Abstract
In iron-replete environments, the Pseudomonas aeruginosa Fur (ferric uptake regulator) protein represses expression of two small regulatory RNAs encoded by prrF1 and prrF2. Here we describe the effects of iron and PrrF regulation on P. aeruginosa physiology. We show that PrrF represses genes encoding enzymes for the degradation of anthranilate (i.e. antABC), a precursor of the Pseudomonas quinolone signal (PQS). Under iron-limiting conditions, PQS production was greatly decreased in a ΔprrF1,2 mutant as compared with wild type. The addition of anthranilate to the growth medium restored PQS production to the ΔprrF1,2 mutant, indicating that its defect in PQS production is a consequence of anthranilate degradation. PA2511 was shown to encode an anthranilate-dependent activator of the ant genes and was subsequently renamed antR. AntR was not required for regulation of antA by PrrF but was required for optimal iron activation of antA. Furthermore, iron was capable of activating both antA and antR in a ΔprrF1,2 mutant, indicating the presence of two distinct yet overlapping pathways for iron activation of antA (AntR-dependent and PrrF-dependent). Additionally, several quorum-sensing regulators, including PqsR, influenced antA expression, demonstrating that regulation of anthranilate metabolism is intimately woven into the quorum-sensing network of P. aeruginosa. Overall, our data illustrate the extensive control that both iron regulation and quorum sensing exercise in basic cellular physiology, underlining how intermediary metabolism can affect the regulation of virulence factors in P. aeruginosa.
Pseudomonas aeruginosa is a Gram-negative opportunistic pathogen that causes serious infections in immuno-compromised individuals, such as burn victims, and in cystic fibrosis (CF)2 patients. To cause disease, P. aeruginosa expresses several virulence factors that allow it to colonize and survive within its host, as well as a variety of systems that allow for the acquisition of nutrients required for metabolism and growth. P. aeruginosa must be able to coordinate the expression of each of these factors to successfully establish and maintain infection. For example, a shortage of iron availability leads to the increased expression of iron acquisition systems and decreased expression of pathways that rely on relatively large amounts of iron. Conversely, the potential for iron toxicity necessitates the tight regulation of iron acquisition in response to iron availability, a function mediated through the action of the ferric uptake regulator (Fur) protein. Under iron-replete conditions, the Fur protein becomes ferrated and binds to a 19-bp consensus sequence, called the Fur box, in the promoters of genes required for iron uptake, thereby preventing their transcription (1, 2). In P. aeruginosa, Fur directly or indirectly controls the expression of a large number of genes and operons involved in iron uptake, as well an assortment of virulence genes (3–6). Fur can also contribute to the increased expression of genes via the repression of two small regulatory RNAs, PrrF1 and PrrF2, which are functionally similar to RyhB in Escherichia coli (7). These small RNAs contribute to iron homeostasis by causing the degradation of mRNAs encoding iron-containing proteins, “sparing” this essential nutrient when intracellular iron concentrations are low (8, 9).
The expression of many virulence factors in P. aeruginosa is controlled by signaling molecules that are synthesized and secreted by this bacterium (10–13). Two signaling systems function through the action of distinct acyl-homoserine lactone molecules, the las system using N-(3-oxododecanoyl) homoserine lactone (3-oxo-C12-HSL) as a signaling molecule (14) and the rhl system using N-butyryl homoserine lactone (C4-HSL) (15, 16). A third system functions through the action of 2-heptyl-3-hydroxyl-4-quinolone, termed Pseudomonas quinolone signal (PQS) (17). PQS, acting as a coinducer for the LysR-type regulator PqsR (MvfR), activates the transcription of several virulence factors and the pqsABCDE operon, the gene products of which direct the synthesis of PQS (18–20). PQS synthesis involves the condensation of a fatty acid with anthranilate, a metabolite that can alternatively be converted by several enzymes to the tricarboxylic acid cycle intermediate succinate. Anthranilate can be acquired from the environment or synthesized by P. aeruginosa via one of two pathways (21). The first of these involves the degradation of tryptophan via the kynurenine pathway (21). Alternatively, anthranilate can be synthesized from chorismate by an anthranilate synthase encoded by phnAB, located just downstream from the PQS biosynthetic operon (22). PQS is found in the lungs of Pseudomonas-infected CF patients (23), and clinical isolates of P. aeruginosa from CF patients produce relatively high levels of PQS (24), indicating that this quorum-sensing molecule may play a significant role in P. aeruginosa lung infection.
This study was undertaken to determine the scope of PrrF regulation in P. aeruginosa. Our findings show that the PrrF RNAs hold extensive control over several aspects of P. aeruginosa physiology, extending beyond the function of maintaining iron homeostasis. We demonstrate that the PrrF RNAs are important for the repression of anthranilate degradation in iron-limiting environments, allowing for PQS biosynthesis. We also show that the genes for anthranilate degradation are regulated in turn by several quorum-sensing regulators, including PqsR. From our data, an intricate regulatory network is proposed in which the utilization of anthranilate for either PQS production or energy is tightly regulated by iron and anthranilate availability, as well as quorum signals.
