Abstract
Changes in the number, size, and shape of dendritic spines are associated with synaptic plasticity, which underlies cognitive functions such as learning and memory. This plasticity is attributed to reorganization of actin, but the molecular signals that regulate this process are poorly understood. In this study, we show neural Wiskott-Aldrich syndrome protein (N-WASP) regulates the formation of dendritic spines and synapses in hippocampal neurons. N-WASP localized to spines and active, functional synapses as shown by loading with FM4–64 dye. Knock down of endogenous N-WASP expression by RNA interference or inhibition of its activity by treatment with a specific inhibitor, wiskostatin, caused a significant decrease in the number of spines and excitatory synapses. Deletion of the C-terminal VCA region of N-WASP, which binds and activates the actin-related protein 2/3 (Arp2/3) complex, dramatically decreased the number of spines and synapses, suggesting activation of the Arp2/3 complex is critical for spine and synapse formation. Consistent with this, Arp3, like N-WASP, was enriched in spines and excitatory synapses and knock down of Arp3 expression impaired spine and synapse formation. A similar defect in spine and synapse formation was observed when expression of an N-WASP activator, Cdc42, was knocked down. Thus, activation of N-WASP and, subsequently, the Arp2/3 complex appears to be an important molecular signal for regulating spines and synapses. Arp2/3-mediated branching of actin could be a mechanism by which dendritic spine heads enlarge and subsequently mature. Collectively, our results point to a critical role for N-WASP and the Arp2/3 complex in spine and synapse formation.
Communication between neurons occurs at highly specialized cell-cell junctions called synapses. These structures are composed of pre- and post-synaptic terminals that allow for propagation of signals between neurons and are the basis for the complex circuitry found in the central nervous system. Postsynaptic terminals of excitatory synapses consist of actin-rich dendritic spines (1) that are small extensions from the dendrite that form connections with axonal terminals. Abnormalities in dendritic spines are associated with various neurological and psychiatric disorders, including mental retardation, schizophrenia, epilepsy, and Alzheimer disease, pointing to the importance of these structures in the central nervous system (2). Morphological changes in dendritic spines due to reorganization of the actin cytoskeleton are thought to be important for synaptic function and, thus, integrating information flow within the brain (3–6). Despite considerable interest, the molecular mechanisms that regulate spine morphology and synapse formation via modulation of the actin cytoskeleton are poorly understood.
Members of the Wiskott-Aldrich syndrome protein (WASP)2 family, including WASP, neural WASP (N-WASP), and WASP-family verprolin homologous proteins (WAVEs), are emerging as critical regulators of the actin cytoskeleton. These proteins initiate the nucleation of new actin filaments through activation of the Arp2/3 complex (7, 8). New filaments generated by the Arp2/3 complex are formed at fixed angles to the mother filament, creating a branched actin network that is commonly found in protrusive regions of cells (9, 10). C-terminal sequences within N-WASP, consisting of a verprolin-like homology domain (V), a central domain (C), and an acidic region (A), mediate binding of G-actin and the Arp2/3 complex to these proteins, which subsequently results in actin nucleation (7, 8, 11, 12).
N-WASP, as its name implies, is highly expressed in the brain, but its function in the nervous system is not well understood (13). In this study, we show a crucial function for N-WASP in the formation of dendritic spines and synapses in hippocampal neurons. This activity of N-WASP is dependent on its C-terminal binding and activation of the Arp2/3 complex. Inhibition of Arp2/3 binding to N-WASP resulted in a significant reduction in the density of spines and synapses. Decreased expression of the N-WASP activator Cdc42 also caused a defect in the formation of spines and synapses. Thus, our results point to a critical role for activated N-WASP and the Arp2/3 complex in the development of dendritic spines and synapses in the central nervous system.
EXPERIMENTAL PROCEDURES
Reagents—SV2 (1:250) and GAD-6 (1:250) monoclonal antibodies were from the Developmental Studies Hybridoma Bank (The University of Iowa, Iowa City, IA). PSD-95 monoclonal antibody was from Chemicon (Temecula, CA). GFP polyclonal antibody was from Invitrogen. β-Actin AC-15 monoclonal antibody, α-tubulin DM 1A monoclonal antibody, and phalloidin-TRITC were from Sigma. N-WASP (1:50) polyclonal antibody was a generous gift from Marc Kirschner (Harvard Medical School, Boston, MA). Arp3 antibody (1:100) was described previously (14). Cdc42 monoclonal antibody (B-8) was from Santa Cruz Biotechnology (Santa Cruz, CA). Alexa Fluor® 488 and 555 anti-mouse, Alexa Fluor® 488 and 555 anti-rabbit for immunohistochemistry, Alexa Fluor® 680 anti-rabbit for Western blotting, and FM4–64 FX were from Molecular Probes (Eugene, OR). IRDye® 800 anti-mouse for Western blotting was from Rockland Immunochemicals (Gilbertsville, PA). Wiskostatin was purchased from Calbiochem.
Plasmids—RNA interference (RNAi) constructs were prepared by ligating annealed sense and antisense 64-mer oligonucleotides into pSUPER vector as described previously (15). The RNAi oligos contained the following 19-nucleotide target sequences: N-WASP RNAi, 5′-GACGAGATGCTCCAAATGG-3′; Arp3 RNAi, 5′-AGGTTTATGGAGCAAGTGA-3′; Cdc42 RNAi, 5′-GGGCAAGAGGATTATGACA-3′; and scrambled RNAi 5′-CAGTCGCGTTTGCGACTGG-3′. The scrambled RNAi target sequence, which had been described previously (16), was used as a control. Full-length GFP-N-WASP and GFP-N-WASP-ΔWA, -WA, and -ΔWH1 were generous gifts from Michael Way (Lincoln's Inn Fields Laboratories, London, UK). GFP-tagged bovine N-WASP was a generous gift from John Condeelis (Albert Einstein College of Medicine, Bronx, NY). GFP-Arp3 has been described previously (17). Myc-tagged WAVE1 was kindly provided by Hiroaki Miki and Tadaomi Takenawa (University of Tokyo, Japan). Myc-tagged dominant negative Cdc42 (Cdc42-N17) was a generous gift from Alan Hall (Memorial Sloan-Kettering Cancer Center, New York, NY).
Cell Culture and Transfection—Hippocampal low density cultures were prepared as described previously (18). Neurons were plated at a density of 70,000 cells/mm2 and transfected by a modified calcium phosphate method (19). HEK-293T cells (ATCC, Manassas, VA) were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum and penicillin/streptomycin. HEK cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Immunohistochemistry and Image Analysis—For most antibodies, neurons were fixed with 4% paraformaldehyde/4% glucose in phosphate-buffered saline (PBS) for 15 min. Coverslips were then permeabilized with 0.2% Triton X-100 for 5 min and washed three times with PBS. For PSD-95 staining, neurons were fixed for 3 min with paraformaldehyde/glucose and then permeabilized for 10 min in cold methanol at -20 °C. Coverslips were then blocked for 1 h with 20% goat serum in PBS. Antibodies were diluted in 5% goat serum in PBS and incubated at the indicated dilutions for 1 h followed by three washes with PBS. Coverslips were mounted with Aqua Poly/Mount (Polysciences, Inc., Warrington, PA). In all experiments, at least 40 dendrites from 15–20 neurons from three different cultures were analyzed to quantify the number of spines, SV2 clusters, and PSD-95 clusters.
