Abstract
The molecular mechanisms underlying differentiation of hematopoietic stem cells into megakaryocytes are poorly understood. Tumor suppressor protein p53 can act as a transcription factor affecting both cell cycle control and apoptosis, and we have previously shown that p53 is activated during terminal megakaryocytic (Mk) differentiation of the CHRF-288-11 (CHRF) cell line. Here, we use RNA interference to reduce p53 expression in CHRF cells and show that reduced p53 activity leads to a greater fraction of polyploid cells, higher mean and maximum ploidy, accelerated DNA synthesis, and delayed apoptosis and cell death upon phorbol 12-myristate 13-acetate-induced Mk differentiation. In contrast, reduced p53 expression did not affect the ploidy or DNA synthesis of CHRF cells in the absence of phorbol 12-myristate 13-acetate stimulation. Furthermore, primary Mk cells from cultures initiated with p53-null mouse bone marrow mononuclear cells displayed higher ploidy compared with wild-type controls. Quantitative reverse transcription-PCR analysis of p53-knockdown CHRF cells, compared with the “scrambled” control CHRF cells, revealed that six known transcriptional targets of p53 (BBC3, BAX, TP53I3, TP53INP1, MDM2, and P21) were down-regulated, whereas BCL2 expression, which is known to be negatively affected by p53, was up-regulated. These studies show that the functional role of the intrinsic activation of p53 during Mk differentiation is to control polyploidization and the transition to endomitosis by impeding cell cycling and promoting apoptosis.
p53 is a widely recognized tumor suppressor protein, which responds to DNA damage by inhibiting cell cycle progression and/or initiating apoptosis. Due to its importance in maintaining genomic stability, p53 is reportedly mutated in over half of human cancers, with the highest prevalence in ovarian, colorectal, and esophageal cancers (1). In addition, p53 has been found to be mutated or deleted in many cancer-derived cell lines (1).
Despite the seemingly normal development of p53–/– mice, mounting evidence suggests that p53 is involved in regulating differentiation, including that in the hematopoietic system (see the recent review by Stiewe (2)). For example, in the lymphoid compartment, several studies have linked p53 to regulation of B-cell differentiation (reviewed by Almog and Rotter (3)). It has also been shown that p53 mediates a key checkpoint in the differentiation of thymocytes from CD4–CD8– to CD4+CD8+ (4). Wild-type p53 expression has also been shown to positively regulate myeloid differentiation of HL-60 (5, 6) and 32D (7) cells and erythrocytic differentiation of K562 cells (8) and Friend erythroleukemia cells (9).
Relatively few studies have examined the role of p53 in megakaryocytic (Mk)4 differentiation and maturation. This is despite the fact that polyploidization and apoptosis, two integral components of Mk differentiation, are commonly associated with p53 in other cell types (10, 11) and that p53 is known to be involved in mediating the so-called tetraploidy checkpoint (12). This raises some interesting questions. What is the role of p53, sometimes referred to as the “guardian of the genome” (13), in cells that inherently undergo multiple rounds of DNA synthesis without cytokinesis? What role might p53 have in governing the constitutive program of apoptosis in such polyploid cells?
Studies in p53-null mice have not reported abnormalities in platelet levels or megakaryopoiesis, although neither has been thoroughly studied. One study that did explicitly examine platelet levels found basal levels to be equivalent between p53–/– and p53+/+ mice (14). However, systemic compensatory mechanisms could mask the effect of p53 on individual steps in Mk differentiation and maturation in steady-state animals. Some evidence of this came from Wlodarski et al. (15), who reported faster recovery of colony-forming Mk progenitors after 5-fluorouracil treatment in p53–/– versus p53+/+ mice.
Other studies have examined p53 using in vitro Mk systems. For example, it has been shown that p53 expression promoted erythroid but not Mk differentiation of the bipotential K562 cell line (16). Furthermore, antisense knockdown of p53 increased Mk colony formation in the presence of interleukin-6, interleukin-3, and erythropoietin in cultures of human peripheral blood mononuclear cells (17). In the human M07e cell line, signaling from c-Mpl, the receptor for the principal Mk cytokine thrombopoietin (Tpo), has been shown to result in a conformational shift in p53 from an antiproliferative to proproliferative state (18). These results implicate p53 in Mk lineage commitment and progenitor expansion.
There have been conflicting reports about the level of p53 expression in maturing endomitotic Mk cells. Datta et al. reported a decrease in total p53 level and a decrease in p53 associated with the CDK-activating kinase complex in phorbol 12-myristate 13-acetate (PMA)-stimulated human erythroleukemia cells (19). In contrast, Baccini et al. (20) reported high expression of p53 in both diploid and polyploid primary human Mk cells.
Recently, we have reported patterns of gene and protein expression in both PMA-stimulated megakaryoblastic CHRF-288-11 (CHRF) cells and Mk-directed primary human mPB CD34+ cell cultures that suggested an increase in p53 activity (21). We initially validated this finding in the CHRF cell line model by demonstrating an increase in p53-DNA binding activity during terminal differentiation (21). Motivated by these preliminary findings, we sought to more rigorously assess the functional role of p53 in terminal Mk differentiation.
EXPERIMENTAL PROCEDURES
All materials were obtained from Sigma unless otherwise noted.
Sequencing of Genomic p53—Genomic DNA was isolated from unstimulated parental CHRF cells using the DNeasy tissue kit (Qiagen, Valencia, CA), following the prescribed protocol for cultured cells. PCR primers were designed using Vector NTI (Invitrogen) to amplify p53 exons 2–10 and their adjacent intronic regions (supplemental Table S1). PCRs were performed using AmpliTaq Gold (Applied Biosystems, Foster City, CA) according to the manufacturer's recommendations with the following parameters: 94 °C for 5 min followed by 35 cycles of 94 °C for 30 s, annealing for 30 s (see Table S1 for temperatures), and 72 °C for 1 min. After a final elongation step at 72 °C for 7 min and column purification (PCR purification kit; Qiagen) PCR products were submitted for sequencing to the Northwestern University Center for Genetic Medicine using the ABI Big Dye version 3.1 and ABI 3730 DNA sequencer (Applied Biosystems).
Design and Production of Lentiviruses Encoding Micro-RNA-adapted shRNAs—Lentiviral vectors for the delivery of micro-RNA-adapted shRNAs (shRNA-mir) were designed and produced using the reagents and protocols included in the BLOCK-iT Lentiviral Pol II miR RNA interference expression system (Invitrogen). Single-stranded DNA oligonucleotides (premicro-RNA inserts) were designed to target the coding regions within the gene of interest using the Invitrogen RNA interference designer. To control against off-target silencing effects, independent premicro-RNA inserts were designed to complement regions at least 200 bases apart in the p53 mRNA. In addition, a scrambled control premicro-RNA, which was designed to not bind any known vertebrate gene, was used as a negative control. The premicro-RNA sequences are provided in Table 1. The premicro-RNA and its reverse complement were annealed and ligated into the pcDNA6.2-GW/EmGFP-miR vector, which contains the full premicro-RNA 5′- and 3′-flanking regions, as well as the co-cistronic Emerald GFP (EmGFP) gene. After sequence verification, the EmGFP-premicro-RNA cassette was transferred to the pLenti6/V5 expression construct using BP/LR recombination reactions. Three micrograms of the resulting pLenti6/EmGFP-premicro-RNA vector, together with 9 μg of ViraPower Packaging mix (a mixture of three plasmids encoding the additional components necessary for virus production), was transfected using Lipofectamine 2000 into 293FT producer cells growing in Dulbecco's modified Eagle's medium with 10% non-heat-inactivated fetal bovine serum (HyClone; Logan, UT), 2 mm l-glutamine, 0.1 mm minimal essential medium nonessential amino acids, and 1 mm minimal essential medium sodium pyruvate. After overnight culture, medium was exchanged to remove transfection reagents. The following day, virus stocks were harvested, centrifuged at 1800 × g for 5 min at 4 °C, and filtered through a 0.45-μm polyvinylidene difluoride syringe filter. Typical virus titers, as determined by serial dilution with HT1080 cells and flow cytometric measurement of EmGFP expression, were ∼1 × 106 transduction units/ml.
TABLE 1.
