for the LDL receptor gene mRNA and promoter. The amount of PCR product generated when a control IgG fraction was used in the immunoprecipitation is also provided (IgG). The binding and expression of Sp1 and Sp3 was also evaluated by ChIP and immunoblotting, respectively. All symbols and notations are as described in the legend for Fig. 2.
For the LDL receptor promoter, the identified SREBP coregulatory proteins are Sp1 and Sp3 (17). There was a modest increase in binding of Sp3 in the Z/L group but no clear changes in the pattern of binding for either Sp1 or Sp3, which correlate with LDL receptor gene expression. There were also no significant changes in total Sp1 or Sp3 levels caused by the feeding conditions.
Fatty-acid synthase (FAS) mRNA was induced 5-fold by Z/L feeding and displayed its signature lipogenic response to fasting and refeeding, where RNA levels declined 10-fold by fasting and increased to 14-fold higher levels than the chow control in the refeeding group (Fig. 7). The binding of both SREBP-1 and SREBP-2 was induced by Z/L treatment, similar in magnitude to the mRNA increase. The association of SREBP-2 with the FAS promoter in the fasting and refeeding group declined in parallel with the decline in total SREBP-2 nuclear protein levels. Interestingly, SREBP-1 binding declined significantly in the fasting group and was elevated to a value 5 times above the control by the high carbohydrate refeeding. The two SREBP co-regulatory proteins identified for the FAS proximal promoter include Sp1/Sp3 and NF-Y (18). Sp3 and NF-Y recruitment was induced by Z/L treatment, whereas there was no change in Sp1 binding across all treatment groups. The binding of Sp3 was not altered by the fasting and refeeding protocol, but the binding of NF-Y was reduced by fasting and returned to the control level by refeeding.
FIGURE 7.
Fatty-acid synthase mRNA and promoter analysis. mRNA expression (left panel) and binding of SREBPs and co-regulatory proteins to the proximal FAS promoter were analyzed with gene-specific primers as described under “Materials and Methods.” All symbols and notations are as described in the legend for Fig. 2.
Squalene synthase mRNA was induced 9-fold by Z/L treatment, repressed by fasting, and rebounded to the control level during the acute refeeding following a fast. The ChIP analyses showed that binding of both SREBPs was induced 4-fold by Z/L. The binding of both SREBPs declined upon fasting; however, only SREBP-1 binding returned to the control value after refeeding, suggesting that SREBP-1 is important for the increase in squalene synthase mRNA expression by refeeding treatment in contrast to the Z/L response, where both SREBP-1 and -2 are involved.
DISCUSSION
Although the existence of gene families and overlapping mRNAs in higher eukaryotes has been known since the 1970s, the extensive clusters of highly related genes and the almost ubiquitous nature of alternative mRNA processing was not fully realized until the sequence of the human genome was reported 7 years ago (36, 37). The sequencing of genomes from several other species has confirmed this as a basic feature of all complex eukaryotic organisms. Thus, a major goal now is to define precisely the unique and common roles for the different proteins produced from overlapping transcripts and for closely related proteins in the same family. This is complicated when the proteins are co-expressed in the same cells and function as dimers/multimers in which the individual molecules can form homo- or heteromers as in the case of the mammalian SREBPs.
The current study was designed to investigate selective binding by SREBP-1 versus SREBP-2 at target gene promoters in vivo and to determine how promoter binding correlates with target gene expression and recruitment of SREBP co-regulatory transcriptional proteins. The use of ChIP with SREBP-selective antibodies, combined with feeding regimens that preferentially alter levels of SREBP-1 or -2 in liver nuclei, has provided several new insights into the molecular mechanism for this key metabolic regulatory system.