Neurons were imaged with a Retiga EXi CCD camera (QImaging, Surrey, British Columbia, Canada) attached to an Olympus IX71 inverted microscope (Melville, NY) with a ×60 objective (numerical aperture 1.45). Image acquisition was controlled by MetaMorph software (Molecular Devices, Sunnyvale, CA) interfaced with a Lambda 10-2 automated controller (Sutter Instruments, Novato, CA). For enhanced GFP and Alexa Fluor® 488, an Endow GFP Bandpass filter cube (excitation HQ470/40, emission HQ525/50, Q495LP dichroic mirror) (Chroma, Brattleboro, VT) was used. Rhodamine and Alexa Fluor® 555 were imaged with a TRITC/Cy3 cube (excitation HQ545/30, emission HQ610/75, Q570LP dichroic mirror).
FM4–64 Loading and Destaining—Day 12 neurons were incubated with 0.5 μm FM4–64 FX in high K+ solution containing 72 mm NaCl, 50 mm KCl, 1 mm NaH2PO4, 26 mm NaHCO3, 1.8 mm CaCl2, 0.8 mm MgSO4, 11 mm d-glucose, and 20 mm HEPES, pH 7.35, for 3 min. Neurons were then washed three times with calcium-free solution, fixed with 4% paraformaldehyde/glucose for 15 min, and visualized in fluorescence.
RESULTS
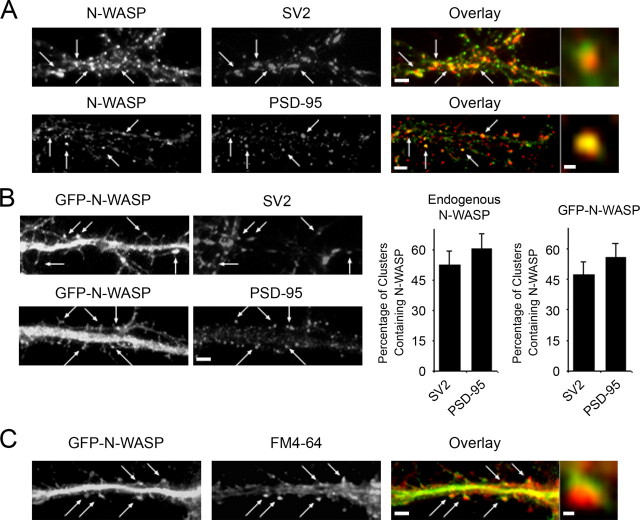
N-WASP Is Enriched in Excitatory Synapses in Hippocampal Neurons—The expression of N-WASP has been shown to increase dramatically in the rat hippocampus during the first several weeks after birth (20), a time when dendritic spines and synapses are developing. This led us to hypothesize that this molecule plays an important role in spine and synapse formation. To begin to test our hypothesis, we examined the subcellular localization of N-WASP in hippocampal neurons by immunostaining low density cultures with N-WASP antibody along with a presynaptic marker, SV2, or a postsynaptic marker, PSD-95. The synaptic vesicle protein SV2 is found in presynaptic terminals of both excitatory and inhibitory synapses, whereas the postsynaptic density protein PSD-95 localizes to the postsynaptic side of shaft and spine excitatory endings. Endogenous N-WASP accumulated in puncta along neuronal processes with both SV2 and PSD-95 (Fig. 1A, arrows). Because it is difficult to determine from the lower magnification images whether N-WASP puncta were completely merged or were in close apposition to the synaptic markers, higher magnification images were generated. As shown in the higher magnification images, N-WASP puncta were in close apposition to the presynaptic marker SV2, suggesting it was in postsynaptic terminals (Fig. 1A). Consistent with this, N-WASP puncta almost completely merged with the postsynaptic marker PSD-95 (Fig. 1A), indicating that N-WASP localizes to the postsynaptic side of excitatory synapses. Although most N-WASP localized to excitatory synapses, a fraction of N-WASP puncta were negative for PSD-95. These puncta might represent N-WASP protein complexes that have been described previously in non-neuronal cells (21).
FIGURE 1.
N-WASP localizes to spines and functional, excitatory synapses in hippocampal neurons. A, hippocampal neurons at day 12 in culture were co-immunostained for endogenous N-WASP and the presynaptic marker SV2 (upper panels) or the postsynaptic marker PSD-95 (lower panels). Endogenous N-WASP accumulated in puncta along neuronal processes with the synaptic markers (Overlays, right panels, arrows). Bar, 2 μm. High magnification images showed that N-WASP puncta were in close apposition to SV2 and completely merged with PSD-95, indicating N-WASP is enriched in the postsynaptic side of excitatory synapses. Bar, 0.4 μm. B, hippocampal neurons were transfected with GFP-N-WASP at day 5 in culture and then fixed and immunostained for SV2 (upper panels) and PSD-95 (lower panels) at day 12 in culture. Like endogenous N-WASP, GFP-N-WASP localized in puncta with the synaptic markers SV2 and PSD-95 (arrows). Quantification of the percentage of SV2 and PSD-95 clusters that contain N-WASP is shown. Error bars represent S.E. of the mean from at least three seperate experiments. C, hippocampal neurons were transfected with GFP-N-WASP at day 5 in culture. At day 12 in culture, neurons were incubated with FM4–64 dye (0.5 μm) in high K+ solution for 3 min, washed, fixed, and viewed in fluorescence. FM4–64, which labels presynaptic terminals of active synapses, was in close apposition to N-WASP puncta (Overlay, right panel, arrows, high magnification image), indicating that N-WASP-containing synapses are functional. Bar, 2 μm. Bar, 0.4 μm for the high magnification image.
The localization of GFP-N-WASP to synapses was examined by transfecting neurons with a GFP-tagged N-WASP construct. The GFP tag was shown previously to have no effect on N-WASP localization (22). Like endogenous N-WASP, GFP-N-WASP co-localized in clusters with PSD-95 and was in close apposition to SV2 puncta (Fig. 1B, arrows, and supplemental Fig. S1), confirming that GFP-N-WASP localizes similarly to the endogenous molecule and is a valid maker for examining N-WASP localization in the neurons.
Our results indicate that N-WASP is enriched in the postsynaptic terminal of excitatory synapses. However, some of the SV2 and PSD-95 puncta lacked N-WASP, which led us to quantify the number of SV2 and PSD-95 clusters that contained N-WASP puncta. Approximately 55% of the SV2 clusters and ∼66% of the PSD-95 clusters contained N-WASP (Fig. 1B), indicating that N-WASP is present in a majority of the SV2 and PSD-95 clusters. Similar results were obtained with GFP-N-WASP. This raised the question as to whether the N-WASP-containing clusters were activity-dependent, functional synapses.
FM4–64 is a member of a family of styryl dyes that can be used as fluorescent probes to visualize active synapses (23, 24). These dyes exhibit prominent fluorescence upon insertion of their hydrophobic tails into a lipid bilayer but can easily be washed out due to their inability to cross bilayers (25). When a cell surface is exposed to an FM dye and then washed with a dye-free solution, only membranes that are no longer surface-exposed retain the fluorescent dye (25). Thus, the activity-dependent uptake of FM dyes into synaptic vesicles provides a useful marker for functional synapses in cultured neurons (26, 27). To determine whether N-WASP-containing clusters were functional synapses, neurons expressing GFP-N-WASP were labeled with FM4–64. Day 12 neurons were incubated with 0.5 μm FM4–64 in high K+ solution for 3 min and then washed three times with calcium-free solution. As expected, FM4–64 puncta, which represented functional synapses, were observed along the dendrites. However, when neurons were incubated with the dye in a buffer lacking K+, FM4–64 puncta were not observed, indicating that FM4–64 uptake is dependent on depolarization. As shown in Fig. 1C, N-WASP puncta localized with FM4–64. Quantification of these results showed that 85.0 ± 3.5% of the FM4–64-labeled synapses contained N-WASP, demonstrating that N-WASP was in most of the active synapses. Thus, our results indicate N-WASP is present in the postsynaptic side of active, functional, excitatory synapses that could undergo changes in morphology mediated by reorganization of the actin cytoskeleton.