Premicro-RNA insert sequences used for construction of lentiviral shRNA-mir vectors
The mature micro-RNA sequence, which is the reverse complement of the target sequence in the target mRNA, is highlighted in boldface type.
| Gene | shRNA-mir | Premicro-RNA insert sequence |
|---|---|---|
| P53 | P53-A | 5′-TGCTGTCAAATCATCCATTGCTTGGGGTTTT GGCCACTGACTGACCCCAAGCAGGATGATTTGA-3′ |
| P53 | P53-B | 5′-TGCTGAATGCAAGAAGCCCAGACGGAGTTTT GGCCACTGACTGACTCCGTCTGCTTCTTGCATT-3′ |
| Scrambled control | Neg-A | 5′-TGCTGAAATGTACTGCGCGTGGAGACGTTTT GGCCACTGACTGACGTCTCCACGCAGTACATTT-3′ |
Transduction of CHRF Cells—CHRF cells (kindly provided by Dr. R. Smith, National Institutes of Health, Bethesda, MD) were cultured in Iscove's modified Dulbecco's medium plus 10% heat-inactivated fetal bovine serum at 37 °C in a fully humidified incubator under 5% CO2 and 95% air. For lentiviral transduction of CHRF cells, 0.5 × 106 CHRF cells were resuspended in 0.5 ml of normal CHRF growth medium and mixed with 0.5 ml of virus stock (multiplicity of infection ∼1). Polybrene was added to a final concentration of 6 μg/ml. After overnight incubation, cells were diluted 3-fold with growth medium and thereafter maintained between 50,000 and 1 × 106 cells/ml. Three to 5 days post-transduction, EmGFP+ cells (typically 20–50% of the total cells) were purified by flow cytometric cell sorting (MoFlo high performance cell sorter; Dako, Fort Collins, CO), resulting in stably transduced subclones (typically >90% EmGFP+).
Induction of Mk Differentiation—For induction of Mk differentiation, on day 0, CHRF cells were seeded at 50,000–75,000 cells/ml and treated with 10 ng/ml PMA in DMSO. As previously reported (21), cells ceased expanding and rapidly adhered to the tissue culture flask upon PMA treatment. At the designated time points, cells were washed with PBS and harvested using either 1× trypsin-EDTA (for apoptosis, ploidy, and Q-RT-PCR analyses) or 1 mm EDTA in PBS (for Western blot or p53-DNA binding enzyme-linked immunosorbant assay (ELISA) or electrophoretic mobility shift assay (EMSA)). Adherent and nonadherent fractions were combined for all analyses.
Isolation and Culture of Mouse Cells—Bone marrow cells were isolated from male 8–9-week-old p53–/– (B6.129S2-Trp53tm1Tyj) mice and age-matched control normal C57BL/6J mice (Jackson Laboratories; Bar Harbor, ME) with approval from the Northwestern University Animal Care and Use Committee. Mice were sacrificed using CO2 and cervical dislocation, and the femurs were isolated. Cells from the bone marrow were collected by flushing the bones with Hanks' balanced salt solution containing 100 IU/ml penicillin and 100 μg/ml streptomycin using a syringe and a 21-gauge needle until the bones appeared white. Bone marrow cells were washed with 2 mm EDTA in PBS and incubated in ACK buffer (0.15 m NH4Cl, 10 mm KHCO3, 0.1 mm Na2EDTA, pH 7.2–7.4) for red cell lysis for 10 min at 4 °C. Next, cells were resuspended in PBS with 2 mm EDTA and passed through a 30-μm preseparation filter (Miltenyi Biotech; Auburn, CA) to remove bone fragments and cell clumps. Cells were seeded in culture flasks at a concentration of 1 × 106 cells/ml in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (HyClone, Logan, UT), 100 IU/ml penicillin, 100 μg/ml streptomycin, and 100 ng/ml human Tpo (Genentech, S. San Francisco, CA). For the histological examination of resident Mk cells in the marrow, sternum bones were isolated from these animals, fixed in 10% formalin, embedded in paraffin, and sectioned using standard protocols.
Flow Cytometric Analysis of Ploidy, DNA Synthesis, and Apoptosis—Flow cytometry data were acquired using LSRII and FACSAria flow cytometers with FACSDiva software (BD Biosciences). Ploidy and apoptosis assays were performed using previously published methods with minor modifications (22, 23). For analysis of murine cell cultures, Mk cells were first labeled with fluorescein isothiocyanate-conjugated rat anti-mouse CD41 IgG1 (BD Biosciences). For ploidy analysis of CHRF and murine Mk cultures, cells were fixed for 15 min at room temperature in 0.5% paraformaldehyde in PBS and permeabilized for 1 h at 4°C in 70% methanol. RNA was digested by 15 min of RNase treatment at 37 °C, and DNA was stained with 50μg/ml propidium iodide in PBS at room temperature. Polyploid CHRF cells are those EmGFP+ events with ≥8 n DNA content. Polyploid murine Mk cells were those events with high forward scatter, positive CD41 expression, and ≥8 n DNA content.
DNA synthesis was assessed using a flow cytometric BrdUrd incorporation assay performed as described by the manufacturer (BD Biosciences). Three different protocols were employed, and they all lead to similar results. Briefly, CHRF cells were incubated with the 10 μm BrdUrd for 12 h and then fixed, permeabilized, treated with DNase to expose incorporated BrdUrd epitopes, incubated with an allophycocyanin (APC)-conjugated anti-BrdUrd antibody, and stained with propidium iodide to measure DNA content (protocol 1) or treated with RNase and then stained with propidium iodide (protocol 2). In a variation of the manufacturer's protocol, where DNA synthesis, ploidy, and apoptosis were assessed simultaneously, the cells were intracellularly labeled with APC-conjugated anti-BrdUrd and PE-conjugated-anti-cleaved caspase-3 antibodies and then treated with RNase and finally counterstained with 7-AAD (protocol 3).
In a separate set of experiments, the active caspase-3 apoptosis kit was employed (BD Biosciences) for caspase-3-based measurement of apoptosis. CHRF cells were fixed and permeabilized and then incubated with a PE-conjugated antibody against active (cleaved) caspase-3 and finally counterstained with TO-PRO-3 (Molecular Probes, Inc., Eugene, OR) to exclude debris. For Annexin V-based apoptosis and viability analysis, CHRF cells were stained for 15 min at room temperature with PE-conjugated Annexin V (AnV) and 7AAD in a calcium-binding buffer to promote AnV binding (Invitrogen). Cells were gated based on forward scatter, side scatter, and EmGFP expression. Apoptosis is reported as the percentage AnV+ cells among all 7AAD– cells.
Quantitative Reverse Transcription-PCR—Q-RT-PCR was performed using the High-Capacity cDNA Archive kit and Assays-on-Demand Taqman kit following the manufacturer's protocols with minor modifications (Applied Biosystems). The Applied Biosystems primer sets used are provided in Table S2. PCRs were scaled down to 25 μl and performed on a Bio-Rad iCycler. A serial dilution of a reference sample (an equal mass mixture of all samples to be tested) was used to verify linearity of the assay. Cycle threshold values were determined and converted to starting mass units (SMU) using the iCycler software. The SMU for each sample was normalized using the average SMU of two housekeeping genes (glucuronidase-β and large ribosomal protein P0) for that sample, as recommended by Applied Biosystems. Normalized SMU were then compared between samples for any given gene.
Western Blot Analysis for p53—For Western blot analysis of p53, cell pellets were resuspended in lysis buffer (Cell Signaling; Danvers, MA) supplemented with 0.1% SDS and 3.4 units/μl aprotinin, vortexed for 30 min at 4 °C, and then centrifuged at 14,000 × g for 30 min to pellet cellular debris. After quantifying protein by a bicinchoninic acid assay (BCA; Pierce), 50 μgof protein was loaded per lane into precast 12% SDS-PAGE ReadyGel (Bio-Rad) and electrophoresed for 1.5 h at 100 V. Proteins were transferred to Bio-Rad polyvinylidene difluoride membranes (1 h, 100 V), blocked with nonfat dry milk, incubated overnight at 4 °C with mouse anti-human-p53 monoclonal antibody (DO-1; Santa Cruz Biotechnology, Inc., Santa Cruz, CA), and then incubated with horseradish peroxidase-conjugated goat anti-mouse-IgG antibody (1 h). After incubation with ECL+ detection reagent (Amersham Biosciences), the chemifluorescent signal was detected using a Storm 860 fluorimager (Amersham Biosciences). Densitometry analysis was performed using local background subtraction and total summed density options within ImageQuant (GE Healthcare).
ELISA-based p53-DNA Binding Activity Assay—DNA-binding activity of p53 was assessed by two assays. The ELISA-based assay used the TransBinding p53 ELISA kit (Panomics, Fremont, CA), following the manufacturer's instructions. Briefly, nuclear extracts were prepared with the Nuclear Extraction kit (Panomics) using 100 μl of nuclear lysis buffer for the cells harvested from one T-150 flask (∼3.5 × 106 starting cells). Protein yields were measured using the Pierce BCA assay per the manufacturer's instructions. Then, for each sample, 4 μgof nuclear extract was incubated with biotinylated p53 consensus-binding sequence oligonucleotides. After immobilization on streptavidin-coated 96-well plates, complexes were detected using a primary anti-p53 antibody and a secondary antibody conjugated to horseradish peroxidase. Samples were tested in duplicate, and assay specificity was verified by preincubation of lysates with nonbiotinylated consensus sequence oligonucleotides, which resulted in an 80–95% reduction in background-subtracted signals.