EXPERIMENTAL PROCEDURES
Growth Conditions—E. coli strains were routinely grown in Luria-Bertani (LB) medium, and P. aeruginosa strains were routinely grown in brain-heart infusion medium. For high and low iron DTSB medium, tryptic soy broth (TSB) was treated with Chelex-100 resin (Bio-Rad) and dialyzed and then supplemented with 50 mm monosodium glutamate and 1% glycerol. FeCl3 was added to a concentration of 50 μgml–1 for iron-replete media. Anthranilate was added at a final concentration of 1 mg ml–1. For quorum-sensing studies, LB broth containing 50 mm MOPS (3-(N-morpholino) propanesulphonic acid) was used. Antibiotics were used at the following concentrations (per milliliter): 100 μg of ampicillin, 15 μg of gentamicin, and 15 μg of tetracycline for E. coli and 750 μg of carbenicillin, 75 μgof gentamicin, and 150 μg of tetracycline for P. aeruginosa.
Bacterial Strains and Genetic Manipulations—The ΔprrF1, ΔprrF2, and ΔprrF1,2 strains of PAO1 were generated previously (8). Unmarked ΔantR and ΔcatR mutants were generated as described previously (25). The ΔantR and ΔcatR mutants were complemented chromosomally as described previously (26) with the antR and catR open reading frames, as well as 200–250 bases upstream from the translational start sites. For inducible expression of antR, the gene was ligated into pUCP18 (27) downstream from the lacO-controlled promoter. For inducible expression of pqsR, the gene was cloned into pJN105 (28) downstream from the araC-controlled promoter. For the antA and pqsA expression reporter fusions, the promoters of antA and pqsA were cloned into pQF50 upstream from a promoter-less lacZ gene (29).
Expression Studies—For microarray analysis, strains were grown at 37 °C for 18 h in DTSB with and without FeCl3 addition. Total RNA was isolated from cultures using RNeasy mini spin columns (Qiagen), and the cDNA probes for microarray analysis were prepared from RNA according to the manufacturer's instructions (Affymetrix). Briefly, cDNA was generated from 5 μg of total RNA and was then fragmented using DNaseI to an approximate length of 200 bases. Fragmented cDNA from each of the RNA samples was biotinylated and hybridized to a GeneChip® P. aeruginosa genome array (Affymetrix), which includes all 5,549 protein-coding sequences of PAO1. The data were then analyzed using GeneSpring® software (Silicon Genetics).
For real-time PCR analysis, strains were grown at 37 °C for 18 h in DTSB and supplemented as indicated with FeCl3 or anthranilate. For the effects of antR overexpression, strain PAO1 carrying either pUCP18 or pUCP-antR was grown at 37 °C for 18 h in DTSB supplemented with 100 μm IPTG to induce antR expression. Total RNA was isolated as described above. RNA was DNase-treated with RNase-free DNaseI (New England Biolabs or Promega). cDNA was prepared from 50 ng of RNA using the InProm II RT system (Promega). Real-time PCR reactions were carried out in a LightCycler® 480 using the LightCycler® 480 RNA master hydrolysis probes master mix (Roche Applied Science) or Applied Biosystems Model 7000 sequence detection system using the SYBR Green PCR amplification master mix (Applied Biosystems). Data were analyzed using the LightCycler® 480 or the 7000 Real-Time PCR system software. Relative amounts of cDNA were normalized by dividing the expression values by the relative amounts of omlA or rplU cDNA in each sample.
Aconitase Assays—To determine aconitase activity, whole cell extracts were prepared from cultures grown for 18 h at 37 °C in DTSB. Aconitase activity was measured as described previously (30). Briefly, 100 μm trisodium citrate, 0.7 units of isocitrate dehydrogenase, and 270 μm NADP+ were added to cleared cell lysates in assay buffer (20 mm Tris-HCl, pH 7.4). Aconitase activity was monitored by following the formation of NADPH (upon conversion of isocitrate to α-ketoglutarate) at 340 nm. Activity was normalized to protein concentration as determined by BCA protein assay (Pierce).
PQS Assays—Bacteria were grown in DTSB for 16 h at 37 °C, with and without FeCl3 or anthranilate as indicated. Each culture was harvested and extracted with acidified ethyl acetate as described by Collier et al. (23). One-half of the resulting organic extract was transferred to a clean tube and evaporated to dryness. Samples were resuspended in 1:1 acidified ethyl acetate: acetonitrile and analyzed by thin-layer chromatography (TLC) (17). The concentration of PQS in extracts was determined by using computer densitometry to compare unknowns with synthetic PQS standards on TLC plates as described by Calfee et al. (20).