The fasting and refeeding protocol to alter expression of SREBP-1c was adapted from Horton et al. (31) and was based on the dramatic regulation of SREBP-1c expression by this protocol with relatively minor effects on SREBP-2. The selection of Zetia (ezetimibe) plus lovastatin feeding to induce SREBP-2 is new and represents an important upgrade to a protocol using a combination of a bile acid sequestrant along with a statin-class HMG-CoA reductase inhibitor that has been known for more than 2 decades to induce cholesterol-regulated genes in the liver (38). This combination was shown to increase SREBP-2 nuclear protein levels but not those for SREBP-1 (30). The bile acid sequestrant prevents bile acid reabsorption and to a lesser extent cholesterol absorption as well. This establishes a metabolic demand for elevated flux through the linked cholesterol to bile acid synthetic pathway. We reasoned that by blocking cholesterol absorption specifically with Zetia, the treatment would be more precise in preventing cholesterol absorption without off-target effects on bile acid metabolism as reported previously (39). Thus, our feeding protocol more accurately reflects changes due to cholesterol depletion without complications resulting from changes in bile acid metabolism.
In response to Z/L, SREBP-2 protein levels increased dramatically in liver nuclei, whereas SREBP-1 protein levels declined. The increase in SREBP-2 was due to cholesterol depletion, whereas the decline in SREBP-1 levels was because cholesterol depletion also reduces the concentration of endogenous cholesterol-derived oxysterol liver X receptor agonists. This reduces transcription from the SREBP-1c promoter, which is the major liver form of SREBP-1 and is a target of liver X receptor signaling (40).
Our experiments first analyzed HMG-CoA reductase in detail, and we then extended the study to include a select handful of other SREBP-regulated genes for comparative purposes. The 35-fold increase in HMG-CoA reductase mRNA mediated by Z/L feeding was accompanied by a 25-fold increase in binding of SREBP-2 to the promoter, detected by ChIP. Additionally, even though the total SREBP-1 protein level declined, there was an almost 4-fold increase in its binding to the promoter. This observation and our other results are unlikely to be due to a cross-reaction of SREBP-1 with the SREBP-2 antibody or vice versa. This is because both antibodies were made from protein fragments that are unique to each SREBP (13), the immunoblotting results with the two antibodies followed a predictable pattern and were very distinct (Fig. 1), and the recruitment of SREBP-1 to the LDL receptor promoter was not enhanced in the Z/L treatment group (Fig. 6).
The reason for the increased binding of SREBP-1 to the HMG-CoA reductase promoter, despite a decrease in its absolute concentration, was unexpected and could be the result of two interesting but difficult to distinguish possibilities. The first is that the robust increase in SREBP-2 protein by Z/L could result in an increase in the concentration and promoter binding of SREBP-1/SREBP-2 heterodimers that would be recognized by the SREBP-1 antibody. Alternatively, there are two closely spaced SREBP binding sites in the HMG-CoA reductase promoter (41), and it is also possible that enhanced SREBP-2 homodimer binding to one recognition site stimulates the binding of a SREBP-1 homodimer to the second motif.
In support of the first model, we have shown that SREBP-1/SREBP-2 heterodimers form in vitro and in vivo, and they are competent to activate gene expression (9). In favor of the second model are the data for SREBP binding to the other promoters revealed in the present study. As in HMG-CoA reductase, there are multiple SREBP recognition sites in both the FAS and squalene synthase promoters (42, 43), and binding of SREBP-1 to these additional promoters was also enhanced by Z/L treatment (Figs. 7 and 8). In contrast, there is a single SREBP recognition site in the LDL receptor promoter (44), where SREBP-1 binding was not enhanced by Z/L (Fig. 6).
FIGURE 8.
Squalene synthase mRNA and SREBP binding. mRNA expression (left panel) and binding of SREBPs to the squalene synthase promoter were analyzed by qPCR and ChIP, with gene-specific primers, as described under “Materials and Methods.” All symbols and notations are as described in the legend for Fig. 2.
In analyses of SREBP-co-regulatory transcriptional regulators, we showed that CREB binding to the HMG-CoA reductase promoter in the Z/L group was induced and that a dominant negative version of CREB blunted the induction of HMG-CoA reductase mRNA by Z/L treatment. Utilizing co-transfection studies, we have also demonstrated that SREBP-CREB synergy on the promoter requires the CREB constitutive activation domain but probably does not require the kinase-inducible domain. Additionally, the KID-selective CREB co-regulatory protein TORC2 was not recruited to the HMG-CoA reductase promoter even though CREB binding was enhanced by Z/L. This is probably because SREBP and TORC both interact with the CREB b-zip domain, and simultaneous interactions are likely to be sterically unfavorable.