EMSA-based p53-DNA Binding Activity Assay—The EMSA was performed using the p53 EMSA kit (Panomics) following the manufacturer's instructions. The nuclear extracts used in these experiments were prepared, and protein yields were quantified as described above. In each sample, 10 μg of nuclear extract proteins were incubated with biotinylated p53 consensus binding sequence oligonucleotides; some samples were preincubated with unlabeled consensus binding sequence oligonucleotides to verify band specificity. Separation of bound and free probe was achieved by electrophoresis using a 6.0% nondenaturing polyacrylamide gel, and the protein-DNA complexes were transferred to a Biodyne B nylon membrane (Pall, East Hills, NY). Protein-DNA complexes were detected using streptavidin-horseradish peroxidase and visualized after exposure to Hyperfilm ECL film (Amersham Biosciences). Each experiment consisted of two biological replicates and was repeated at least twice. Densitometry analysis was performed on the region encompassing bands specific to our p53 consensus binding sequence oligonucleotides using ImageJ version 1.38 software (available on the World Wide Web).
Statistical Analysis—Unless otherwise noted, tests for statistical significance were performed using Student's t test for independent samples applied to individual time points.
RESULTS
CHRF Cells Have the Wild-type p53 Gene—The p53 gene is commonly mutated in leukemic cell lines (24), but the p53 status of CHRF cells has not, to our knowledge, been reported. We sequenced exons 2–10 of p53 and their surrounding intronic boundaries in genomic DNA isolated from CHRF cells. The sequences matched those deposited in NCBI (NC_0000017.9) except that the CHRF cells encode the P72R polymorphism, which is the more common form in persons of European descent (1). These data coupled with our previous comparisons of CHRF cells and primary hematopoietic stem and progenitor cell (HSPC) culture-derived Mk cells, in which we showed that the PMA-stimulated CHRF cells recapitulated many of the transcriptional changes exhibited by the differentiating primary Mk cells (21), reinforce the use of CHRF cells as a model system to study Mk differentiation and the role of p53 therein.
Lentiviral Delivery of p53-targeting shRNA-mir Effectively Decreases p53 Expression and Transactivation Activity—Recent studies have shown that high efficiency RNA interference can be accomplished by overexpressing an exogenous primary micro-RNA that has been engineered to encode a 21-mer sequence that perfectly complements a segment of the gene targeted for knockdown (shRNA-mir) (25–27). Replication-incompetent lentiviruses were used to stably express shRNA-mir in CHRF cells. Two independent targeting sequences (p53-A and p53-B) were designed to control for potential off-target silencing effects. In addition, a scrambled control shRNA-mir sequence (neg-A) that was designed to not target any known vertebrate gene was employed as a negative control. Once integrated in the infected CHRF cells, the shRNA-mir is co-cistronically expressed with EmGFP under the control of the human cytomegalovirus immediate early promoter. EmGFP+ cells were selected by FACS sorting to obtain stably transduced CHRF subclones (referred to hereafter as CHRF-p53-A, CHRF-neg-A, etc.).
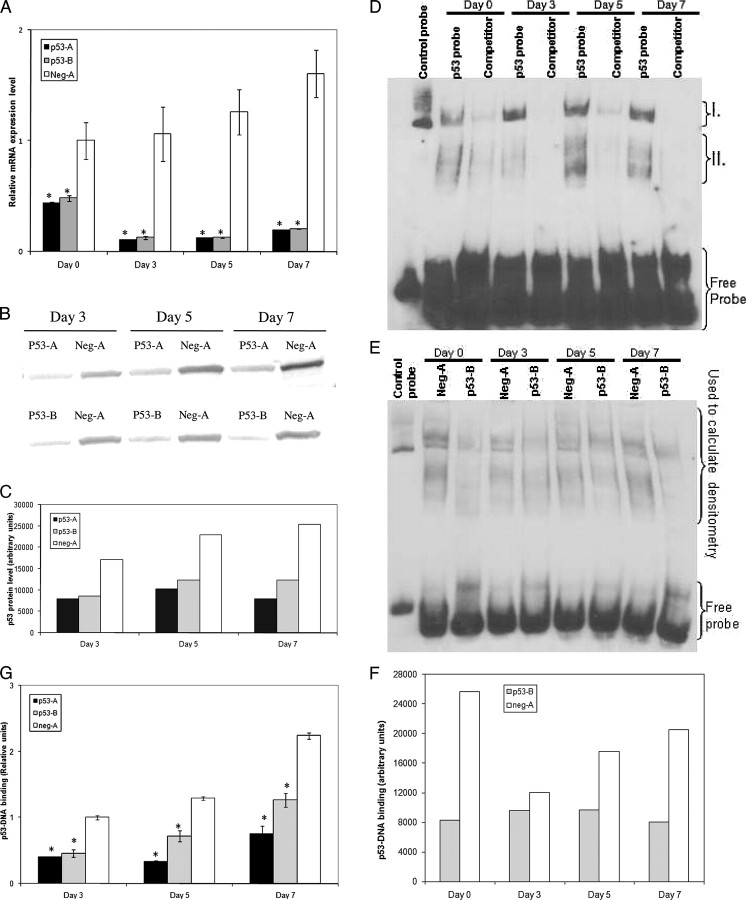
The efficiency of p53 mRNA level knockdown was verified by Q-RT-PCR in CHRF-p53-A and CHRF-p53-B cells compared with CHRF-neg-A cells both in their normal growth state and after PMA stimulation (Fig. 1A). Both targeting sequences effectively decreased p53 mRNA expression by 50–91% (p < 0.01 at all time points). We also examined p53 protein expression by Western blots in PMA-treated CHRF-p53-A, CHRF-p53-B, and CHRF-neg-A cells (Fig. 1, B and C). Protein levels of p53 increased slightly in the CHRF-neg-A cells over the time course of PMA stimulation. However, expression was significantly lower in both the CHRF-p53-A and CHRF-p53-B cells when compared with the scrambled micro-RNA-expressing control cultures.
FIGURE 1.
Expression of p53-A and p53-B shRNA-mir effectively decreases p53 expression and activity. A, CHRF cells stably expressing p53-A (black), p53-B (gray), or neg-A (white) shRNA-mir were analyzed by Q-RT-PCR for p53 mRNA abundance. The p53 mRNA expression data were normalized to those of the CHRF-neg-A unstimulated controls. Error bars, S.E. (n = 3–6). B, Western blot analysis of p53 in whole cell lysates from CHRF cells stably expressing the given shRNA-mir at the designated time points after PMA stimulation. C, densitometry of the Western blot in B shown with arbitrary units. D, EMSA of nuclear extracts from CHRF-neg-A cells using biotinylated p53 consensus binding sequence oligonucleotides. Regions I and II (discussed under “Results”) represent low and high mobility p53-DNA complexes, respectively. Competitor lanes were preincubated with unlabeled p53 consensus binding sequence oligonucleotides to verify band specificity. E, EMSA of differentiating CHRF-neg-A and CHRF-p53-B nuclear extracts incubated with biotinylated p53 consensus binding sequence oligonucleotides. F, densitometry of the indicated region from EMSA in E, shown with arbitrary units. G, the binding of p53 protein from nuclear lysates to p53 consensus-binding sequence DNA oligonucleotides was measured by ELISA as described under “Experimental Procedures.” Binding level is shown in arbitrary units normalized to CHRF-neg-A cells from day 3. Error bars, S.E. (n = 2–4). A and G, an asterisk denotes statistically significant difference versus corresponding neg-A-expressing CHRF cell sample from the same time point. A, p < 0.01; G, p < 0.05.
Because some residual p53 protein is expressed in the CHRF-p53-A and CHRF-p53-B cells, we performed EMSAs to verify that p53 binding of its consensus DNA sequence was indeed reduced in the p53 knockdown CHRF cells and to investigate p53 binding activity upon PMA-induced differentiation. Our experiments (n = 3) identified a low mobility DNA-protein complex that increased during CHRF cell differentiation (Fig. 1D, region I) as well as a region of higher mobility complexes (Fig. 1D, region II). Both regions I and II were used in subsequent analyses, because the binding affinity of both regions varied during the course of CHRF cell differentiation and were specific to the p53 consensus binding sequence oligonucleotides used during our experiments. As we have previously reported for the parental CHRF cells (21), p53-DNA binding activity increased along the course of PMA-induced differentiation of CHRF-neg-A cells (Fig. 1, E and F). Additionally, we demonstrated reduced p53-DNA binding in both CHRF-p53-A (data not shown) and CHRF-p53-B cells (Fig. 1, E and F) upon PMA-induced CHRF cell differentiation. Furthermore, we employed a ELISA-based DNA-binding assay in an independent effort (using a separate set of experiments and nuclear extracts) to assess p53 transactivation potential upon PMA-induced CHRF cell differentiation (Fig. 1G). This assay has been assessed to be analogous to the traditional EMSA in that it measures the ability of p53 from nuclear lysates to bind to a p53 consensus-binding sequence and has been validated extensively (28). As expected, the data from this assay (Fig. 1G) show that when compared with CHRF-neg-A controls at the same time points after PMA stimulation, transactivation potential was reduced by 60–74% and 43–55% in the CHRF-p53-A and CHRF-p53-B cells, respectively (p < 0.05 at all time points). These results are in agreement with the EMSA data and verify that the p53 knockdown CHRF cells have reduced, but still detectable, levels of p53 transactivation activity.