Expression and Purification of His-tagged AntR (AntRhis)— AntRhis overexpression and purification were done as described previously (31). For construction of the AntRhis expression vector pJLAhis, the antR open reading frame was cloned into pET16b. The resulting plasmid was transformed into P. aeruginosa strain PAO-T7, which is genetically modified to express T7 RNA polymerase (26). PAO-T7 carrying pJLAhis was grown at 37 °C to an A600 of 0.5. IPTG was added to induce AntRhis expression, and the culture was grown another 16 h at 16 °C. Cells were harvested at 4 °C, and subsequent AntR purification procedures were performed at 0–4 °C. Harvested cells were suspended in binding buffer (20 mm Tris-HCl, 500 mm NaCl, and 5 mm imidazole, pH 7.9) and lysed by sonication. The lysate was cleared by centrifugation, and the supernatant was fractionated by nickel-nitrilotriacetic acid agarose column chromatography (Qiagen). Bound protein was washed with a buffer containing 20 mm Tris-HCl, 500 mm NaCl, and 68 mm imidazole, pH 7.9, and eluted by increasing concentrations of imidazole. Fractions containing AntRhis were pooled, dialyzed in 100 mm KCl, 50 mm NaCl, 2 mm EDTA, 0.5% Tween 20, 20% glycerol, and 50 mm Tris-HCl (pH 7.0), and stored at –80 °C.
Electrophoretic Mobility Shift Assays—Electrophoretic mobility shift assays were performed as described previously (31). Briefly, DNA probes were prepared by PCR amplification of the 198-bp antA promoter (–21 → –219 relative to the +1 site) and the 153-bp mini-CTX (32) multiple cloning site as a nonspecific competitor. The PCR products were end-labeled with [γ-32P]ATP using T4 nucleotide kinase. Binding reactions contained 10–30 pm of both the antA promoter and the mini-CTX multiple cloning site in 20 μl of DNA binding buffer (50 mm KCl, 1 mm EDTA, 1 mm dithiothreitol, 0.1 mg/ml bovine serum albumin, and 5% glycerol and 20 mm Tris, pH 7.5). Purified AntRhis and 0.1 mm anthranilate were added as indicated, and the binding reactions were incubated at room temperature for 20 min. The reaction mixtures were then separated by electrophoresis on a native 5% Tris-glycine-EDTA polyacrylamide gel, and radioactivity was detected using a Typhoon model 8600 PhosphorImager with ImageQuant software (GE Healthcare).
β-Galactosidase Assays—Strain PAO1 carrying the indicated promoter-lacZ fusion and pqsR overexpression construct was supplemented with 0.4% l-arabinose for induction of PqsR. β-galactosidase activity was assayed using the Galacto-Light Plus™ kit (Tropix). Results are given in units of β-galactosidase activity per A600.
RESULTS
The PrrF RNAs Influence Expression of a Wide Range of Genes—A previous study examined the effect of iron on global gene expression in P. aeruginosa. Although 333 genes showed decreased expression under high iron conditions, the expression of over 460 genes was induced at least 2-fold by high iron (3). Among the genes induced by high iron were bfrB, encoding a bacterioferritin, and sodB, encoding iron superoxide dismutase.3 After the identification of the iron-regulated PrrF RNAs, several of the previously identified iron-induced genes were also shown to be derepressed in a ΔprrF1,2 mutant grown in low iron conditions (8). These included the genes for iron superoxide dismutase (sodB), iron aconitase A (acnA), and succinate dehydrogenase (sdh-CDAB), all of which are also repressed by RyhB in E. coli. This was a minimal list of candidate PrrF-regulated genes, however, and a more extensive analysis of PrrF-regulated gene expression would be needed to understand the full scope of PrrF regulation.