The association of NF-Y was also enhanced by Z/L feeding, consistent with an important role for this heterotrimeric protein in activation of HMG-CoA reductase expression in vivo. There was also a more modest yet significant increase in acetylation of promoter-proximal histone H3. The binding studies for SREBPs are novel and support interesting interactions between SREBP-1 and SREBP-2 in promoter activation. The observations for CREB, NF-Y, and H3 are consistent with our original study in cultured cells, where cholesterol depletion resulted in an increase in the binding of both SREBP co-regulatory proteins and an increase in H3 acetylation at the HMG-CoA reductase promoter (32). Along with the new information noted above, these similarities reinforce the validity of evaluating the mechanism for cholesterol regulation in cultured cells as a predictor of regulatory mechanisms that operate to maintain homeostasis in an intact animal model.
The fasting/refeeding protocol was designed to analyze the role of SREBPs in which the expression of SREBP-1c fluctuates significantly (31, 45). HMG-CoA reductase mRNA declined and rebounded to the control level following fasting and refeeding, respectively, consistent with previous observations (31). This was accompanied by a similar biphasic association of SREBP-1, but not SREBP-2, with the promoter. Binding of NF-Y followed a decline and rebound similar to the mRNA level and binding of SREBP-1. However, CREB association did not go down during fasting but did decline slightly during the acute high carbohydrate refeeding phase.
These results suggest that SREBP-1 synergy with NF-Y is responsible for the fasting/refeeding response of HMG-CoA reductase. Thus, the ChIP results, with samples from the different feeding studies, have uncovered interesting differences in the molecular mechanisms for activating the HMG-CoA reductase promoter in response to Z/L feeding versus fasting/refeeding.
Induction of LDL receptor mRNA by Z/L treatment was much more modest compared with HMG-CoA reductase. This was accompanied by a similar more modest increase in binding of SREBP-2 to its promoter. However, SREBP-1 binding was not different between the control and Z/L groups. The comparison with HMG-CoA reductase demonstrates for the first time that the differing relative strengths of SREBP binding correlate with the difference in magnitude of gene activation. Also, the lack of induction of SREBP-1 binding to the LDL receptor promoter has revealed a significant difference between SREBP regulation of HMG-CoA reductase and LDL receptor genes in the liver. Binding of the co-regulatory protein Sp3 to the LDL receptor promoter was also stimulated by Z/L, but the binding of Sp1 did not change, suggesting that the Z/L treatment increased expression of the LDL receptor mRNA through the concerted action of SREBP-2 and Sp3.
LDL receptor mRNA expression did not change significantly following the fasting/refeeding protocol, despite a significant decrease in binding of SREBP-1 to the promoter by fasting and a superinduction to a level above the control following the high carbohydrate refeeding phase (Fig. 6). It is possible that even though the binding was increased, the RNA level would not change because SREBP-1c, the major SREBP-1 isoform induced during refeeding, is a weak transcription factor (7). Additionally, it is possible that the LDL receptor mRNA would not decline during fasting because the mRNA is relatively stable, and the more acute changes in promoter occupancy by SREBP-1 is insufficient to change the total mRNA level.
The analysis of FAS showed some intriguing differences with both the LDL receptor and HMG-CoA reductase promoters. FAS mRNA was induced 5-fold by Z/L treatment, and there was a similar increase in binding of both SREBP-1 and SREBP-2. As mentioned above, we could not distinguish between SREBP-1/SREBP-2 homo- and heterodimer binding to the different closely spaced recognition sites. The elevated mRNA expression and SREBP binding by Z/L was also accompanied by a similar increase in the association of NF-Y and Sp3 at the promoter. Similar to the LDL receptor promoter, binding of Sp1 did not change. These results suggest that both SREBP-1 and -2 are responsible for activation of the FAS promoter by Z/L and that the two key co-regulatory proteins for mRNA induction under these conditions are NF-Y and Sp3.