Knockdown of p53 Increases Polyploidization of PMA-stimulated CHRF Cells—We next examined the effects of p53-A and p53-B micro-RNA expression on Mk differentiation after PMA stimulation. We and others have previously reported that, in response to PMA stimulation, CHRF cells stop expanding, increase in size, undergo polyploidization, extend proplatelet-like processes, and ultimately succumb to apoptosis (21, 29). Based on its known involvement in cell cycle regulation as well as stress and DNA-damage-induced apoptosis, we hypothesized that knockdown of p53 would affect the ploidy and cell cycle kinetics of CHRF cells when stimulated to differentiate into megakaryocytes. To examine this, we treated the shRNA-mir-expressing CHRF cells with 10 ng/ml PMA and measured ploidy, apoptosis, and viability as the cells differentiated over the 7–9-day cultures. Replicate experiments were conducted from separate CHRF cell transductions to control for possible clonal variations.
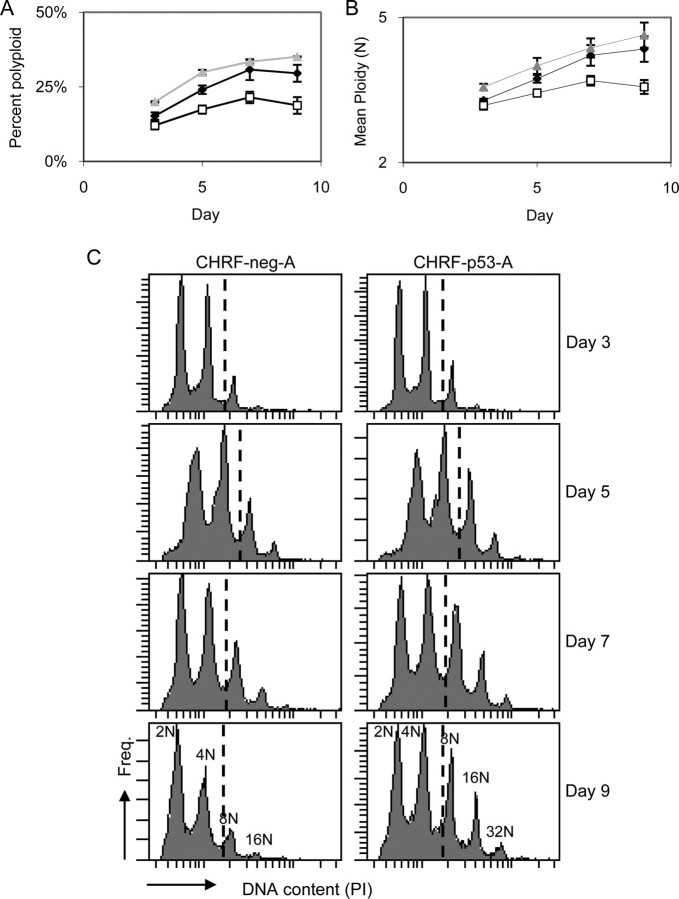
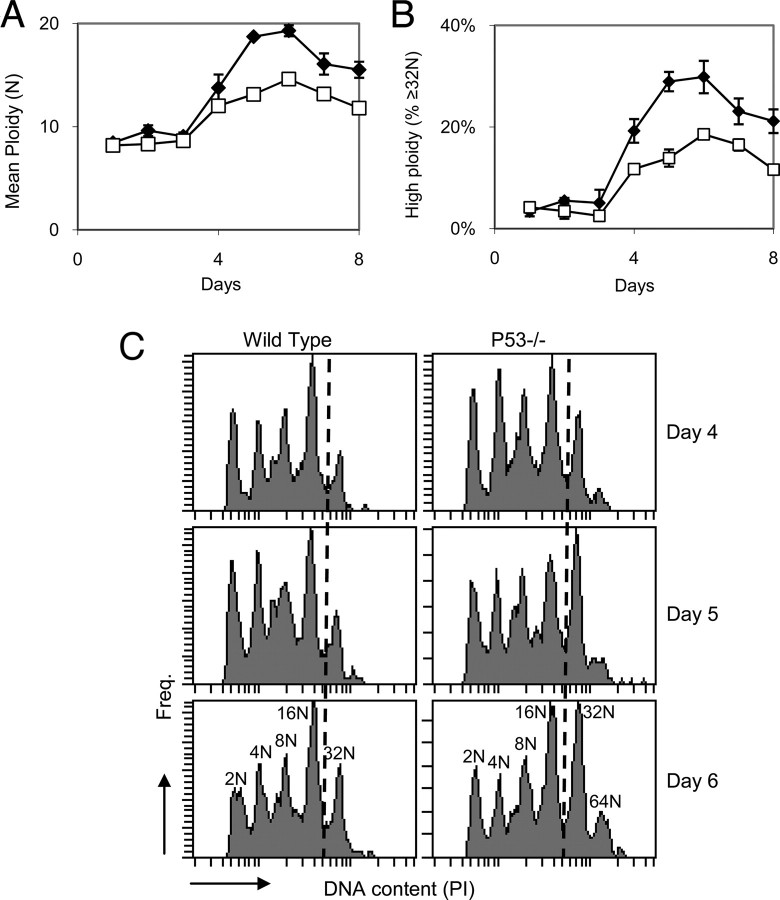
Indeed, upon PMA treatment, the fraction of CHRF-p53-A and CHRF-p53-B cells that reached high ploidy (≥8 n) increased by 26–57% and 27–43%, respectively, over 9-day cultures when compared with the control CHRF-neg-A cells (Fig. 2A; p < 0.05). In addition, knockdown of p53 expression resulted in an increase in mean ploidy upon PMA stimulation (Fig. 2B; p < 0.05). The ploidy distributions were clearly shifted toward higher ploidy in the p53 knockdown CHRF cell cultures, and the p53 knockdown cells reached higher ploidy classes than the control cells at the later time points (Fig. 2C). These data show that reducing p53 activity during terminal CHRF cell megakaryopoiesis results in greater overall polyploidization.
FIGURE 2.
Knockdown of p53 expression in CHRF cells leads to increased polyploidization. CHRF-neg-A (open squares), CHRF-p53-A (black diamonds), and CHRF-p53-B (gray triangles) cells were treated with PMA to induce Mk differentiation and evaluated by flow cytometry for DNA content. The percentage of high ploidy (≥8 n) cells among all cells (A) and the mean DNA content per cell (B) are shown for each condition. Error bars, S.E. (n = 2–6). C, DNA histograms from representative samples at the designated time points. The dashed lines indicate the ≥8 n polyploid cells.
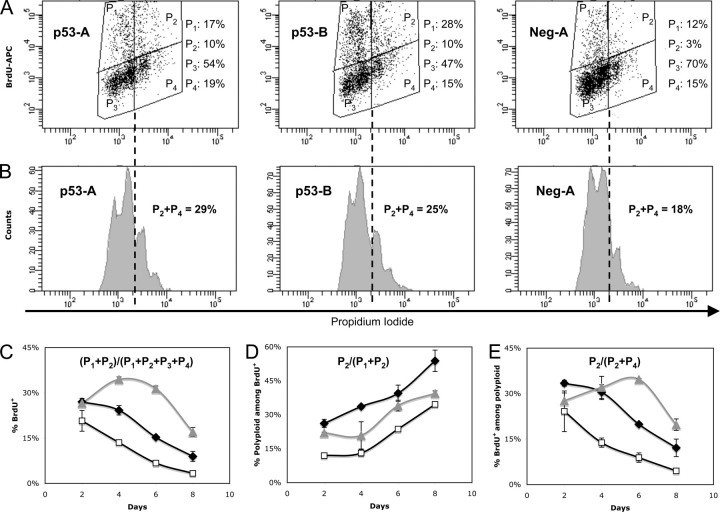
p53 Down-regulation Leads to Increased DNA Synthesis in PMA-stimulated CHRF Cells—In order to examine the role of p53 down-regulation on the cell cycle of PMA-stimulated CHRF cells, we employed a flow cytometric BrdUrd incorporation assay performed on PMA-stimulated CHRF-p53-A, CHRF-p53-B, and CHRF-neg-A control cells (Fig. 3). Silencing of p53 led to increased DNA synthesis in PMA-stimulated CHRF-p53-A and CHRF-p53-B cells between days 4 and 8 after PMA treatment. We recorded differences of 79–127% (p < 0.05) or 155–365% (p < 0.05) on days 4 and 6 and up to 167 or 409% on day 8 between CHRF-p53-A and CHRF-neg-A or CHRF-p53-B and CHRF-neg-A cells, respectively (Fig. 3C). Furthermore, and most interestingly, we noted that among the actively cycling cells, which incorporate BrdUrd, the split between endomitotic (cells in the P2 window) and mitotic cells (those in the P1 window) was generally higher in the stimulated p53-deficient than in the CHRF-neg-A cells (Fig. 3D). Additionally, a closer examination of the polyploid cells (≥8 n) revealed that DNA synthesis was significantly higher in the p53-deficient cells than the CHRF-neg-A cells. We noted differences of 120–123% (p < 0.05) or 132–290% (p < 0.05) on days 4 and 6 and up to 168 or 336% on day 8 between CHRF-p53-A and CHRF-neg-A or CHRF-p53-B and CHRF-neg-A cells, respectively (Fig. 3E). Based on these data, we conclude that reduced p53 activity leads to acceleration in DNA synthesis and promotes the transition from mitosis to endomitosis.