N-WASP Regulates Dendritic Spine Morphology and the Formation of Excitatory Synapses—To examine the function of N-WASP in the neurons, we treated cultures with a specific inhibitor of N-WASP, wiskostatin, and determined the density of synapses and spines by staining with SV2 and phalloidin, respectively. Wiskostatin stabilizes the autoinhibited form of the protein so that the C-terminal VCA domains are not available to bind G-actin and the Arp2/3 complex to initiate new actin filaments (28). When neurons were treated with wiskostatin at day 7 and then fixed and immunostained at day 12, a dose-dependent decrease in the number of spines and synapses was observed (Fig. 2A). Wiskostatin did not adversely affect the health of the neurons, because they were observed to develop normally and no detectable effects on the growth of axons or dendrites were seen at any of the wiskostatin concentrations used.
FIGURE 2.
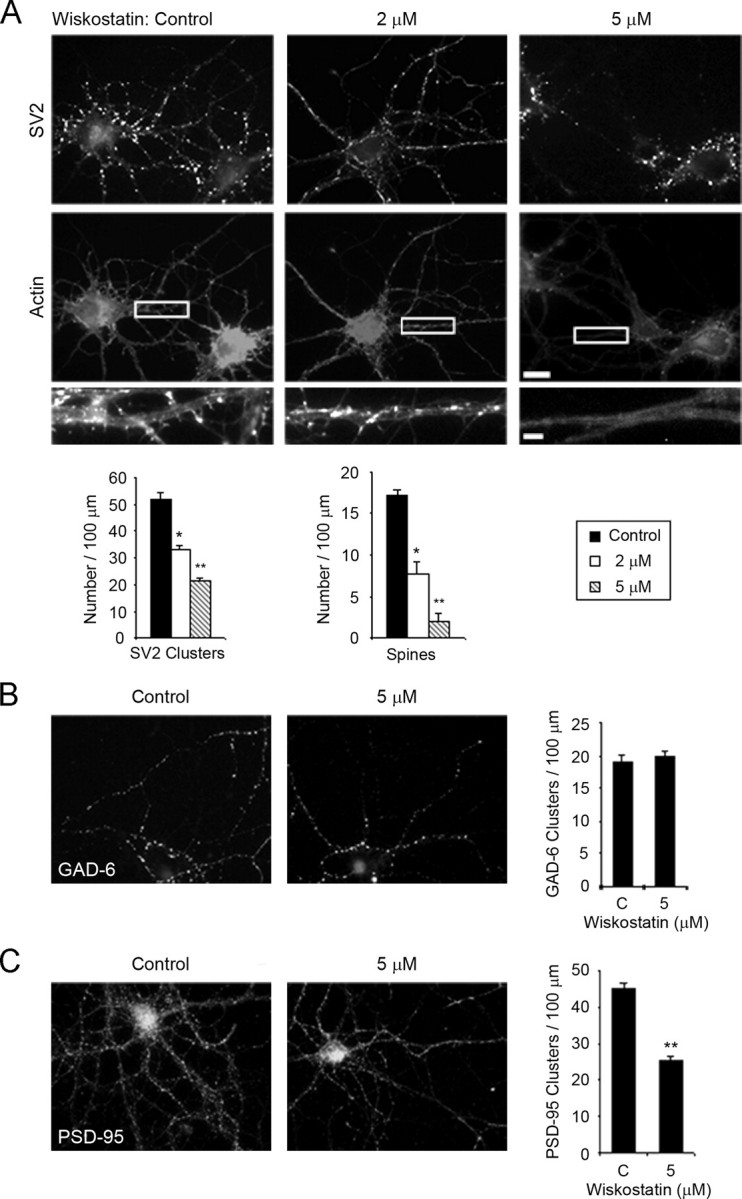
Inhibition of N-WASP activity by wiskostatin affects excitatory, but not inhibitory, synapses. A, hippocampal neurons were treated with either DMSO (Control) or an N-WASP-specific inhibitor, wiskostatin (2 or 5 μm), at day 7 in culture. At day 12 in culture, neurons were fixed and stained for SV2 (upper panels) and actin using rhodamine-conjugated phalloidin (lower panels). Bar, 10 μm. Enlargements of the boxed regions are shown below the panels. Bar, 2 μm. Differences between Control-2 μm, Control-5 μm, and Control 2–5 μm were statistically significant as determined by Student's t test (for SV2 clusters and spines, * and **, p < 0.0001). B, wiskostatin, even at the highest concentration (5 μm), did not significantly affect the number of inhibitory synapses as shown by immunostaining for glutamic acid decarboxylase (GAD-6). C, in contrast, a significant decrease in the number of excitatory synapses (**, p < 0.0001), as shown by a reduction in the density of PSD-95 clusters, was seen with 5 μm wiskostatin treatment. Error bars represent S.E. from at least three separate experiments.
Interestingly, we noticed that at the highest concentration of wiskostatin (5 μm) many of the remaining synapses were concentrated around the neuronal cell bodies. It has been reported previously that inhibitory GABAergic synapses are most dense around the cell body and form directly on the dendrite or cell soma surface without a spine (29), which led us to look further at the type of synapses that were regulated by N-WASP activity. To examine the effect of wiskostatin treatment on inhibitory synapses, we used GAD-6 antibody to immunostain for glutamic acid decarboxylase. Glutamic acid decarboxylase localizes to the presynaptic side of inhibitory synapses and catalyzes the synthesis of the inhibitory neurotransmitter γ-aminobutyric acid (GABA). There was no significant difference in the number of inhibitory GABAergic synapses with wiskostatin treatment (Fig. 2B), indicating that N-WASP activation is not necessary for the formation of these synapses.
In contrast, wiskostatin treatment had a significant effect on the density of excitatory glutamatergic synapses as the number of PSD-95 puncta was decreased by >50% (Fig. 2C). Because excitatory synapses can form on dendritic spines or on the dendritic shaft, we assessed the effects of wiskostatin treatment on each of these types of excitatory synapses. Wiskostatin treatment decreased the number of excitatory synapses that formed on spines by almost 90% (20.4 ± 1.7 in controls versus 2.3 ± 0.6 in wiskostatin-treated) but had no significant effect on shaft synapses (20.2 ± 1.7 in controls versus 19.6 ± 1.6 in wiskostatin-treated). In these experiments, neurons were treated for 5 days with wiskostatin to ensure that N-WASP activity was inhibited during a critical time when many spines and synapses form. However, treatment of the neurons for a shorter period of time also resulted in a significant decrease in the number of spines and synapses. We added 5 μm wiskostatin to the neuronal cultures at day 7, washed out the wiskostatin at day 8, and stained for PSD-95 and phalloidin at day 12. The 1-day wiskostatin treatment caused a 51% decrease in the number of spines (17.9 ± 1.0 in controls versus 8.7 ± 0.6 in wiskostatin-treated) and a 30% reduction in the number of synapses (37.5 ± 1.8 in controls versus 26.2 ± 1.7 in wiskostatin-treated). Taken together, our results suggest that activation of N-WASP is important for the regulation of spine morphogenesis and the formation of excitatory glutamatergic synapses.