Consistent with many other studies, FAS mRNA declined 10-fold following the fasting and was superinduced to a level more than 10-fold higher than the chow-fed control group during the refeeding program. The binding of SREBP-1 but not SREBP-2 closely followed this pattern of mRNA expression. Importantly, the mRNA “overshoot” by the acute high carbohydrate refeeding phase has been observed in other studies (46) but a mechanism for this effect has not been uncovered. Our studies suggest that this overshoot phenomenon is likely due to the dramatic increase in the binding of SREBP-1.
We also have shown that the binding of NF-Y to the FAS promoter declines with fasting and returns to the control level by refeeding. Neither Sp1 nor Sp3 exhibited significant changes during the fasting/refeeding process. These results indicate that SREBP-1 and NF-Y play key roles in the fasting/refeeding response of FAS. Because NF-Y binding did not exhibit an overshoot, it is possible that additional proteins such as upstream stimulatory factor and/or carbohydrate response element-binding protein (CHREBP) are involved, as these two proteins have been shown to play key roles in the fasting/refeeding response of FAS in prior studies (47, 48).
Overall, these studies reveal that there are alternative molecular mechanisms that utilize different SREBPs in combination with distinct co-regulatory proteins to activate the corresponding target genes in response to the different dietary challenges evoked by Zetia/lovastatin versus fasting/refeeding. This underscores the mechanistic flexibility that has evolved at the individual gene/promoter level to maintain metabolic homeostasis in response to shifting nutritional states and environmental fluctuations.
Acknowledgments
We thank Jay Horton, Marc Montminy, and Paul Brindle for invaluable reagents and Cherryl Nugas-Selby and Andrew Bourgeois for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant HL48044. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: HMG, 3-hydroxy-3-methylglutaryl; SREBP, sterol regulatory element-binding protein; bHLH, basic helix-loop-helix; ChIP, chromatin immunoprecipitation; NF-Y, nuclear factor Y; CREB, cyclic AMP regulatory element-binding protein; CBP, CREB-binding protein; GFP, green fluorescent protein; qPCR, quantitative PCR; Z/L, lovastatin and Zetia; LDL, low density lipoprotein; GFP, green fluorescent protein; PEPCK, phosphoenolpyruvate carboxykinase; KID, kinase-inducible domain; FAS, fatty-acid synthase; TORC, transducer of regulated CREB.
M. K. Bennett, Y.-K. Seo, S. Datta, D.-J. Shin, and T. F. Osborne, unpublished observations.
References
- 1.Brown, M. S., and Goldstein, J. L. (1980) J. Lipid Res. 21 505–517 [PubMed] [Google Scholar]
- 2.Goldstein, J., and Brown, M. (1990) Nature 343 425–430 [DOI] [PubMed] [Google Scholar]
- 3.Brown, M. S., and Goldstein, J. L. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 11041–11048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osborne, T. F., and Rosenfeld, J. R. (1998) Curr. Opin. Lipidol. 9 137–140 [DOI] [PubMed] [Google Scholar]