FIGURE 3.
Knockdown of p53 expression increases DNA synthesis in CHRF cells undergoing terminal Mk differentiation. CHRF-p53-A (black diamonds), CHRF-p53-B (gray triangles), and CHRF-neg-A (open squares) cells were stimulated with PMA, and DNA synthesis was assessed by flow cytometry after a 12-h incubation with 10 μm BrdUrd followed by intracellular staining of the cells with an APC-conjugated anti-BrdUrd antibody, RNase treatment and counterstaining with propidium iodide to evaluate DNA content. Representative cell cycle/DNA synthesis (A) and ploidy among total cells (B) data from day 4 after PMA stimulation are shown for CHRF-p53-A, CHRF-p53-B, and CHRF-Neg-A cells. The sum of 2 n/4 n cells gated by P1 and 8 n/higher cells gated by P2 represents the total number of cells undergoing DNA synthesis (BrdUrd+). Gates P3 and P4 represent the noncycling 2 n/4 n and 8 n or higher cells, respectively, and constitute the BrdUrd– cells. DNA synthesis was assayed on days 2, 4, 6, and 8 after PMA treatment. DNA synthesis assessed among total cells (C). Polyploidy assessed among cells undergoing DNA synthesis (D). DNA synthesis measured among polyploid cells (E). Error bars, S.E. (n = 2). All data presented in this figure were acquired using protocol 2 for analysis of DNA synthesis as described under “Experimental Procedures.“
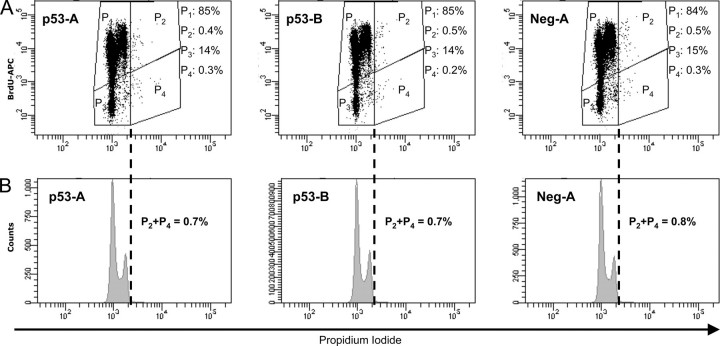
p53 Down-regulation Does Not Affect Cell Cycle Kinetics and Polyploidization of CHRF Cells in the Absence of PMA-induced Mk Differentiation—Loss of p53 can lead to hyperploidy in various cell types that are not physiologically polyploid (10, 30). To exclude the possibility that the effect of p53 on polyploidization and the cell cycle kinetics is not directly related to Mk differentiation, cell cycle kinetics and ploidy were assessed by the flow cytometric BrdUrd incorporation assay performed on unstimulated CHRF-p53-A, CHRF-p53-B, and CHRF-neg-A control cells (Fig. 4). Without PMA stimulation, CHRF-p53-A, CHRF-p53-B, and CHRF-neg-A cells are normoploid (Fig. 4B) irrespective of the level of expression of p53. The same is true for the parental CHRF cells (data not shown). Furthermore, silencing of p53 without PMA stimulation had no effect on DNA synthesis (Fig. 4A). In contrast, knockdown of p53 increased polyploidization (Fig. 2) and accelerated cell cycle progression of PMA-stimulated CHRF-p53-A and CHRF-p53-B cells (Fig. 3). We conclude that the increased polyploid phenotype upon p53 silencing and PMA stimulation reflects the putative role of p53 in the megakaryocytic differentiation program. In the absence of signaling to the cells to differentiate into megakaryocytes, loss of p53 is incapable of promoting polyploidization.
FIGURE 4.
Knockdown of p53 expression in CHRF cells has little impact on cell cycle kinetics and does not affect spontaneous polyploidization in the absence of PMA treatment. Unstimulated CHRF-p53-A, CHRF-p53-B, and CHRF-neg-A cells were incubated for 12 h with 10μm BrdUrd and then stained intracellularly with an APC-conjugated anti-BrdUrd antibody to assess the amount of synthesized DNA and counterstained with PI to evaluate DNA content. The sum of 2 n/4 n cells gated by P1 and 8 n/higher cells gated by P2 represents the total number of cells undergoing DNA synthesis (BrdUrd+). Gates P3 and P4 represent the noncycling 2 n/4 n and 8 n or higher cells, respectively, and constitute the BrdUrd– cells. Three independent biological experiments were carried out. Representative cell cycle/DNA synthesis (A) and ploidy among total cells (B) data are shown for CHRF-p53-A, CHRF-p53-B and CHRF-neg-A cells. In all three cases, only ∼0.7–0.8% of the total number of cells are shown to undergo spontaneous polyploidization. All data presented in this figure were acquired using protocol 2 for analysis of DNA synthesis as described under “Experimental Procedures.”
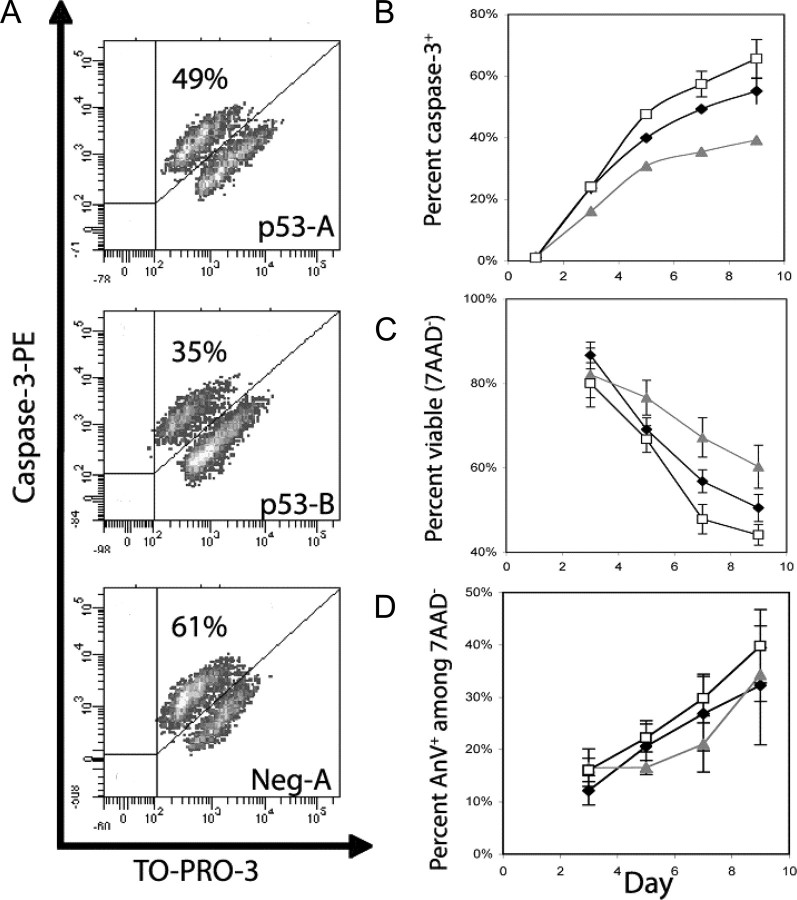
Knockdown of p53 Delays Apoptosis and Increases Viability of PMA-stimulated CHRF Cells—The proteolytic cleavage of effector caspase-3 is required for its activation, which is an early event in Mk apoptosis (31). Activation of caspase-3 was reduced by 14–16% and 35–40% in CHRF-p53-A and CHRF-p53-B cells, respectively, relative to the CHRF-neg-A controls, after day 5 of PMA stimulation (Fig. 5, A and B, p < 0.01). On a separate set of experiments, where activation of caspase-3 was examined together with DNA synthesis (via BrdUrd incorporation) and ploidy using protocol 3 as described under “Experimental Procedures,” apoptosis among total cells was reduced by 28 and 124% in CHRF-p53-A and CHRF-p53-B cells, respectively, relative to CHRF-neg-A cells, by day 8 of PMA (data not shown). Among cycling (BrdUrd+) cells, apoptosis was reduced by 36 and 218% in CHRF-p53-A and CHRF-p53-B cells, respectively, relative to CHRF-neg-A cells, by day 8 of PMA stimulation (data not shown). Finally, among polyploid cells, apoptosis was reduced by 32 and 96% in CHRF-p53-A and CHRF-p53-B cells, respectively, relative to CHRF-neg-A cells, by day 8 of PMA stimulation (data not shown). Therefore, we conclude that loss of p53 delays apoptosis in CHRF cells undergoing polyploidization. To further strengthen this finding, we assayed the CHRF cells for Annexin V binding/7AAD uptake. This apoptosis assay is performed on live cells that have been freshly harvested from culture and thus differs substantially from the cleaved caspase-3 assay, which is performed on fixed and permeabilized cells. Annexin V binds phosphatidylserine exposed on the surface membranes of apoptotic cells. When assayed for Annexin V binding/7AAD uptake, the p53 knockdown CHRF cells also exhibited a trend toward higher viability and lower apoptosis, particularly on days 7 and 9 (Fig. 5, C and D). By day 9, knockdown of p53 reduced the fraction of apoptotic cells by 14–19% and increased the fraction of viable cells by 14–36% compared with CHRF-neg-A controls. In contrast to the caspase-3, because there was more culture-to-culture variability in the progression of apoptosis and cell death in this Annexin V/7AAD assay (Fig. 5, C and D), we utilized a pairwise t test (testing each p53 knockdown culture versus its concurrent negative control culture and aggregating the results across time points and experiments) to assess statistical significance. Applying this test, we found a statistically significant decrease in apoptosis (p < 0.001 and p < 0.05 for CHRF-p53-A and CHRF-p53-B cells, respectively) and increase in viability (p < 0.001 for both cell lines) compared with the CHRF-neg-A cells. In summary, although less profound than the changes in polyploidization, these statistically significant differences in apoptosis and viability are biologically relevant and further illuminate the impact of p53 activation in Mk differentiation.