We generated an RNAi construct to knock down expression of endogenous N-WASP to further explore the role of N-WASP in dendritic spine and synapse formation. The RNAi sequence had been shown previously to be specific for N-WASP and to almost completely knock down expression of the protein (30–32). However, we confirmed the ability of the RNAi construct to specifically knock down rat N-WASP expression by transiently transfecting it into HEK-293T cells along with GFP-tagged rat N-WASP or WAVE1. As determined by immunoblot analysis, the N-WASP RNAi construct decreased expression of N-WASP by >85% compared with empty pSUPER vector (Fig. 3A, left panels). In contrast, a scrambled RNAi control had no effect on expression of N-WASP (Fig. 3A, right panels). The N-WASP RNAi construct did not affect expression of WAVE1, another WASP family member, indicating its specificity for N-WASP (Fig. 3A, middle panels). The RNAi construct was similarly effective in decreasing expression of N-WASP in the neurons. Transfection of neurons with N-WASP RNAi resulted in an 87.2 ± 4.8% decrease in N-WASP expression compared with control cultures. When neurons were transfected with the N-WASP RNAi construct at day 6 and then fixed and stained for SV2 and PSD-95 at day 12, a 2- to 3-fold decrease in the number of dendritic spines and synapses was observed compared with cells expressing a scrambled RNAi construct or empty pSUPER vector (Fig. 3, B and C, and supplemental Fig. S2). To further show that the effects of N-WASP RNAi were specifically attributable to the loss of N-WASP, rescue experiments were performed in which bovine N-WASP was co-expressed with N-WASP RNAi. The rat N-WASP RNAi target sequence does not significantly affect expression of bovine N-WASP due to several nucleotide mismatches (30). Expression of bovine N-WASP completely rescued the N-WASP RNAi-mediated defect in spine and synapse formation (Fig. 3C). These results suggest that the defect is due to the loss of endogenous N-WASP and point to a critical role for N-WASP in spine and synapse formation.
FIGURE 3.
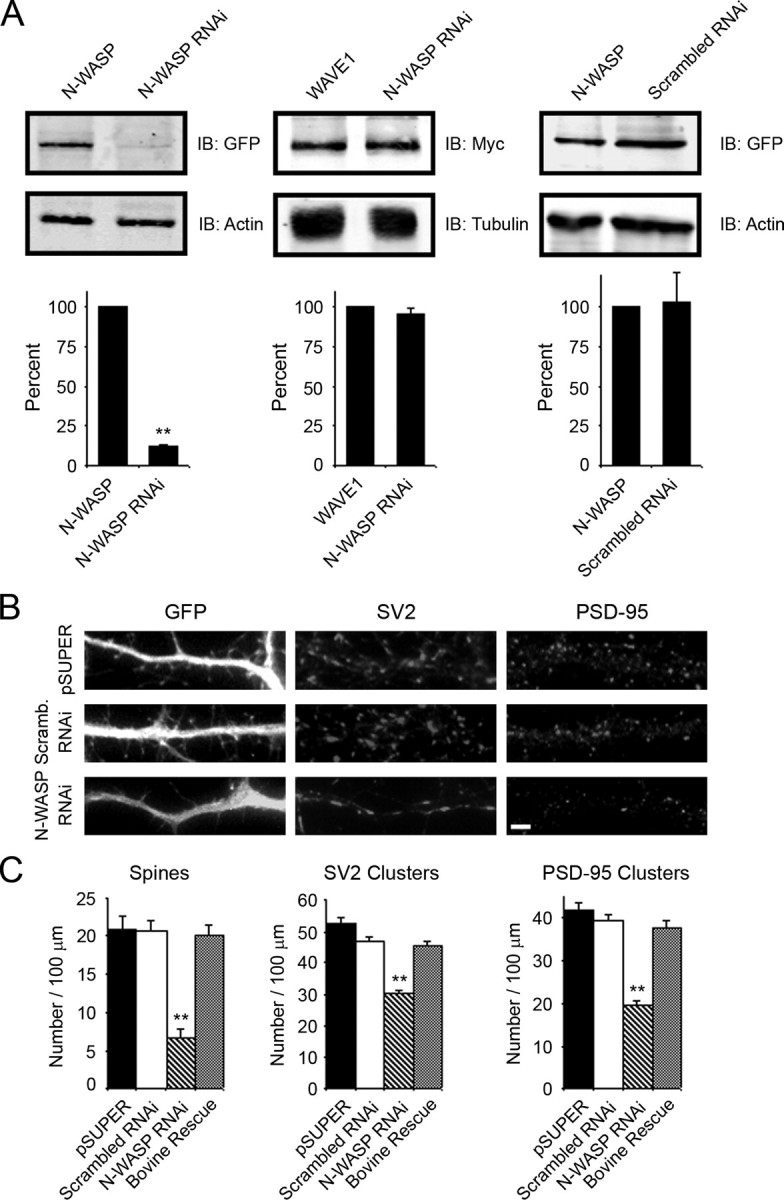
Knock down of endogenous N-WASP affects spines and synapses. A, HEK-293T cells were co-transfected with N-WASP RNAi or empty pSUPER vector and either GFP-N-WASP or Myc-WAVE1. For a nonsilencing control, a scrambled RNAi construct was prepared and tested by co-transfection with GFP-N-WASP into HEK-293T cells. The lysates were blotted for GFP and β-actin or Myc and α-tubulin. The N-WASP RNAi construct specifically decreased N-WASP expression by 85% (left panels) but did not alter expression of WAVE1 (middle panels), another WASP family member. The scrambled RNAi construct did not affect expression of N-WASP (right panels). Quantification of blots from three separate experiments is shown (lower panels). B, hippocampal neurons were co-transfected with GFP and either empty pSUPER vector, scrambled RNAi, or the N-WASP RNAi construct at day 6 in culture and then fixed at day 12 in culture and stained for SV2 (middle panels) and PSD-95 (right panels). A significant decrease in the number of spines and synapses was observed in neurons expressing N-WASP RNAi compared with either scrambled RNAi or pSUPER controls. Bar, 2 μm. C, quantification of the number of spines and synapses (SV2 and PSD-95 clusters) in neurons transfected with empty pSUPER vector, N-WASP RNAi, or scrambled RNAi is shown (**, p value <0.0001). The N-WASP RNAi-mediated defects in spine and synapse formation were rescued by expression of bovine N-WASP (Bovine Rescue). Error bars represent S.E. from at least three separate experiments.
The Actin and Arp2/3 Binding Domains of N-WASP Are Critical for Its Function in Regulating Spine and Synapse Formation—We next used several deletion constructs of N-WASP to determine the domains of the protein necessary for spine and synapse formation. For these experiments, the synaptic density was determined by staining for both SV2 and PSD-95 and the number of spines was assessed by GFP fluorescence as well as staining with phalloidin. Phalloidin, which binds F-actin and is commonly used to visualize dendritic spines (3, 33), was used to assess spine density because it is possible that the GFP-tagged fragments of N-WASP differentially localize to spines. Interestingly, similar results for the spine density were observed with both methods.
The C terminus of N-WASP is composed of two verprolin homology (V) domains, which bind G-actin; a central (C) domain; and an acidic (A) region that binds the Arp 2/3 complex (7, 8). The VCA region of N-WASP, which we refer to as “WA,” is sufficient to stimulate the nucleation of new actin filaments by the Arp2/3 complex (7, 8). Expression of GFP-N-WASP caused a small but significant increase in the density of dendritic spines and synapses compared with GFP alone (Fig. 4, B and C). Expression of N-WASP lacking the C-terminal VCA region (ΔWA) significantly decreased the number of spines and synapses compared with control neurons expressing GFP, but protrusions were still observed along the dendrites of N-WASP-ΔWA-expressing neurons (Fig. 4, B and C, and supplemental Fig. S3). Protrusions were defined as extensions from the dendrites without associated presynaptic endings whereas dendritic spines were in contact with presynaptic terminals. In contrast, expression of N-WASP lacking the N-terminal WH1 region (ΔWH1) did not significantly affect the density of spines or synapses as compared with controls (Fig. 4, B and C, and supplemental Fig. S3). These results suggest that activation of the Arp2/3 complex by N-WASP promotes spine and synapse formation in the neurons and could be a mechanism by which dendritic spine heads enlarge and subsequently mature.