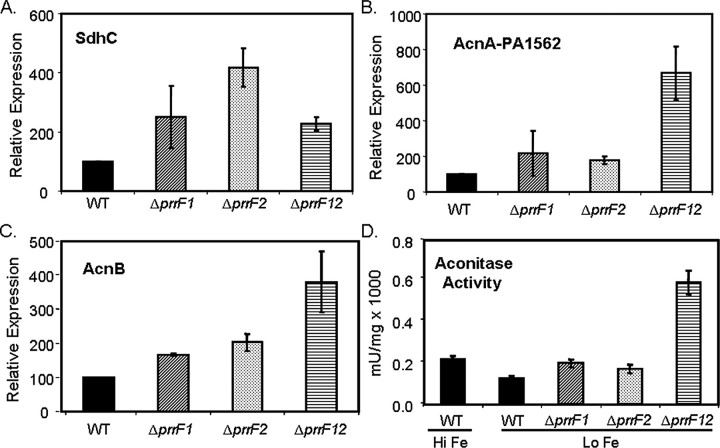
In the present study, an in-depth analysis of PrrF-regulated gene expression revealed a much broader scope of PrrF regulation than was initially appreciated (see supplemental data, Table S1, for a complete list of PrrF-repressed genes). Consistent with previous studies, this analysis identified several genes involved in iron storage and oxidative stress protection (Table 1). The majority of PrrF-repressed genes that were identified, however, encode enzymes that participate in aerobic and anaerobic metabolism, several of which make up the tricarboxylic acid cycle (Table 1). Among these were genes encoding aconitase A (PA1562), aconitase B (PA1787), and succinate dehydrogenase (sdhCDAB), which have previously been identified as RyhB-repressed in E. coli (7, 33). Real-time PCR confirmed that sdhC, acnA, and acnB are all derepressed in the ΔprrF1,2 double mutant (Fig. 1, A–C). Microarray analysis performed on single prrF mutants, in which only one of the prrF genes at a time were deleted, showed little or no expression change in genes identified in the double ΔprrF1,2 mutant (Table 1 and Ref. 8). In agreement with these data, real-time PCR showed that acnA and acnB were induced very little by individual deletion of either prrF gene as compared with the deletion of both prrF genes (Fig. 1, B and D). We also tested aconitase activity in the wild type and ΔprrF mutants. As was shown previously (34), aconitase activity increased in PAO1 cells grown in high iron versus low iron conditions (Fig. 1D). Furthermore, the levels of aconitase activity were greatly increased in the ΔprrF1,2 mutant as compared with wild type, demonstrating that the loss of aconitase activity in low iron is due to the activity of the PrrF RNAs. In contrast with our microarray data, real-time PCR analysis revealed a large increase in sdhC expression in both the single and the double prrF mutants (Fig. 1A). The reason for this difference is not clear but may reflect the limitations of using microarray analysis to dissect the regulatory interplay of PrrF1 and PrrF2.
TABLE 1.
Selected genes induced in the Δ prrF1,2 mutant
WT, wild type.
|
Gene ID |
Function of gene product |
WT high iron vs. WT low
irona |
prrF mutants vs. WT, low
ironb |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ΔprrF1 | ΔprrF2 | ΔprrF1,2 | |||||||||
| Tricarboxylic acid cycle | |||||||||||
| PA1562 acnA | Aconitase A | 5.3 | 4.6 | 3.7 | 3.5 | NC | 1.9 | 8.6 | 2.5 | 6.5 | 9.8 |
| PA1581 sdhC | Succinate dehydrogenase | 8.6c | 5.7c | 1.9 | 2.1 | NCc | NCc | 2.1c | NCc | 1.7 | 2.0 |
| PA1582 sdhD | 6.1c | 6.1c | 2.0 | 2.5 | NCc | NCc | 3.2c | NCc | 2.0 | 2.5 | |
| PA1583 sdhA | 4.0c | 4.6c | 2.1 | 2.8 | NCc | NCc | 3.7c | NCc | 2.1 | 2.5 | |
| PA1584 sdhB | 2.8c | 3.5c | NC | 2.1 | NCc | NCc | 4.0c | NCc | NC | NC | |
|
PA1787 acnB |
Aconitase B
|
4.9
|
5.7
|
3.2
|
1.6
|
NC
|
NC
|
NC
|
NC
|
2.3
|
2.5
|
| Anthranilate metabolism | |||||||||||
| PA2507 catA | Catechol degradation | 5.7 | 7.0 | 32.0 | 32.0 | NC | NC | 5.7 | 2.8 | 3.0 | 3.7 |
| PA2508 catC | 6.1 | 4.9 | 27.9 | 24.3 | NC | NC | 64.0 | 1.9 | 2.6 | 3.2 | |
| PA2509 catB | 2.3 | 3.2 | 8.0 | 12.1 | NC | NC | 4.6 | NC | 3.5 | 2.5 | |
| PA2511 antR | AraC-type regulator | 168.9c | 6.5c | 8.6 | 22.6 | 2.3c | NCc | 104.0c | 13.0c | 4.0 | 4.3 |
| PA2512 antA | Anthranilate degradation | 52.0c | 238.9c | 104.0 | 238.9 | NCc | NCc | 512.0c | 111.4c | 2.3 | 2.1 |
| PA2513 antB | 68.6c | 207.9c | 194.0 | 222.9 | 1.9c | NCc | 137.2c | 73.5c | 2.6 | 2.3 | |
|
PA2514 antC |
52.0c |
73.5c |
168.9
|
59.7
|
1.7c |
1.7c |
55.7c |
13.9c |
3.7
|
3.0
|
|
| Anthranilate biosynthesis | |||||||||||
| PA2080 kynU | Kynurenine pathway | NC | NC | 2.0 | 4.9 | NC | NC | 1.5 | NC | 2.3 | 1.9 |
| PA2081 kynB | NC | NC | NC | NC | NC | 1.5 | NC | 0.4 | NC | NC | |
|
PA2579 kynA |
2.3
|
NC
|
NC
|
1.7
|
NC
|
NC
|
NC
|
NC
|
1.7
|
NC
|
|
| Iron storage and oxidative stress | |||||||||||
| PA3531 bfrB | Bacterioferritin | 168.9c | 24.3c | 9.2 | 26.0 | 1.6c | NCc | 3.0c | NCc | 0.7 | 0.6 |
| PA4236 katA | Catalase | 7.5c | 7.0c | 3.0 | 4.0 | NCc | 1.6c | 5.7c | 3.2c | 5.3 | 6.1 |
| PA4366 sodB | Iron superoxide dismutase | 5.3c | 9.8c | 5.7 | 1.4 | NCc | NCc | 4.6c | 1.9c | 4.6 | 4.0 |
| PA4880 | Probable bacterioferritin | 13.0c | 7.5c | 5.3 | 2.3 | NCc | 2.5c | 14.9c | 2.1c | 13.9 | 21.1 |
Ratio of expression in wild type PAO1 grown in iron-replete conditions vs. wild type PAO1 grown in iron-depleted conditions from four independent microarray experiments. p < 0.0001 for the expression ratio of each gene in each experiment as determined by GeneSpring® analysis software, or indicated as NC (no change).