FIGURE 5.
Knockdown of p53 expression in CHRF cells delays apoptosis and cell death. CHRF-p53-A (black diamonds), CHRF-p53-B (gray triangles), and CHRF-Neg-A (open squares) cells were stimulated with PMA and assayed for cleaved caspase-3 (A and B). Representative data are shown from one experiment on day 7 after PMA stimulation (A). The cells were fixed and permeabilized, gating was done on TO-PRO-3+ events to exclude debris, and binding of a PE-conjugated antibody against active caspase-3 was assessed via flow cytometry (A). B, summary of the active caspase-3 detection based apoptosis assay. Error bars represent the S.E. (n = 2). Cell viability was assayed by the 7AAD exclusion assay (C), and apoptosis among viable (7AAD–) cells was measured by co-staining with Annexin V-PE and 7AAD (D). Error bars, S.E. (n = 2–6).
Polyploidization Is Enhanced in Mk Cell Cultures Initiated with Bone Marrow Mononuclear Cells from p53–/– Mice—To further examine the effects of p53 in terminal megakaryopoiesis, bone marrow mononuclear cells were obtained from p53–/– and wild type mice and were cultured with recombinant human Tpo to induce Mk differentiation. We found that there was a significant increase in the mean DNA content among Mk cells from p53–/– versus wild-type cultures (Fig. 6A, p < 0.05). Specifically, Tpo culture-derived p53–/– Mk cells clearly reached higher levels of polyploidy, particularly after day 3, as measured by the fraction of cells reaching ≥32 n DNA content when compared with wild type controls (Fig. 6B, p < 0.05). Furthermore, examination of the DNA histograms from the ploidy analysis revealed that cells from p53–/– mice reached higher ploidy levels than did wild type cells (Fig. 6C). This validates the findings from our CHRF cell cultures and further strengthens the argument that p53 is involved in regulating terminal Mk differentiation. It should be noted, however, that histological analysis coupled with ploidy analysis of mouse bone marrow Mk cells revealed no difference in the steady-state number or ploidy of Mk cells between p53–/– and wild type mice. This is consistent with the findings discussed in the Introduction, whereby no abnormalities have been reported in platelet levels of p53–/– mice. This issue is further addressed under “Discussion.”
FIGURE 6.
Polyploidization is increased in p53–/– murine Mk cell cultures. Bone marrow mononuclear cells from p53–/– (black diamonds) and wild type (white squares) mice were cultured in the presence of recombinant human thrombopoietin for 8 days and assayed daily by flow cytometry for DNA content. Mk cells from p53–/– mice reached higher mean ploidy levels than did cells from wild type mice (A) and were more likely to be highly polyploid (≥32 n)(B). Error bars, S.E. for two biological experiments. C, ploidy distributions are shown for the designated time points from one representative experiment for each genotype. The dashed lines indicate the ≥32 n polyploid megakaryocytes.
p53 Transcriptionally Regulates Multiple Genes during Mk Differentiation—Using samples from the PMA-stimulated cultures of shRNA-mir-expressing CHRF cells, Q-RT-PCR was performed on eight p53 target genes (Fig. 7). These eight genes were chosen either for their canonical association with the p53 regulon (MDM2, BAX, BCL2, and P21) or based on their reported up-regulation in our previous gene expression microarray profiling of Mk differentiation (BBC3, TP53I3, TP53INP1, and GADD45A) (21).
FIGURE 7.
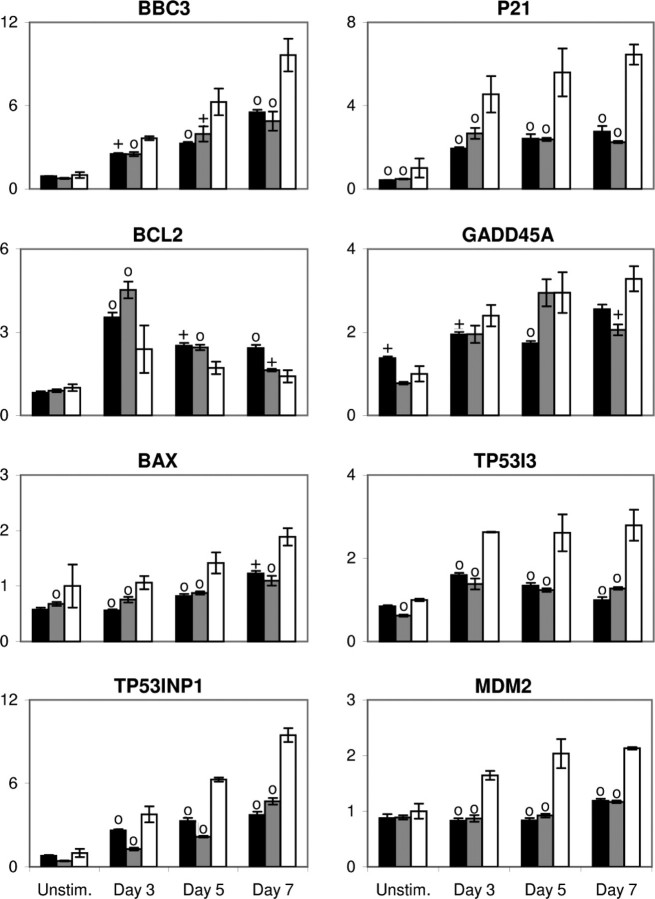
Modulation of p53 expression alters gene expression in differentiating CHRF cells. Q-RT-PCR analysis of mRNA abundance in PMA-induced cultures of CHRF cells expressing the p53-A (black), p53-B (gray), or neg-A control (white) shRNA-mir are shown for the designated genes. Data were normalized across samples using the average of two housekeeping genes (GUSB and RPLP0). For each gene, data were standardized to the expression in unstimulated CHRF-neg-A cells. Error bars, S.E. (n = 3–6). Crosses and circles denote statistically significant differences (p < 0.05 and p < 0.01, respectively) versus the corresponding neg-A-expressing CHRF cell sample from the same time point.
As expected, BBC3, P21, TP53I3 (also known as PIG3), TP53INP1, MDM2, BAX, BCL2, and GADD45A were all up-regulated upon PMA stimulation in the control CHRF-neg-A cells (Fig. 7). These data are in agreement with our previously published microarray study of PMA-induced CHRF cell differentiation (21). Knockdown of p53 partially attenuated BBC3, P21, TP53INP1, and BAX up-regulation and nearly eliminated TP53I3 and MDM2 up-regulation (Fig. 7; p < 0.05, most time points p < 0.01). These genes are all known to be transcriptionally activated by p53. It is not possible to ascertain from these data whether the residual up-regulation of these genes in the p53 knockdown cells is due to incomplete p53 knockdown or p53-independent transcriptional regulation. However, this does show that p53, at least partially, regulates the expression of these genes during PMA-induced CHRF cell differentiation. Similarly, BCL2, which is negatively regulated by p53, was more up-regulated in p53 knockdown cells than in the CHRF-neg-A controls (Fig. 7, p < 0.05, most time points p < 0.01). On the contrary, GADD45A expression was only marginally attenuated by p53 knockdown (Fig. 7), suggesting that this gene is probably not part of the core p53 regulon in terminal megakaryopoiesis. However, this does not rule out a potential role for GADD45A in Mk differentiation, as we discuss below.