FIGURE 4.
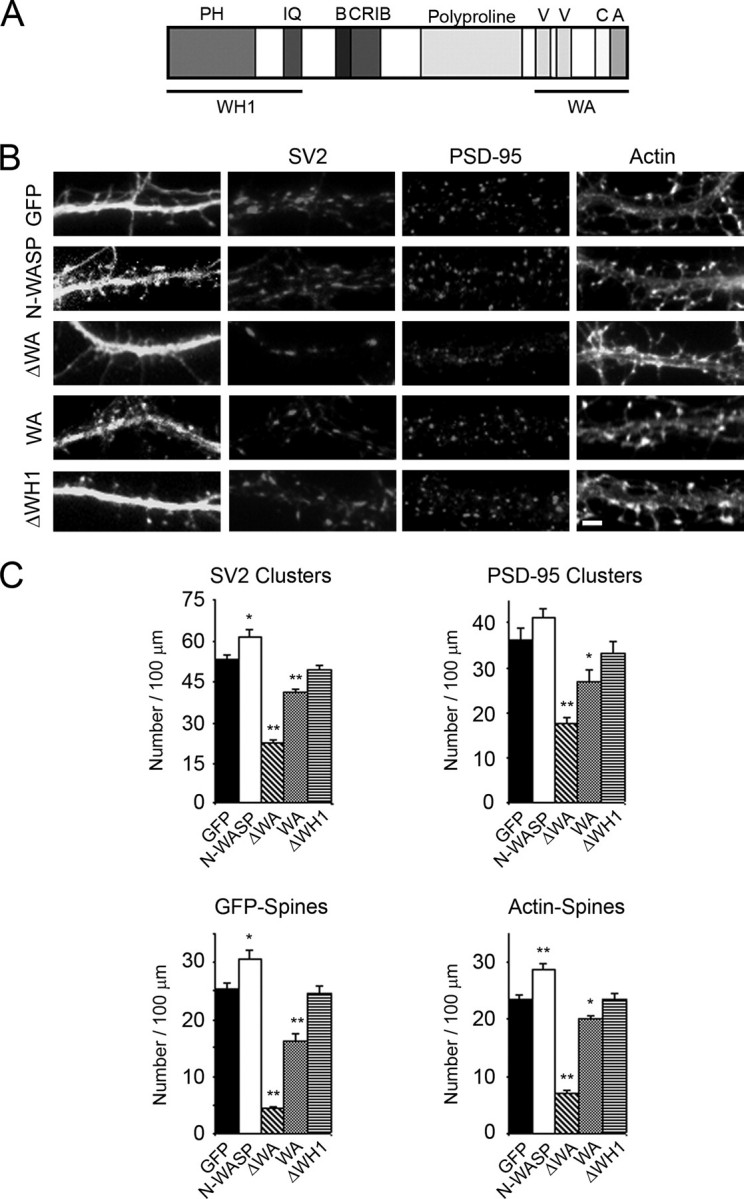
N-WASP binding to actin and the Arp 2/3 complex is important for dendritic spine and synapse formation. A, domain structure of N-WASP. Pleckstrin homology (PH), calmodulin binding (IQ), basic (B), Cdc42/Ras interactive binding (CRIB), verprolin homology (V), central (C), and acidic (A) domains are shown. B, hippocampal neurons were transfected with GFP, GFP-N-WASP, GFP-N-WASP-ΔWA, GFP-N-WASP-WA, and GFP-N-WASP-ΔWH at day 5 in culture and then fixed and immunostained for SV2, PSD-95, and actin (phalloidin) at day 12 in culture. Bar, 2 μm. C, quantification of SV2 and PSD-95 clusters and dendritic spines using GFP fluorescence and by staining for actin (phalloidin) from transfected neurons is shown. Asterisks denote statistically significant differences when compared with control GFP expressing neurons (*, p < 0.025; **, p < 0.0001). Error bars represent S.E. from at least three seperate experiments.
To further explore the function of the WA region in spine and synapse formation, we transfected neurons with a construct encoding the C-terminal region (WA) of N-WASP. Expression of the WA region resulted in a slight decrease in the density of spines and synapses. In neurons expressing the WA region, a 14% decrease in the number of spines, as determined by staining with phalloidin, and a 25% reduction in the synaptic density, as determined by staining with PSD-95, was observed when compared with cells expressing GFP alone (Fig. 4, B and C). However, significantly more spines and synapses were observed in neurons expressing the WA region than with cells expressing truncated N-WASP that lacked the C-terminal WA region, suggesting that the WA region of N-WASP is critical for its function in spine and synapse formation. Taken together, our results strongly suggest that activation of the Arp2/3 complex by N-WASP is critical for the formation of dendritic spines and synapses.
Cdc42 Plays a Role in the Development of Dendritic Spines and Synapses—Because the Rho family GTPase Cdc42 is a known activator of N-WASP (34), we explored its function in regulating spine and synapse formation by preparing an RNAi construct to knock down expression of Cdc42 in neurons. We tested the ability of the Cdc42 RNAi construct to knock down expression of the endogenous protein by transfecting it into Rat2 fibroblasts. Immunoblot analysis showed that the RNAi construct decreased expression of Cdc42 by >75% compared with control pSUPER vector (Fig. 5A, left panels). In contrast, a scrambled RNAi construct did not significantly affect expression of Cdc42 (Fig. 5A, right panels). When neurons were transfected with the Cdc42 RNAi construct, a significant decrease in the number of spines and synapses was observed compared with cells transfected with a scrambled RNAi construct or empty pSUPER vector (Fig. 5, B and C, and supplemental Fig. S4). A similar decrease in the density of spines and synapses was observed in neurons expressing dominant negative Cdc42 (Fig. 5, B and C). The Cdc42 RNAi-mediated defect in spine and synapse formation could be rescued, at least partially, by expression of the WA region of N-WASP, linking Cdc42 to N-WASP activation in the development of spines and synapses (Fig. 5, B and C).
FIGURE 5.