Ratio of expression in the indicated Δ prrF mutant grown in iron-depleted conditions vs. wild type PAO1 grown in iron-depleted conditions from four independent microarray experiments. p < 0.0001 for the expression ratio of each gene in each experiment as determined by GeneSpring® analysis software, or indicated as NC (no change).
Microarray data previously published by Wilderman, et al (8) that has been reanalyzed in conjunction with newly acquired microarray data.
FIGURE 1.
Regulation of tricarboxylic acid cycle enzymes by PrrF. RT-PCR was used to measure expression of sdhC (A), acnA (B), and acnB (C) mRNA from the indicated strains grown at 37 °C for 18 h in DTSB. Error bars show the standard deviation of three independent experiments performed in triplicate. WT, wild type. D, the indicated strains were grown at 37 °C for 18 h in DTSB. To determine aconitase activity, whole cell extracts were prepared from cultures grown for 18 h at 37 °C in DTSB. Aconitase activity was measured as described under “Experimental Procedures.” Error bars show the standard deviation of three independent experiments performed in duplicate. Hi Fe, high iron; Lo Fe, low iron.
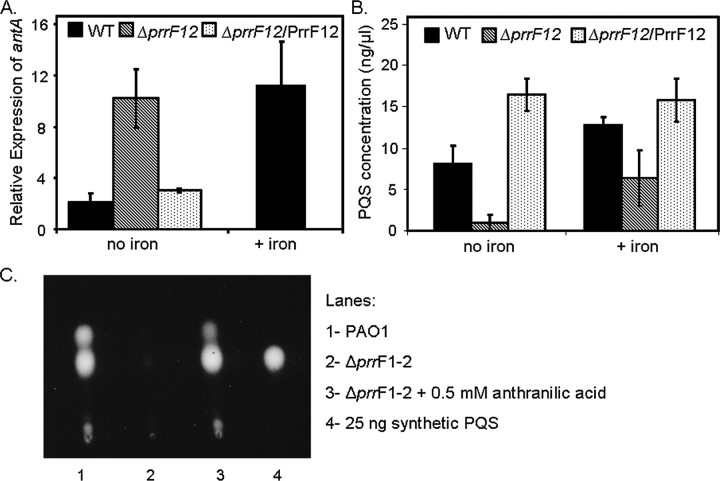
Our microarray analysis also revealed that PrrF represses genes for the degradation of anthranilate (antABC and catBCA, Table 1), a precursor of PQS. Real-time PCR analysis confirmed that antA is derepressed in the ΔprrF1,2 mutant grown in low iron as compared with wild type (∼5-fold), and this phenotype was complemented by the expression of prrF1 and prrF2 from a plasmid (Fig. 2A). It should be noted that although antA was consistently shown to be derepressed in the ΔprrF1,2 mutant, the amount of derepression varied between different microarray and real-time PCR experiments; possible reasons for this variation are presented in the discussion. Deletion of either prrF gene individually led to no significant increase in antA expression (data not shown), indicating that both PrrF RNAs contribute to the regulation of antA expression. These data provided an interesting connection between iron regulation and quorum sensing, a link that will be the subject of the remainder of this report.
FIGURE 2.
PrrF allows for PQS production by repressing the genes for anthranilate degradation. A, RT-PCR was used to measure antA mRNA from wild type (WT), ΔprrF1,2, and the complemented ΔprrF1,2 (ΔprrF1,2/PrrF1,2) strains grown at 37 °C for 18 h in DTSB with and without FeCl3 as indicated. Error bars show the standard deviation of three independent experiments performed in triplicate. B and C, bacteria were grown at 37 °C for 16 h in DTSB with and without FeCl3 as indicated. Cultures were then prepared and analyzed for PQS synthesis by TLC as described under “Experimental Procedures.” Error bars show the standard deviation of four independent experiments performed in duplicate.