- 5.Kim, J. B., Spotts, G. D., Halvorsen, Y.-D., Shih, H.-M., Ellenberger, T., Towle, H. C., and Spiegelman, B. M. (1995) Mol. Cell. Biol. 15 2582–2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Athanikar, J. N., and Osborne, T. F. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 4935–4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toth, J. I., Datta, S., Athanikar, J. N., Freedman, L. P., and Osborne, T. F. (2004) Mol. Cell. Biol. 24 8288–8300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hannah, V. C., Ou, J., Luong, A., Goldstein, J. L., and Brown, M. S. (2001) J. Biol. Chem. 276 4365–4372 [DOI] [PubMed] [Google Scholar]
- 9.Datta, S., and Osborne, T. F. (2005) J. Biol. Chem. 280 3338–3345 [DOI] [PubMed] [Google Scholar]
- 10.Shin, D.-J., Campos, J. A., Gil, G., and Osborne, T. F. (2003) J. Biol. Chem. 278 50047–50052 [DOI] [PubMed] [Google Scholar]
- 11.Herzig, S., Long, F., Jhala, U. S., Hedrick, S., Quinn, R., Bauer, A., Rudolph, D., Schutz, G., Yoon, C., Puigserver, P., Spiegelman, B., and Montminy, M. (2001) Nature 413 179–183 [DOI] [PubMed] [Google Scholar]
- 12.Shin, D. J., Plateroti, M., Samarut, J., and Osborne, T. F. (2006) Nucleic Acids Res. 34 3853–3861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang, G., Yang, J., Horton, J. D., Hammer, R. E., Goldstein, J. L., and Brown, M. S. (2002) J. Biol. Chem. 277 9520–9528 [DOI] [PubMed] [Google Scholar]
- 14.Shin, D. J., and Osborne, T. F. (2003) J. Biol. Chem. 278 34114–34118 [DOI] [PubMed] [Google Scholar]
- 15.Bennett, M., Toth, J. I., and Osborne, T. F. (2004) J. Biol. Chem. 279 37360–37367 [DOI] [PubMed] [Google Scholar]
- 16.Sanchez, H. B., Yieh, L., and Osborne, T. F. (1995) J. Biol. Chem. 270 1161–1169 [DOI] [PubMed] [Google Scholar]
- 17.Athanikar, J. N., Sanchez, H. B., and Osborne, T. F. (1997) Mol. Cell. Biol. 17 5193–5200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magaña, M. M., Koo, S.-H., Towle, H. C., and Osborne, T. F. (2000) J. Biol. Chem. 275 4726–4733 [DOI] [PubMed] [Google Scholar]
- 19.Dooley, K. A., Bennett, M. K., and Osborne, T. F. (1999) J. Biol. Chem. 274 5285–5291 [DOI] [PubMed] [Google Scholar]
- 20.Ferreri, K., Gill, G., and Montminy, M. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 1210–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez, G. A., and Montminy, M. R. (1989) Cell 59 675–680 [DOI] [PubMed] [Google Scholar]
- 22.Ngo, T. T., Bennett, M. K., Bourgeois, A. L., Toth, J. I., and Osborne, T. F. (2002) J. Biol. Chem. 277 33901–33905 [DOI] [PubMed] [Google Scholar]
- 23.Datta, S., Wang, L., Moore, D. D., and Osborne, T. F. (2006) J. Biol. Chem. 281 807–812 [DOI] [PubMed] [Google Scholar]
- 24.Ravnskjaer, K., Kester, H., Liu, Y., Zhang, X., Lee, D., Yates, J. R., 3rd, and Montminy, M. (2007) EMBO J. 26 2880–2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu, W., Kasper, L. H., Lerach, S., Jeevan, T., and Brindle, P. K. (2007) EMBO J. 26 2890–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia-Calvo, M., Lisnock, J., Bull, H. G., Hawes, B. E., Burnett, D. A., Braun, M. P., Crona, J. H., Davis, H. R., Jr., Dean, D. C., Detmers, P. A., Graziano, M. P., Hughes, M., Macintyre, D. E., Ogawa, A., O'Neill, K. A., Iyer, S. P., Shevell, D. E., Smith, M. M., Tang, Y. S., Makarewicz, A. M., Ujjainwalla, F., Altmann, S. W., Chapman, K. T., and Thornberry, N. A. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 8132–8137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yokoyama, C., Wang, X., Briggs, M. R., Admon, A., Wu, J., Hua, X., Goldstein, J. L., and Brown, M. S. (1993) Cell 75 185–197 [PubMed] [Google Scholar]