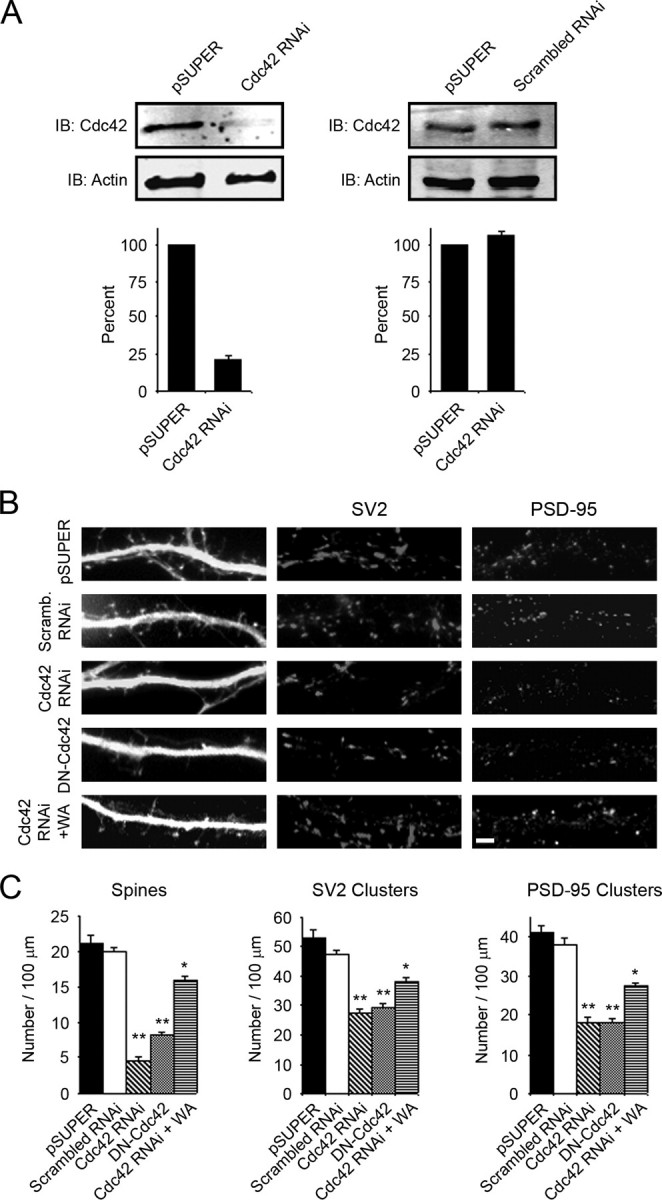
Cdc42 plays a role in the development of dendritic spines and synapses. A, Rat2 fibroblasts were co-transfected with GFP and either Cdc42 RNAi, scrambled RNAi, or empty pSUPER vector. The lysates were blotted for Cdc42 and actin as a loading control. Expression of the Cdc42 RNAi construct decreased endogenous levels of Cdc42 by >75% (left panels) whereas the scrambled RNAi construct did not affect Cdc42 expression (right panels). Quantification of blots from three separate experiments is shown (lower panels). B, hippocampal neurons were co-transfected with GFP and either empty pSUPER vector, scrambled RNAi, Cdc42 RNAi, or dominant negative Cdc42 (DN-Cdc42) and stained for SV2 (middle panels) and PSD-95 (right panels). Bar, 2 μm. C, quantification of the number of spines and synapses (SV2 and PSD-95 clusters) in neurons transfected with empty pSUPER vector, scrambled RNAi, Cdc42 RNAi, DN-Cdc42, or Cdc42 RNAi + GFP-WA is shown. Differences between control-Cdc42 RNAi, control-DN-Cdc42 (**, p < 0.0001) and Cdc42 RNAi-Cdc42RNAi + GFP-WA (*, p < 0.0001) were statistically significant. Error bars in A and C represent S.E. of three seperate experiments.
Arp3 Regulates the Formation of Spines and Synapses in Hippocampal Neurons—Because our results strongly implicated the Arp2/3 complex as a downstream effector of N-WASP in spine and synapse formation, we examined its subcellular localization in hippocampal neurons. GFP-tagged Arp3, one of the seven proteins found in the Arp2/3 complex, localized in puncta along neuronal processes with SV2 and PSD-95, suggesting that it was synaptic (Fig. 6A, arrows). A similar localization was observed by immunostaining for endogenous Arp3 and SV2 (Fig. 6A). Higher magnification images showed that Arp3 puncta were in close apposition to SV2 clusters and completely merged with PSD-95 puncta, indicating that Arp3, like N-WASP, is enriched on the postsynaptic side of excitatory synapses (Fig. 6A).
FIGURE 6.
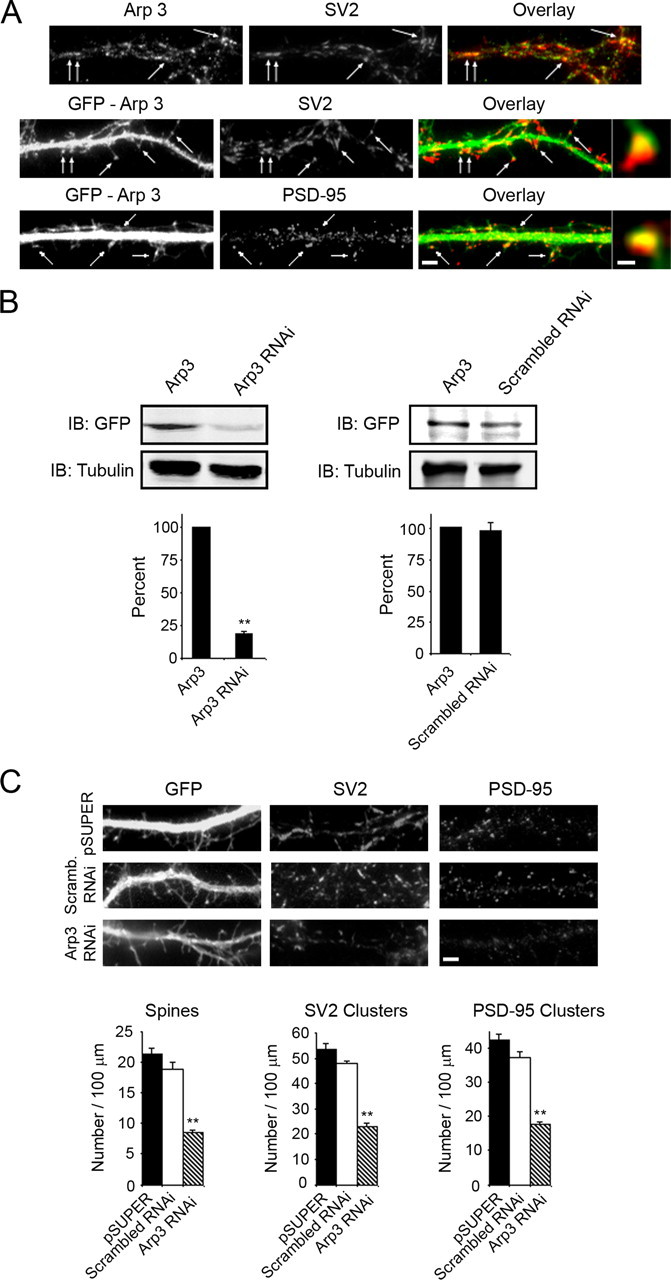
The Arp2/3 complex regulates the formation of dendritic spines and synapses in hippocampal neurons. A, hippocampal neurons at day 14 in culture were co-immunostained for endogenous Arp3 (upper panels) and the synaptic marker SV2. Endogenous Arp3 accumulated in puncta with SV2 (Overlay, arrows). Hippocampal neurons were transfected with GFP-Arp3 at day 5 in culture and then fixed and immunostained for SV2 (middle panels) and PSD-95 (lower panels) at day 12 in culture. GFP-Arp3 localized in puncta along neuronal processes with SV2 and PSD-95 (Overlays, arrows). Bar, 2 μm. High magnification images showed Arp3 puncta were in close apposition to SV2 and completely merged with PSD-95, indicating that Arp3 is enriched in the postsynaptic side of excitatory synapses. Bar, 0.4 μm. B, HEK-293T cells were co-transfected with Arp3 RNAi, scrambled RNAi, or empty pSUPER vector and GFP-Arp3. The lysates were blotted for GFP and α-tubulin. The Arp3 RNAi construct decreased expression of Arp3 by almost 80% (left panels) whereas the scrambled RNAi did not alter Arp3 expression (right panels). Quantification of blots from three separate experiments is shown (lower panels). C, hippocampal neurons were co-transfected with GFP and either empty pSUPER vector, scrambled RNAi, or the Arp3 RNAi construct at day 6 in culture and then fixed and stained at day 12 for the synaptic markers SV2 (middle panels) and PSD-95 (right panels). Bar, 2 μm. Quantification of the number of spines and synapses (SV2 and PSD-95 clusters) in neurons transfected with empty pSUPER vector, scrambled RNAi, or Arp3 RNAi (**, p < 0.0001) is shown. Error bars in B and C represent S.E. of three seperate experiments.