The PrrF RNAs Are Required for Optimal PQS Synthesis in Low Iron Conditions—The microarray and real-time PCR data discussed above indicate that PrrF represses the genes for anthranilate degradation (Table 1 and Fig. 2A). Since anthranilate serves as a precursor for PQS synthesis, it was of interest to see how repression of anthranilate degradation by the PrrF RNAs affects production of this signaling molecule. Therefore, the effects of iron and prrF1,2 deletion on PQS production were examined more closely. Wild type and ΔprrF1,2 mutant strains were grown in DTSB medium with and without supplementation of iron. After 16 h, cells were harvested, and the extracts were assayed for PQS production by TLC. Ample amounts of PQS were produced by the wild type strain grown in low iron conditions, whereas PQS production was very low in the double ΔprrF1,2 mutant grown under the same conditions (Fig. 2B). The addition of anthranilic acid to the growth medium restored PQS production to the ΔprrF1,2 mutant (Fig. 2C), indicating that the loss of PQS production in this mutant was due to depletion of anthranilate because of the increased expression of the antABC genes. Single mutants, in which either of the prrF genes was deleted, showed only very minor defects in PQS production (data not shown), demonstrating that repression of antA by both PrrF RNAs contributes to this effect.
Surprisingly, the addition of iron did not cause a decrease in PQS production in the wild type, and instead, led to an increase in PQS production (Fig. 2B). Furthermore, the levels of PQS increased more than 6-fold in the ΔprrF1,2 mutant when iron was added (Fig. 2B). Since the addition of exogenous anthranilate was able to restore PQS production to the ΔprrF1,2 mutant in low iron (Fig. 2C), we explored the possibility that an endogenous source of anthranilate could restore PQS production to wild type and ΔprrF1,2 strains in high iron conditions. Two genes encoding enzymes of the kynurenine pathway, which provides anthranilate for the production of PQS via the degradation of tryptophan (21), were induced by high iron as compared with low iron in wild type PAO1 (between 1.7- and 4.9-fold in some microarray experiments, Table 1). Furthermore, an increased utilization of tryptophan in high iron as compared with low iron was previously observed for wild type PAO1 using BIOLOG phenotypic arrays.4 These data suggest that the kynurenine pathway may supply anthranilate for PQS synthesis in high iron conditions. Overall, these results demonstrate that PrrF, under iron-limiting conditions, spares anthranilate for PQS synthesis by repressing the genes for anthranilate degradation.
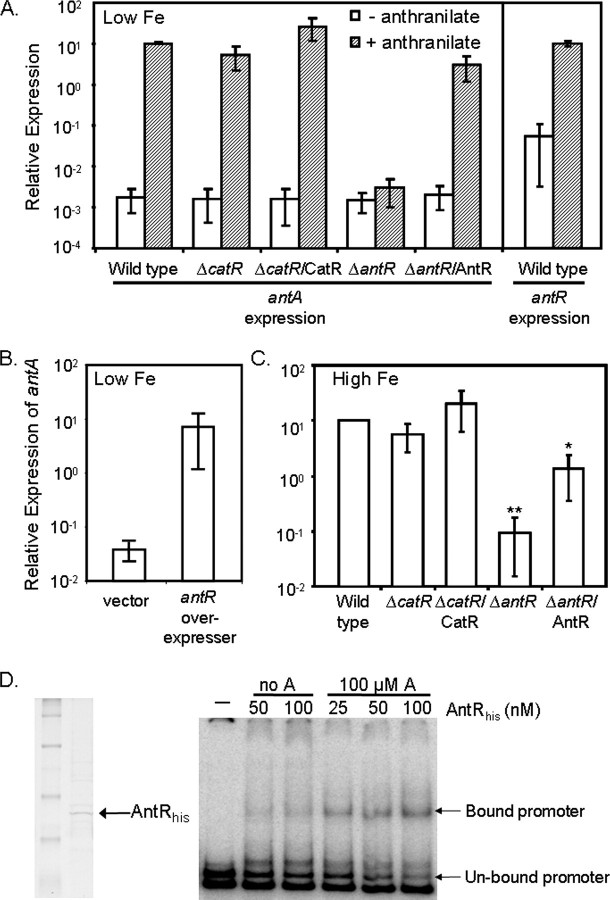
Anthranilate Induces Expression of antA via Activation of the AraC-type Regulator Encoded by PA2511—To determine how PrrF affects regulation of the ant genes, we first attempted to clarify what regulators directly controlled expression of these genes. Anthranilate is known to activate expression of the antABC genes on the carbazole-degradative plasmid pCAR1 of Pseudomonas resinovorans via the AraC-type regulator AntR (35). Because of the close homology between P. resinovorans and P. aeruginosa, we hypothesized that strain PAO1 may similarly modulate antA expression in response to anthranilate. The addition of anthranilate to wild type cultures of strain PAO1 grown in DTSB without iron supplementation had a substantial impact on antA expression, inducing it by over 10,000-fold (Fig. 3A); a similar level of induction by anthranilate was observed when wild type PAO1 was grown in iron-replete conditions (data not shown). These results establish that anthranilate modulates the expression of the genes for anthranilate degradation in PAO1.