We conclude that the role of p53 in Mk differentiation suggested by our data is to control endomitosis and polyploidization indirectly by decelerating cell cycling and promoting apoptosis rather than by directly inhibiting endomitosis and polyploidization. In the absence of signaling to the CHRF cells to differentiate into megakaryocytes, p53 is incapable of altering endomitosis or affecting its kinetics. This would suggest that the signals that promote megakaryocytic differentiation also lead to p53 activation in a delayed fashion (as shown in Fig. 1 and in Ref. 21). The role of p53 then is that of a gatekeeper of Mk endomitosis and polyploidization.
DISCUSSION
The involvement of p53 in regulating cell cycle progression, polyploidization, and apoptosis is well established in many cell types, and loss of p53 activity is commonly associated with aneuploidy or polyploidy (10, 32). However, we have shown, for the first time, that p53 activity increases during terminal Mk differentiation and that this increase is functionally associated with cessation of endomitosis and the initiation of apoptosis. Such a role for p53 in differentiation and development has been shown in numerous other cell types (see the Introduction). The apparent contradiction between our in vitro data and the studies that have shown that p53 is dispensable for in vivo murine megakaryopoiesis may be explained in several ways. For example, other p53 family members, including p63 and p73, may provide functional compensation in p53 knock-out mice, as has been suggested for other cases in which p53 has been linked to differentiation programs (2). Alternatively, steady-state compensatory mechanisms, such as altered Tpo levels or platelet clearance rates, or additional effects of p53 elimination, such as alterations in the physiology of the hematopoietic stem cell compartment, may act to mask changes in steady-state megakaryopoiesis. Future studies will be required to understand the mechanisms governing p53 activation in Mk cells.
Datta et al. (19) studied the role of MDM2 and p53 in the polyploidization of human erythroleukemia cells in response to PMA and concluded that p53 activity is reduced during polyploidization. They reported an increase in CDK-activating kinase activity in PMA-treated versus unstimulated HEL cells. This was linked to increased phosphorylation of CDK2 and increased kinase activity of CDK complexes with cyclins E, D3, and A in PMA-treated polyploid cells. Finally, they showed that p53 forms a complex with CDK-activating kinase, reducing CDK-activating kinase-specific kinase activity, and that the amount of p53 complexed with CDK-activating kinase decreased in polyploid cells. However, it is important to note that they reported a strong decrease in CDK-activating kinase activity, which would suggest increased p53 activity, with increasing ploidy class. This fits with our hypothesis that p53 activation in terminal Mk differentiation is linked to the cessation of polyploidization and the switch to apoptosis.
Although the complete molecular mechanism by which p53 impacts Mk polyploidization remains to be defined, we have confirmed several genes to be part of the Mk p53 regulon. Some of these genes have been previously studied in Mk cells, whereas others were first associated with Mk cells in our previous publication (21). Studies are presently under way in our laboratory to assess the specific function of these p53 target genes in Mk differentiation and to identify the upstream signaling pathway(s) activating p53 in Mk cells.
The CDK inhibitor p21 has been extensively studied in Mk cells, and high expression of p21 is generally reported in maturing Mk cells, although its functional role remains unclear and appears to be dependent on expression level (33). An interesting study by Baccini et al. (20) reported that p21–/– mice have normal platelet levels and normal steady-state Mk cell ploidy. They also found only slightly altered ploidy in Mk cells derived from the culture of p21–/– mouse HSPCs. However, they reported decreased Mk endomitosis when p21 was over expressed in p21–/– mouse cultures and saw the same effect by overexpressing p21 in wild-type mouse cultures. Overexpression of p21 in cultured human Mk cells also decreased polyploidization (20). Most importantly for this study, p21 was found to be expressed in Mk cells derived from the culture of p53–/– mouse bone marrow HSPCs (20). This is in agreement with our observation of attenuated, but not eliminated, p21 up-regulation in p53 knockdown cultures.
Of the p53 target genes we tested, only MDM2 up-regulation was completely blocked by p53 knockdown. MDM2 expression is often used as a marker of p53 transcriptional activity, and therefore its up-regulation in control CHRF cell cultures upon PMA treatment supports the hypothesis of p53 activation in these cells. MDM2 is also known to participate in a negative feedback loop by decreasing p53 transactivation activity and targeting it for degradation via ubiquitination. In the aforementioned study, Datta et al. (19) showed an increased association between p53 and MDM2 in polyploid HEL cells. Further studies are necessary to understand the role of this feedback loop in Mk differentiation.
BCL2 is a well characterized antiapoptotic regulator. Overexpression of BCL2 throughout the murine hematopoietic compartment led to a 50% reduction in platelet levels with no change in Mk numbers (34). Expression of BCL2 is decreased in response to PMA in the UT7 megakaryoblastic cell line and remained low and unchanged during Tpo-driven Mk differentiation of cord blood-derived CD34+ cells (35). We observed suppression of BCL2 expression by p53 in terminal Mk maturation. This suggests one molecular mechanism by which p53 may trigger Mk apoptosis.
Among the Mk p53 target genes identified, BBC3, TP53I3, and TP53INP1 were first linked to Mk cells by our recent global gene expression analyses (21). BBC3 is a BH3-only member of the BCL2 gene family that mediates both p53-dependent and p53-independent apoptosis (36). Given the strong residual up-regulation of BBC3 in the p53 knockdown cells (Fig. 7), it is likely that BBC3 is transcriptionally regulated by other factors in addition to p53. TP53I3 is associated with p53-induced cell death (37) and has significant homology to an NADPH-quinine oxidoreductase (38). Although it has been proposed that TP53I3 participates in reactive oxygen species generation during apoptosis (38), its actual molecular role in apoptosis has not been firmly established. TP53INP1 is both a p53 transcriptional target and an enhancer of p53 activity (39). TP53INP1 is associated with both cell cycle arrest and apoptosis upon p53 activation (40). Further study of both TP53I3 and TP53INP1 in the Mk compartment may increase our understanding of terminal Mk differentiation and better elucidate the molecular function of these genes.
We have recently shown that GADD45a is up-regulated in human megakaryocytic cells undergoing terminal differentiation (41). However, we have not yet validated a functional role of GADD45a on megakaryocytic differentiation. Although GADD45a is often referred to as a transcriptional target of p53, it is also known to be regulated independently of p53 (42). One should also note the enormous heterogeneity of the p53 binding sites and how such binding sites may be related to the functional classification (apoptosis, cell cycle, DNA repair, etc.) of the target gene (43). It is perhaps noteworthy that among the genes in Fig. 7, only GADD45A is classified under “DNA repair”; the rest are classified under “apoptosis” and “cell cycle” (43). It is therefore possible that in this case its control by p53 is not a strong one.
This study employed a model Mk differentiation system based on the CHRF-288-11 cell line. As with any model system, the CHRF cells are not a perfect representation of in vivo physiology. However, we have shown, based on global transcriptional analysis, that CHRF cells exhibit many of the transcriptional features of differentiating human primary Mk cells in vitro (21). The strongest evidence that our observations regarding the activation of p53 during terminal Mk differentiation are not artifacts of the cell line system comes from our studies of Mk differentiation of p53–/– murine bone marrow cells. Despite the absence of gross platelet or Mk cell defects in steady-state p53–/– mice, we have shown that the loss of p53 results in more highly polyploid murine Mk cells in vitro. Therefore, since Mk DNA content has been shown to be correlated with platelet production (44), modulation of p53 could affect platelet production in vitro and possibly in vivo.
Taken together, these data support a model in which the tumor suppressor p53 plays a role in terminating endomitosis and initiating cell death during terminal megakaryopoiesis. This is consistent with the known proapoptotic and cell cycleinhibitory functions of p53 in normal cells. These findings provide a new direction for efforts to increase Mk maturation, particularly the extent of polyploidization, in cultured megakaryocytes.
Supplementary Material
Acknowledgments
We acknowledge the use of instruments in the Northwestern University Flow Cytometry Core, Keck Biophysics, and the Pathology Core facilities. We gratefully acknowledge Genentech for the donation of recombinant human thrombopoietin. We thank Dr. Hiro Kiyokawa, Dr. Dipankar Ray, Brian Zwecker, and Tom O'Grady for technical assistance. We thank Dr. Mohamed Eldibany for assistance in analyzing the histology slides. We also thank Dr. Hiroaki Kiyokawa and Dr. Elizabeth Eklund for insightful comments and suggestions.
This work was supported, in whole or in part, by National Institutes of Health Grant HL48276. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2.
Footnotes
The abbreviations used are: Mk, megakaryocytic; Tpo, thrombopoietin; PMA, phorbol 12-myristate 13-acetate; CHRF, CHRF-288-11; shRNA, short hairpin RNA; shRNA-mir, micro-RNA-adapted shRNA; EmGFP, Emerald green fluorescent protein; AnV, Annexin V; SMU, starting mass units; HSPC, hematopoietic stem and progenitor cell; CDK, cyclin-dependent kinase; PBS, phosphate-buffered saline; Q-RT-PCR, quantitative reverse transcription-PCR; BrdUrd, bromodeoxyuridine; PE, phycoerythrin; 7AAD, 7-amino-actinomycin D; ELISA, enzyme-linked immunosorbent assay; EMSA, electrophoretic mobility shift assay; APC, allophycocyanin.