Because Arp3 localized to spines and excitatory synapses, we generated an RNAi construct to examine its function in regulating the formation of these structures. Although the RNAi sequence had been shown previously to effectively knock down expression of Arp3 (35), we tested its ability to knock down rat Arp3 by transfecting it into HEK-293T cells with GFP-tagged Arp3. The Arp3 RNAi construct decreased expression of Arp3 by almost 80% whereas a scrambled RNAi control did not affect Arp3 expression (Fig. 6B). When neurons were transfected with the Arp3 RNAi construct, a significant decrease in the number of spines and synapses was observed compared with control cultures expressing scrambled RNAi or empty pSUPER vector (Fig. 6C and supplemental Fig. S5). The decrease in the density of spines and synapses in neurons expressing the Arp3 RNAi construct was similar to that observed when N-WASP expression was knocked down in these cells. Thus, our results suggest that N-WASP and the Arp2/3 complex are important regulators in the formation of dendritic spines and synapses.
DISCUSSION
Our results reveal a novel mechanism by which N-WASP activation of the Arp2/3 complex regulates the formation of dendritic spines and synapses in hippocampal neurons. N-WASP is enriched in spines and excitatory synapses that are active and functional as determined by loading with FM4–64 dye. Knock down of endogenous N-WASP resulted in a significant decrease in the density of spines and synapses. The C-terminal region of the protein, which binds and activates the Arp2/3 complex, is critical for this function of N-WASP. Knock down of Arp3, an important protein in the Arp2/3 complex, caused a similar defect in the formation of spines and synapses, pointing to the significance of activation of the Arp2/3 complex in this process. An activator of N-WASP, Cdc42, is also involved in regulating spine and synapse formation. Knock down of Cdc42 expression resulted in a significant decrease in the number of spines and synapses and expression of the WA region of N-WASP rescued the Cdc42-mediated defect in spine and synapse formation, suggesting that activation of N-WASP by Cdc42 is important in this process. Thus, our study provides a molecular mechanism by which N-WASP and an activator and effector regulate the development of dendritic spines and synapses.
The function of Cdc42 in regulating the development of spines and synapses is presently unclear. Recent studies have indicated a role for Cdc42 in dendritic morphogenesis in Drosophila and learning-related synaptic growth in Aplysia sensory neurons (36, 37). In addition, activation of Cdc42 is proposed to be an important event in converting nonfunctional contacts into functional synapses (38). However, others have suggested that Cdc42 does not have a significant effect on the maintenance of dendritic spine morphology in hippocampal neurons (39, 40). Our results are consistent with Cdc42 regulating the formation of dendritic spines and synapses. The previous studies in hippocampal neurons relied on expression of constitutively active and dominant negative Cdc42 mutants after many spines and synapses had formed (day 10–12 in culture), whereas our study used RNAi to specifically knock down Cdc42 expression during a time when many spines and synapses are actively forming (day 5–6 in culture). It may be difficult to see a dramatic effect of Cdc42 on spines and synapses after a significant number of them have already formed.
Our results show that the spine and synaptic density in the WA-expressing neurons was slightly decreased compared with neurons expressing full-length N-WASP. It was reported previously that the WA region alone is much more potent in stimulating actin polymerization by the Arp2/3 complex than full-length N-WASP (8). Delocalized reorganization of actin may limit the amount of G-actin and Arp2/3 complex available for actin assembly at synaptic sites, which could diminish the ability of spines to stabilize and mature. Consistent with this, aberrant actin reorganization has been reported to result in a decrease in the density of spines and synapses in neurons (19, 41).
The process of spine formation has received a great deal of interest because of the importance of these structures in cognitive function, but the molecular mechanisms that regulate spine formation still remain poorly understood. Our results suggest that activation of the Arp2/3 complex by N-WASP is critical for the formation of dendritic spines and synapses. This raises the question of how the Arp2/3 complex functions in spine and synapse formation. Although the exact steps in spinogenesis are not clear, it appears that spines can either form from dendritic protrusions or extend directly from the dendritic shaft (42–46). In both cases, the spine head enlarges or expands as the spine matures, and the Arp2/3 complex-mediated branching of actin may play an important role in this process. When activation of the Arp2/3 complex by N-WASP is inhibited, the formation of dendritic spines is impaired. Activation of the Arp2/3 complex may form a branched actin network that promotes and supports the enlargement and maturation of the spine head. This branched actin network may also be important for the morphological changes in spines associated with synaptic plasticity. Future studies are needed to better understand the complex regulation of actin and its contribution to the formation and plasticity of spines and synapses.
In a proteomic analysis of rat brain, members of the Arp2/3 complex were shown to be present in the postsynaptic density (28). This is consistent with our results showing that the Arp2/3 complex localizes to the postsynaptic side of excitatory synapses where activation by N-WASP and Cdc42 supports enlargement of spine heads. Although the guanine nucleotide exchange factors (GEFs) that mediate activation of Cdc42 and, subsequently, branching of actin through the Arp2/3 complex are unknown, several Cdc42 GEFs could function in this capacity. Both α- and β-p-21-activated kinase-interacting exchange factor, which are Cdc42 and Rac GEFs, localize to the postsynaptic terminals of excitatory synapses where they regulate spine morphogenesis (19, 47, 48). Another Cdc42 GEF, intersectin, localizes to dendritic spines and plays a role in their development (49). Alternatively, other Cdc42 GEFs may contribute to this process.
In summary, we show that activation of N-WASP regulates the formation of dendritic spines and synapses through the Arp2/3 complex. The activation of the Arp2/3 complex could stimulate spine development by forming a branched actin network that promotes the enlargement of spine heads and leads to their maturation.
Supplementary Material
Acknowledgments
We thank Marc Kirschner, Michael Way, John Condeelis, Hiroaki Miki, Tadaomi Takenawa, and Alan Hall for generously providing reagents.
This work was supported, in whole or in part, by National Institutes of Health Grant MH071674 (to D. J. W.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
Footnotes
The abbreviations used are: WASP, Wiskott-Aldrich syndrome protein; N-WASP, neural WASP; GFP, green fluorescent protein; RNAi, RNA interference; HEK, human embryonic kidney; PBS, phosphate-buffered saline; GEF, guanine nucleotide exchange factor; TRITC, tetramethylrhodamine isothiocyanate.