- 28.Shimomura, I., Shimano, H., Horton, J. D., Goldstein, J. L., and Brown, M. S. (1997) J. Clin. Investig. 99 838–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sato, R., Inoue, J., Kawabe, Y., Kodama, T., Takano, T., and Maeda, M. (1996) J. Biol. Chem. 271 26461–26464 [DOI] [PubMed] [Google Scholar]
- 30.Sheng, Z., Otani, H., Brown, M. S., and Goldstein, J. L. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 935–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horton, J. D., Bashmakov, Y., Shimomura, I., and Shimano, H. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 5987–5992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bennett, M. K., and Osborne, T. F. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 6340–6344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chrivia, J. C., Kwok, R. P., Lamb, N., Hagiwara, M., Montminy, M. R., and Goodman, R. H. (1993) Nature 365 855–859 [DOI] [PubMed] [Google Scholar]
- 34.Gonzalez, G. A., Menzel, P., Leonard, J., Fischer, W. H., and Montminy, M. R. (1991) Mol. Cell. Biol. 11 1306–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parker, D., Ferreri, K., Nakajima, T., LaMorte, V. J., Evans, R., Koerber, S. C., Hoeger, C., and Montminy, M. R. (1996) Mol. Cell. Biol. 16 694–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Venter, J. C., Adams, M. D., Myers, E. W., Li, P. W., Mural, R. J., Sutton, G. G., Smith, H. O., Yandell, M., Evans, C. A., Holt, R. A., Gocayne, J. D., Amanatides, P., Ballew, R. M., Huson, D. H., Wortman, J. R., Zhang, Q., Kodira, C. D., Zheng, X. H., Chen, L., Skupski, M., Subramanian, G., Thomas, P. D., Zhang, J., Gabor Miklos, G. L., Nelson, C., Broder, S., Clark, A. G., Nadeau, J., McKusick, V. A., Zinder, N., Levine, A. J., Roberts, R. J., Simon, M., Slayman, C., Hunkapiller, M., Bolanos, R., Delcher, A., Dew, I., Fasulo, D., Flanigan, M., Florea, L., Halpern, A., Hannenhalli, S., Kravitz, S., Levy, S., Mobarry, C., Reinert, K., Remington, K., Abu-Threideh, J., Beasley, E., Biddick, K., Bonazzi, V., Brandon, R., Cargill, M., Chandramouliswaran, I., Charlab, R., Chaturvedi, K., Deng, Z., Di Francesco, V., Dunn, P., Eilbeck, K., Evangelista, C., Gabrielian, A. E., Gan, W., Ge, W., Gong, F., Gu, Z., Guan, P., Heiman, T. J., Higgins, M. E., Ji, R. R., Ke, Z., Ketchum, K. A., Lai, Z., Lei, Y., Li, Z., Li, J., Liang, Y., Lin, X., Lu, F., Merkulov, G. V., Milshina, N., Moore, H. M., Naik, A. K., Narayan, V. A., Neelam, B., Nusskern, D., Rusch, D. B., Salzberg, S., Shao, W., Shue, B., Sun, J., Wang, Z., Wang, A., Wang, X., Wang, J., Wei, M., Wides, R., Xiao, C., Yan, C., Yao, A., Ye, J., Zhan, M., Zhang, W., Zhang, H., Zhao, Q., Zheng, L., Zhong, F., Zhong, W., Zhu, S., Zhao, S., Gilbert, D., Baumhueter, S., Spier, G., Carter, C., Cravchik, A., Woodage, T., Ali, F., An, H., Awe, A., Baldwin, D., Baden, H., Barnstead, M., Barrow, I., Beeson, K., Busam, D., Carver, A., Center, A., Cheng, M. L., Curry, L., Danaher, S., Davenport, L., Desilets, R., Dietz, S., Dodson, K., Doup, L., Ferriera, S., Garg, N., Gluecksmann, A., Hart, B., Haynes, J., Haynes, C., Heiner, C., Hladun, S., Hostin, D., Houck, J., Howland, T., Ibegwam, C., Johnson, J., Kalush, F., Kline, L., Koduru, S., Love, A., Mann, F., May, D., McCawley, S., McIntosh, T., McMullen, I., Moy, M., Moy, L., Murphy, B., Nelson, K., Pfannkoch, C., Pratts, E., Puri, V., Qureshi, H., Reardon, M., Rodriguez, R., Rogers, Y. H., Romblad, D., Ruhfel, B., Scott, R., Sitter, C., Smallwood, M., Stewart, E., Strong, R., Suh, E., Thomas, R., Tint, N. N., Tse, S., Vech, C., Wang, G., Wetter, J., Williams, S., Williams, M., Windsor, S., Winn-Deen, E., Wolfe, K., Zaveri, J., Zaveri, K., Abril, J. F., Guigo, R., Campbell, M. J., Sjolander, K. V., Karlak, B., Kejariwal, A., Mi, H., Lazareva, B., Hatton, T., Narechania, A., Diemer, K., Muruganujan, A., Guo, N., Sato, S., Bafna, V., Istrail, S., Lippert, R., Schwartz, R., Walenz, B., Yooseph, S., Allen, D., Basu, A., Baxendale, J., Blick, L., Caminha, M., Carnes-Stine, J., Caulk, P., Chiang, Y. H., Coyne, M., Dahlke, C., Mays, A., Dombroski, M., Donnelly, M., Ely, D., Esparham, S., Fosler, C., Gire, H., Glanowski, S., Glasser, K., Glodek, A., Gorokhov, M., Graham, K., Gropman, B., Harris, M., Heil, J., Henderson, S., Hoover, J., Jennings, D., Jordan, C., Jordan, J., Kasha, J., Kagan, L., Kraft, C., Levitsky, A., Lewis, M., Liu, X., Lopez, J., Ma, D., Majoros, W., McDaniel, J., Murphy, S., Newman, M., Nguyen, T., Nguyen, N., Nodell, M., Pan, S., Peck, J., Peterson, M., Rowe, W., Sanders, R., Scott, J., Simpson, M., Smith, T., Sprague, A., Stockwell, T., Turner, R., Venter, E., Wang, M., Wen, M., Wu, D., Wu, M., Xia, A., Zandieh, A., and Zhu, X. (2001) Science 291 1304–135111181995 [Google Scholar]
- 37.Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., Funke, R., Gage, D., Harris, K., Heaford, A., Howland, J., Kann, L., Lehoczky, J., LeVine, R., McEwan, P., McKernan, K., Meldrim, J., Mesirov, J. P., Miranda, C., Morris, W., Naylor, J., Raymond, C., Rosetti, M., Santos, R., Sheridan, A., Sougnez, C., Stange-Thomann, N., Stojanovic, N., Subramanian, A., Wyman, D., Rogers, J., Sulston, J., Ainscough, R., Beck, S., Bentley, D., Burton, J., Clee, C., Carter, N., Coulson, A., Deadman, R., Deloukas, P., Dunham, A., Dunham, I., Durbin, R., French, L., Grafham, D., Gregory, S., Hubbard, T., Humphray, S., Hunt, A., Jones, M., Lloyd, C., McMurray, A., Matthews, L., Mercer, S., Milne, S., Mullikin, J. C., Mungall, A., Plumb, R., Ross, M., Shownkeen, R., Sims, S., Waterston, R. H., Wilson, R. K., Hillier, L. W., McPherson, J. D., Marra, M. A., Mardis, E. R., Fulton, L. A., Chinwalla, A. T., Pepin, K. H., Gish, W. R., Chissoe, S. L., Wendl, M. C., Delehaunty, K. D., Miner, T. L., Delehaunty, A., Kramer, J. B., Cook, L. L., Fulton, R. S., Johnson, D. L., Minx, P. J., Clifton, S. W., Hawkins, T., Branscomb, E., Predki, P., Richardson, P., Wenning, S., Slezak, T., Doggett, N., Cheng, J. F., Olsen, A., Lucas, S., Elkin, C., Uberbacher, E., Frazier, M., Gibbs, R. A., Muzny, D. M., Scherer, S. E., Bouck, J. B., Sodergren, E. J., Worley, K. C., Rives, C. M., Gorrell, J. H., Metzker, M. L., Naylor, S. L., Kucherlapati, R. S., Nelson, D. L., Weinstock, G. M., Sakaki, Y., Fujiyama, A., Hattori, M., Yada, T., Toyoda, A., Itoh, T., Kawagoe, C., Watanabe, H., Totoki, Y., Taylor, T., Weissenbach, J., Heilig, R., Saurin, W., Artiguenave, F., Brottier, P., Bruls, T., Pelletier, E., Robert, C., Wincker, P., Smith, D. R., Doucette-Stamm, L., Rubenfield, M