References
- 1.Olivier, M., Eeles, R., Hollstein, M., Khan, M. A., Harris, C. C., and Hainaut, P. (2002) Hum. Mutat. 19 607–614 [DOI] [PubMed] [Google Scholar]
- 2.Stiewe, T. (2007) Nat. Rev. Cancer 7 165–168 [DOI] [PubMed] [Google Scholar]
- 3.Almog, N., and Rotter, V. (1997) Biochim. Biophys. Acta 1333 F1–27 [DOI] [PubMed] [Google Scholar]
- 4.Jiang, D., Lenardo, M. J., and Zuniga-Pflucker, C. (1996) J. Exp. Med. 183 1923–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soddu, S., Blandino, G., Citro, G., Scardigli, R., Piaggio, G., Ferber, A., Calabretta, B., and Sacchi, A. (1994) Blood 83 2230–2237 [PubMed] [Google Scholar]
- 6.Banerjee, D., Lenz, H. J., Schnieders, B., Manno, D. J., Ju, J. F., Spears, C. P., Hochhauser, D., Danenberg, K., Danenberg, P., and Bertino, J. R. (1995) Cell Growth Differ. 6 1405–1413 [PubMed] [Google Scholar]
- 7.Soddu, S., Blandino, G., Scardigli, R., Coen, S., Marchetti, A., Rizzo, M. G., Bossi, G., Cimino, L., Crescenzi, M., and Sacchi, A. (1996) J. Cell Biol. 134 193–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehinger, M., Nilsson, E., Persson, A. M., Olsson, I., and Gullberg, U. (1995) Cell Growth Differ. 6 9–17 [PubMed] [Google Scholar]
- 9.Johnson, P., Chung, S., and Benchimol, S. (1993) Mol. Cell. Biol. 13 1456–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duensing, A., and Duensing, S. (2005) Biochem. Biophys. Res. Commun. 331 694–700 [DOI] [PubMed] [Google Scholar]
- 11.Vousden, K. H., and Lu, X. (2002) Nat. Rev. Cancer 2 594–604 [DOI] [PubMed] [Google Scholar]
- 12.Aylon, Y., Michael, D., Shmueli, A., Yabuta, N., Nojima, H., and Oren, M. (2006) Genes Dev. 20 2687–2700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lane, D. P. (1992) Nature 358 15–16 [DOI] [PubMed] [Google Scholar]
- 14.Horie, K., Kubo, K., and Yonezawa, M. (2002) J. Radiat. Res. (Tokyo) 43 353–360 [DOI] [PubMed] [Google Scholar]
- 15.Wlodarski, P., Wasik, M., Ratajczak, M. Z., Sevignani, C., Hoser, G., Kawiak, J., Gewirtz, A. M., Calabretta, B., and Skorski, T. (1998) Blood 91 2998–3006 [PubMed] [Google Scholar]
- 16.Chylicki, K., Ehinger, M., Svedberg, H., Bergh, G., Olsson, I., and Gullberg, U. (2000) Cell Growth Differ. 11 315–324 [PubMed] [Google Scholar]
- 17.Mahdi, T., Brizard, A., Millet, C., Dore, P., Tanzer, J., and Kitzis, A. (1995) J. Cell Sci. 108 1287–1293 [DOI] [PubMed] [Google Scholar]
- 18.Ritchie, A., Vadhan-Raj, S., and Broxmeyer, H. E. (1996) Stem Cells 14 330–336 [DOI] [PubMed] [Google Scholar]
- 19.Datta, N. S., and Long, M. W. (2002) Exp. Hematol. 30 158–165 [DOI] [PubMed] [Google Scholar]
- 20.Baccini, V., Roy, L., Vitrat, N., Chagraoui, H., Sabri, S., Le Couedic, J. P., Debili, N., Wendling, F., and Vainchenker, W. (2001) Blood 98 3274–3282 [DOI] [PubMed] [Google Scholar]
- 21.Fuhrken, P. G., Chen, C., Miller, W. M., and Papoutsakis, E. T. (2007) Exp. Hematol. 35 476–489 [DOI] [PubMed] [Google Scholar]
- 22.Herault, O., Colombat, P., Domenech, J., Degenne, M., Bremond, J. L., Sensebe, L., Bernard, M. C., and Binet, C. (1999) Br. J. Haematol. 104 530–537 [DOI] [PubMed] [Google Scholar]
- 23.Yang, H., Miller, W. M., and Papoutsakis, E. T. (2002) Stem Cells 20 320–328 [DOI] [PubMed] [Google Scholar]
- 24.Sugimoto, K., Toyoshima, H., Sakai, R., Miyagawa, K., Hagiwara, K., Ishikawa, F., Takaku, F., Yazaki, Y., and Hirai, H. (1992) Blood 79 2378–2383 [PubMed] [Google Scholar]
- 25.Boden, D., Pusch, O., Silbermann, R., Lee, F., Tucker, L., and Ramratnam, B. (2004) Nucleic Acids Res. 32 1154–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McManus, M. T., Petersen, C. P., Haines, B. B., Chen, J., and Sharp, P. A. (2002) RNA 8 842–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeng, Y., Wagner, E. J., and Cullen, B. R. (2002) Mol. Cell 9 1327–1333 [DOI] [PubMed] [Google Scholar]
- 28.Jagelska, E., Brazda, V., Pospisilova, S., Vojtesek, B., and Palecek, E. (2002) J. Immunol. Methods 267 227–235 [DOI] [PubMed] [Google Scholar]
- 29.Jiang, F., Jia, Y., and Cohen, I. (2002) Blood 99 3579–3584 [DOI] [PubMed] [Google Scholar]
- 30.Kramer, A., Neben, K., and Ho, A. D. (2002) Leukemia 16 767–775 [DOI] [PubMed] [Google Scholar]
- 31.De Botton, S., Sabri, S., Daugas, E., Zermati, Y., Guidotti, J. E., Hermine, O., Kroemer, G., Vainchenker, W., and Debili, N. (2002) Blood 100 1310–1317 [DOI] [PubMed] [Google Scholar]
- 32.Margolis, R. L., Lohez, O. D., and Andreassen, P. R. (2003) J. Cell. Biochem. 88 673–683 [DOI] [PubMed] [Google Scholar]
- 33.Ravid, K., Lu, J., Zimmet, J. M., and Jones, M. R. (2002) J. Cell. Physiol. 190 7–20 [DOI] [PubMed] [Google Scholar]
- 34.Ogilvy, S., Metcalf, D., Print, C. G., Bath, M. L., Harris, A. W., and Adams, J. M. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 14943–14948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanz, C., Benet, I., Richard, C., Badia, B., Andreu, E. J., Prosper, F., and Fernandez-Luna, J. L. (2001) Exp. Hematol. 29 728–735 [DOI] [PubMed] [Google Scholar]
- 36.Yu, J., and Zhang, L. (2003) Cancer Cell 4 248–249 [DOI] [PubMed] [Google Scholar]
- 37.Campomenosi, P., Monti, P., Aprile, A., Abbondandolo, A., Frebourg, T., Gold, B., Crook, T., Inga, A., Resnick, M. A., Iggo, R., and Fronza, G. (2001) Oncogene 20 3573–3579 [DOI] [PubMed] [Google Scholar]
- 38.Polyak, K., Xia, Y., Zweier, J. L., Kinzler, K. W., and Vogelstein, B. (1997) Nature 389 300–305 [DOI] [PubMed] [Google Scholar]
- 39.Tomasini, R., Samir, A. A., Carrier, A., Isnardon, D., Cecchinelli, B., Soddu, S., Malissen, B., Dagorn, J. C., Iovanna, J. L., and Dusetti, N. J. (2003) J. Biol. Chem. 278 37722–37729 [DOI] [PubMed] [Google Scholar]
- 40.Tomasini, R., Samir, A. A., Vaccaro, M. I., Pebusque, M.-J., Dagorn, J. C., Iovanna, J. L., and Dusetti, N. J. (2001) J. Biol. Chem. 276 44185–44192 [DOI] [PubMed] [Google Scholar]
- 41.Chen, C., Fuhrken, P.G., Huang L.T, Paredes, C. J., Miller, W. M., and Papoutsakis, E. T. (2007) BMC Genomics 8 384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhan, Q. (2005) Mutat. Res. 569 133–143 [DOI] [PubMed] [Google Scholar]
- 43.Horvath, M. M., Wang, X., Resnick, M. A., and Bell, D. A. (2007) PLoS Genet. 3 e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mattia, G., Vulcano, F., Milazzo, L., Barca, A., Macioce, G., Giampaolo, A., and Hassan, H. J. (2002) Blood 99 888–897 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.