References
- 1.Matus, A., Ackermann, M., Pehling, G., Byers, H. R., and Fujiwara, K. (1982) Proc. Natl. Acad. Sci. U. S. A. 79 7590-7594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fiala, J. C., Spacek, J., and Harris, K. M. (2002) Brain Res. Brain Res. Rev. 39 29-54 [DOI] [PubMed] [Google Scholar]
- 3.Fischer, M., Kaech, S., Knutti, D., and Matus, A. (1998) Neuron 20 847-854 [DOI] [PubMed] [Google Scholar]
- 4.Matus, A. (2000) Science 290 754-758 [DOI] [PubMed] [Google Scholar]
- 5.Dunaevsky, A., Tashiro, A., Majewska, A., Mason, C., and Yuste, R. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 13438-13443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris, K. M. (1999) Curr. Opin. Neurobiol. 9 343-348 [DOI] [PubMed] [Google Scholar]
- 7.Machesky, L. M., Mullins, R. D., Higgs, H. N., Kaiser, D. A., Blanchoin, L., May, R. C., Hall, M. E., and Pollard, T. D. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 3739-3744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rohatgi, R., Ma, L., Miki, H., Lopez, M., Kirchhausen, T., Takenawa, T., and Kirschner, M. W. (1999) Cell 97 221-231 [DOI] [PubMed] [Google Scholar]
- 9.Mullins, R. D., Heuser, J. A., and Pollard, T. D. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 6181-6186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bailly, M., Macaluso, F., Cammer, M., Chan, A., Segall, J. E., and Condeelis, J. S. (1999) J. Cell Biol. 145 331-345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blanchoin, L., Amann, K. J., Higgs, H. N., Marchand, J. B., Kaiser, D. A., and Pollard, T. D. (2000) Nature 404 1007-1011 [DOI] [PubMed] [Google Scholar]
- 12.Yamaguchi, H., Miki, H., Suetsugu, S., Ma, L., Kirschner, M. W., and Takenawa, T. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 12631-12636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miki, H., Miura, K., and Takenawa, T. (1996) EMBO J. 15 5326-5335 [PMC free article] [PubMed] [Google Scholar]
- 14.Yarar, D., To, W., Abo, A., and Welch, M. D. (1999) Curr. Biol. 9 555-558 [DOI] [PubMed] [Google Scholar]
- 15.Zhang, H., and Macara, I. G. (2008) Dev. Cell 14 216-226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saito, T., Jones, C. C., Huang, S., Czech, M. P., and Pilch, P. F. (2007) J. Biol. Chem. 282 32280-32287 [DOI] [PubMed] [Google Scholar]
- 17.Welch, M. D., Iwamatsu, A., and Mitchison, T. J. (1997) Nature 385 265-269 [DOI] [PubMed] [Google Scholar]
- 18.Goslin, K., Asmussen, H., and Banker, G. (1998) in Rat Hippocampal Neurons in Low-density Culture (Banker, G., and Goslin, K., eds) pp. 339-370, MIT Press, Cambridge, MA
- 19.Zhang, H., Webb, D. J., Asmussen, H., and Horwitz, A. F. (2003) J. Cell Biol. 161 131-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsuchiya, D., Kitamura, Y., Takata, K., Sugisaki, T., Taniguchi, T., Uemura, K., Miki, H., Takenawa, T., and Shimohama, S. (2006) Neurosci. Res. 56 459-469 [DOI] [PubMed] [Google Scholar]
- 21.Takenawa, T., and Suetsugu, S. (2007) Nat. Rev. Mol. Cell. Biol. 8 37-48 [DOI] [PubMed] [Google Scholar]
- 22.Moreau, V., Frischknecht, F., Reckmann, I., Vincentelli, R., Rabut, G., Stewart, D., and Way, M. (2000) Nat. Cell Biol. 2 441-448 [DOI] [PubMed] [Google Scholar]
- 23.Betz, W. J., Mao, F., and Smith, C. B. (1996) Curr. Opin. Neurobiol. 6 365-371 [DOI] [PubMed] [Google Scholar]
- 24.Betz, W. J., and Bewick, G. S. (1992) Science 255 200-203 [DOI] [PubMed] [Google Scholar]
- 25.Cochilla, A. J., Angleson, J. K., and Betz, W. J. (1999) Annu. Rev. Neurosci. 22 1-10 [DOI] [PubMed] [Google Scholar]
- 26.Betz, W. J., Mao, F., and Bewick, G. S. (1992) J. Neurosci. 12 363-375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryan, T. A., Reuter, H., Wendland, B., Schweizer, F. E., Tsien, R. W., and Smith, S. J. (1993) Neuron 11 713-724 [DOI] [PubMed] [Google Scholar]
- 28.Peterson, J. R., Bickford, L. C., Morgan, D., Kim, A. S., Ouerfelli, O., Kirschner, M. W., and Rosen, M. K. (2004) Nat. Struct. Mol. Biol. 11 747-755 [DOI] [PubMed] [Google Scholar]
- 29.Craig, A. M., Blackstone, C. D., Huganir, R. L., and Banker, G. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 12373-12377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamaguchi, H., Lorenz, M., Kempiak, S., Sarmiento, C., Coniglio, S., Symons, M., Segall, J., Eddy, R., Miki, H., Takenawa, T., and Condeelis, J. (2005) J. Cell Biol. 168 441-452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kempiak, S. J., Yamaguchi, H., Sarmiento, C., Sidani, M., Ghosh, M., Eddy, R. J., Desmarais, V., Way, M., Condeelis, J., and Segall, J. E. (2005) J. Biol. Chem. 280 5836-5842 [DOI] [PubMed] [Google Scholar]
- 32.Kawamura, K., Takano, K., Suetsugu, S., Kurisu, S., Yamazaki, D., Miki, H., Takenawa, T., and Endo, T. (2004) J. Biol. Chem. 279 54862-54871 [DOI] [PubMed] [Google Scholar]
- 33.Allison, D. W., Gelfand, V. I., Spector, I., and Craig, A. M. (1998) J. Neurosci. 18 2423-2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Symons, M., Derry, J. M., Karlak, B., Jiang, S., Lemahieu, V., McCormick, F., Francke, U., and Abo, A. (1996) Cell 84 723-734 [DOI] [PubMed] [Google Scholar]
- 35.Steffen, A., Faix, J., Resch, G. P., Linkner, J., Wehland, J., Small, J. V., Rottner, K., and Stradal, T. E. (2006) Mol. Biol. Cell 17 2581-2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scott, E. K., Reuter, J. E., and Luo, L. (2003) J. Neurosci. 23 3118-3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Udo, H., Jin, I., Kim, J. H., Li, H. L., Youn, T., Hawkins, R. D., Kandel, E. R., and Bailey, C. H. (2005) Neuron 45 887-901 [DOI] [PubMed] [Google Scholar]
- 38.Shen, W., Wu, B., Zhang, Z., Dou, Y., Rao, Z. R., Chen, Y. R., and Duan, S. (2006) Neuron 50 401-414 [DOI] [PubMed] [Google Scholar]
- 39.Tashiro, A., Minden, A., and Yuste, R. (2000) Cereb. Cortex 10 927-938 [DOI] [PubMed] [Google Scholar]
- 40.Govek, E. E., Newey, S. E., Akerman, C. J., Cross, J. R., Van der Veken, L., and Van Aelst, L. (2004) Nat. Neurosci. 7 364-372 [DOI] [PubMed] [Google Scholar]
- 41.Zhang, H., Webb, D. J., Asmussen, H., Niu, S., and Horwitz, A. F. (2005) J. Neurosci. 25 3379-3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dailey, M. E., and Smith, S. J. (1996) J. Neurosci. 16 2983-2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ziv, N. E., and Smith, S. J. (1996) Neuron 17 91-102 [DOI] [PubMed] [Google Scholar]
- 44.Fiala, J. C., Feinberg, M., Popov, V., and Harris, K. M. (1998) J. Neurosci. 18 8900-8911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ethell, I. M., and Pasquale, E. B. (2005) Prog. Neurobiol. (Oxf.) 75 161-205 [DOI] [PubMed] [Google Scholar]
- 46.Marrs, G. S., Green, S. H., and Dailey, M. E. (2001) Nat. Neurosci. 4 1006-1013 [DOI] [PubMed] [Google Scholar]
- 47.Node-Langlois, R., Muller, D., and Boda, B. (2006) J. Cell Sci. 119 4986-4993 [DOI] [PubMed] [Google Scholar]
- 48.Park, E., Na, M., Choi, J., Kim, S., Lee, J. R., Yoon, J., Park, D., Sheng, M., and Kim, E. (2003) J. Biol. Chem. 278 19220-19229 [DOI] [PubMed] [Google Scholar]
- 49.Nishimura, T., Yamaguchi, T., Tokunaga, A., Hara, A., Hamaguchi, T., Kato, K., Iwamatsu, A., Okano, H., and Kaibuchi, K. (2006) Mol. Biol. Cell 17 1273-1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.