., Weinstock, K., Lee, H. M., Dubois, J., Rosenthal, A., Platzer, M., Nyakatura, G., Taudien, S., Rump, A., Yang, H., Yu, J., Wang, J., Huang, G., Gu, J., Hood, L., Rowen, L., Madan, A., Qin, S., Davis, R. W., Federspiel, N. A., Abola, A. P., Proctor, M. J., Myers, R. M., Schmutz, J., Dickson, M., Grimwood, J., Cox, D. R., Olson, M. V., Kaul, R., Raymond, C., Shimizu, N., Kawasaki, K., Minoshima, S., Evans, G. A., Athanasiou, M., Schultz, R., Roe, B. A., Chen, F., Pan, H., Ramser, J., Lehrach, H., Reinhardt, R., McCombie, W. R., de la Bastide, M., Dedhia, N., Blocker, H., Hornischer, K., Nordsiek, G., Agarwala, R., Aravind, L., Bailey, J. A., Bateman, A., Batzoglou, S., Birney, E., Bork, P., Brown, D. G., Burge, C. B., Cerutti, L., Chen, H. C., Church, D., Clamp, M., Copley, R. R., Doerks, T., Eddy, S. R., Eichler, E. E., Furey, T. S., Galagan, J., Gilbert, J. G., Harmon, C., Hayashizaki, Y., Haussler, D., Hermjakob, H., Hokamp, K., Jang, W., Johnson, L. S., Jones, T. A., Kasif, S., Kaspryzk, A., Kennedy, S., Kent, W. J., Kitts, P., Koonin, E. V., Korf, I., Kulp, D., Lancet, D., Lowe, T. M., McLysaght, A., Mikkelsen, T., Moran, J. V., Mulder, N., Pollara, V. J., Ponting, C. P., Schuler, G., Schultz, J., Slater, G., Smit, A. F., Stupka, E., Szustakowski, J., Thierry-Mieg, D., Thierry-Mieg, J., Wagner, L., Wallis, J., Wheeler, R., Williams, A., Wolf, Y. I., Wolfe, K. H., Yang, S. P., Yeh, R. F., Collins, F., Guyer, M. S., Peterson, J., Felsenfeld, A., Wetterstrand, K. A., Patrinos, A., Morgan, M. J., de Jong, P., Catanese, J. J., Osoegawa, K., Shizuya, H., Choi, S., and Chen, Y. J. (2001) Nature 409 860–92111237011 [Google Scholar]
- 38.Liscum, L., Luskey, K. L., Chin, D. J., Ho, Y. K., Goldstein, J. L., and Brown, M. S. (1983) J. Biol. Chem. 258 8450–8455 [PubMed] [Google Scholar]
- 39.Repa, J. J., Dietschy, J. M., and Turley, S. D. (2002) J. Lipid Res. 43 1864–1874 [DOI] [PubMed] [Google Scholar]
- 40.Repa, J. J., Liang, G., Ou, J., Bashmakov, Y., Lobaccaro, J. M., Shimomura, I., Shan, B., Brown, M. S., Goldstein, J. L., and Mangelsdorf, D. J. (2000) Genes Dev. 14 2819–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Millinder-Vallett, S., Sanchez, H. B., Rosenfeld, J. M., and Osborne, T. F. (1996) J. Biol. Chem. 271 12247–12253 [DOI] [PubMed] [Google Scholar]
- 42.Bennett, M. K., Lopez, J. M., Sanchez, H. B., and Osborne, T. F. (1995) J. Biol. Chem. 270 25578–25583 [DOI] [PubMed] [Google Scholar]
- 43.Guan, G., Dai, P.-H., Osborne, T. F., Kim, J. B., and Shechter, I. (1997) J. Biol. Chem. 272 10295–10302 [DOI] [PubMed] [Google Scholar]
- 44.Briggs, M. R., Yokoyama, C., Wang, X., Brown, M. S., and Goldstein, J. L. (1993) J. Biol. Chem. 268 14490–14496 [PubMed] [Google Scholar]
- 45.Kim, J. B., Sarraf, P., Wright, M., Yao, K. M., Mueller, E., Solanes, G., Lowell, B. B., and Speigelman, B. M. (1998) J. Clin. Investig. 101 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim, T. S., and Freake, H. C. (1996) J. Nutr. 126 611–617 [DOI] [PubMed] [Google Scholar]
- 47.Ishii, S., Iizuka, K., Miller, B. C., and Uyeda, K. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 15597–15602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Casado, M., Vallet, V. S., Kahn, A., and Vaulont, S. (1999) J. Biol. Chem. 274 2009–2013 [DOI] [PubMed] [Google Scholar]