FIGURE 3.
Role of anthranilate, AntR, and CatR in the regulation of antA. A–C, RT-PCR was used to measure expression of antA and antR mRNA from the indicated strains grown at 37 °C for 18 h in DTSB, with or without iron or anthranilate supplementation as indicated. Low Fe, low iron. B, for the effects of antR overexpression, strain PAO1 carrying either pUCP18 or pUCP-antR was grown at 37 °C for 18 h in DTSB supplemented with 100μm IPTG to induce antR expression. Error bars show the standard deviation of three independent experiments performed in triplicate. *, p < 0.05 between ΔantR/AntR and ΔantR by Student's t test. **, p < 0.0001 between ΔantR and wild type by Student's t test. High Fe, high iron. D, His-tagged AntR was purified over a nickel column as described under “Experimental Procedures,” dialyzed, concentrated, and run on an SDS-PAGE gel. Purified AntR was hybridized to the labeled antR promoter in the presence (A) or absence (no A) of anthranilate as indicated. The resulting hybridization reactions were run on a non-denaturing polyacrylamide gel. The arrows point to the shifted antR promoter fragments.
We next sought to identify the regulator responsible for anthranilate-induced expression of antA in P. aeruginosa. The PA2511 gene, which is transcribed divergently from the antABC operon in P. aeruginosa strain PAO1, encodes a putative AraC-type regulator sharing 59% identity with P. resinovorans AntR. Expression of the PA2511 gene was induced in the ΔprrF1,2 mutant along with antABC (Table 1), suggesting that these genes may be in the same regulatory pathway. Real-time PCR showed a 100-fold increase in antA expression upon overexpression of PA2511 (Fig. 3B), indicating that the protein encoded by this gene activates ant expression in P. aeruginosa. Interestingly, induction of PA2511 gene expression did not cause a decrease in PQS production (data not shown), which may be a result of alternative anthranilate biosynthesis pathways supplying this PQS precursor. The expression of PA2511, as with that of antA, was also induced over 1,000-fold in response to the addition of anthranilate (Fig. 3A). Therefore, a deletion mutant for the PA2511 gene was constructed and tested by real-time PCR. The expression of antA in the ΔPA2511 mutant was similar to that of wild type in DTSB (Fig. 3A) but was reduced ∼100-fold in ΔPA2511 as compared with wild type when iron was added to the medium, allowing for more optimal expression levels of antA in the wild type strain (Fig. 3C). In addition, anthranilate-induced expression of antA was completely abolished in the ΔPA2511 mutant (Fig. 3A), indicating that this gene is absolutely required for this response. Furthermore, purified PA2511-encoded protein bound specifically to the antA promoter in electrophoretic mobility shift assays. This binding was enhanced in the presence of anthranilate (Fig. 3D), indicating that anthranilate serves as a co-factor for its activation of antA. These results demonstrate that the regulator encoded by PA2511 activates expression of the ant genes in response to anthranilate, similarly to AntR in P. resinovorans, and we have therefore named this gene antR.
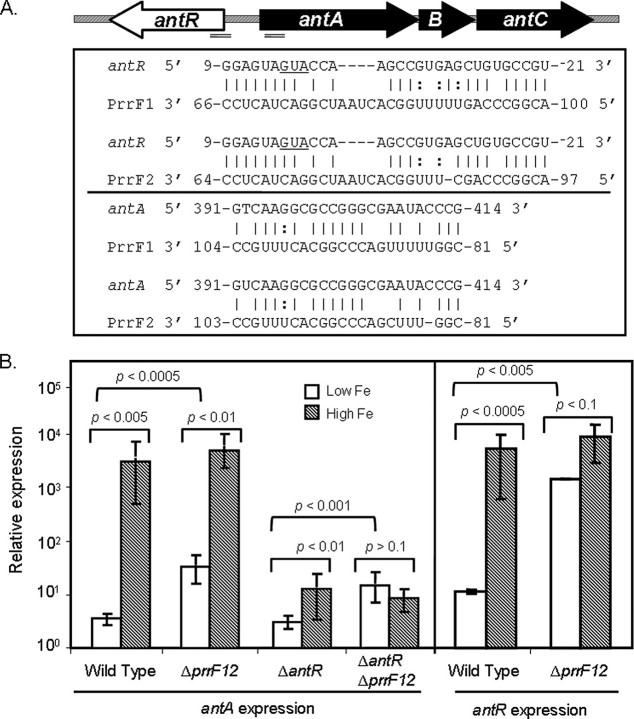
Neither AntR nor CatR Are Required for PrrF-regulated Expression of antA—A region of significant complementarity was identified between both PrrF RNAs and the translation initiation site of antR (Fig. 4A and supplemental Table S2), which led to the hypothesis that an interaction between PrrF and the antR mRNA is responsible for the repression of antA by PrrF. If this were true, then deletion of antR should result in a loss of PrrF-regulated expression of antA. Therefore, the antR deletion was moved into the ΔprrF1,2 mutant, and the levels of antA expression in the ΔantR and ΔantRΔprrF1,2 mutants were measured by real-time PCR. Surprisingly, expression of antA was repressed by PrrF in the ΔantR mutant almost as strongly as in wild type (Fig. 4B). Furthermore, PQS production was not restored to the ΔprrF1,2 mutant upon deletion of antR (data not shown). We tested the idea that CatR was also involved in the regulatory pathway between PrrF and antA, somehow masking the effect of the ΔantR deletion. Deletion of catR, in conjunction with the antR deletion or on its own, had no effect on the expression of antA (Fig. 3, A and C) or on the ability of PrrF to regulate antA expression (data not shown). Although these data do not rule out the possibility that the PrrF RNAs directly regulate the expression of antR, they do show that antR is not required for this regulation.
FIGURE 4.
AntR is not required for PrrF regulation of antA expression. A, map of the antR-antABC region on the PAO1 genome. Double lines indicate the approximate region of the shown complementarity between the antR/antA mRNAs and the PrrF RNAs; the map of the ant gene region was not drawn to scale. B, RT-PCR was used to measure expression of antA and antR mRNA from the indicated strains grown at 37 °C for 18 h in DTSB. Error bars show the standard deviation of three independent experiments performed in triplicate. The indicated p values were determined by Student's t test for comparisons between high iron (High Fe) and low iron (Low Fe) for each strain and for comparisons between wild type and deleted prrF1,2 strains grown in low iron.
In our examination of the ΔantRΔprrF1,2 double mutant, we unexpectedly found that iron-dependent regulation of antA was greatly reduced upon deletion of antR and completely abolished in the ΔantRΔprrF1,2 mutant (Fig. 4B). Additionally, the ΔprrF1,2 mutant still demonstrated a substantial, albeit reduced as compared with wild type, induction of antA expression by iron (Fig. 4B). The expression of antR was also induced by iron in the ΔprrF1,2 mutant, also at a reduced level as compared with the wild type strain (Fig. 4B). These data indicate that two distinct regulatory mechanisms lead to the induction of antA by iron. One of these involves the PrrF-independent iron activation of antR, which subsequently activates expression of antA. A second mechanism involves the PrrF RNAs, which may be able to bypass AntR and repress antA directly. A region of the antA open reading frame was found to share complementarity to the PrrF RNAs (Fig. 4A and supplemental Table S2), suggesting the possibility that PrrF directly causes the degradation of both the antA and the antR mRNAs, a scenario that is currently under investigation. It is also possible that several additional factors involved in the regulation of the ant genes, as discussed below, may be masking any involvement of AntR in the PrrF-mediated regulation of antA.
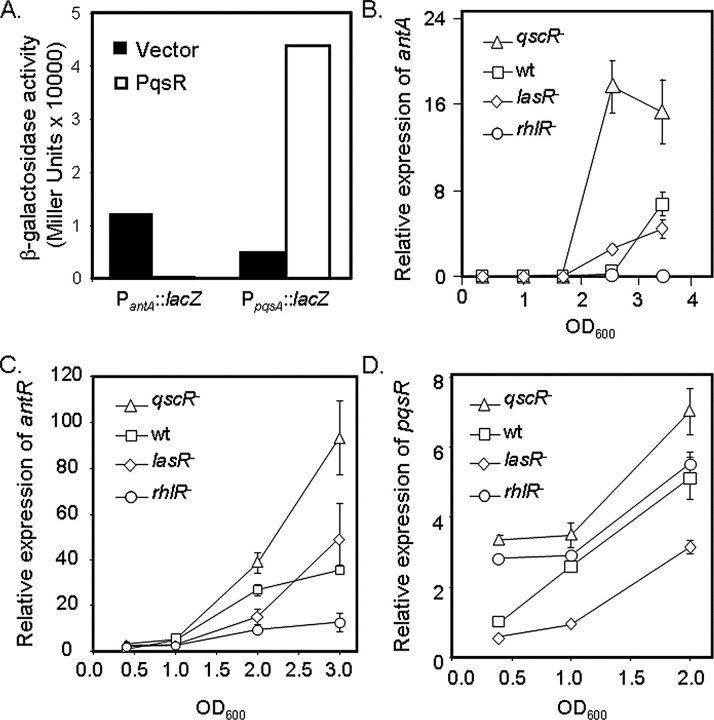
Regulation of antABC by Quorum Sensing—Our data demonstrate that degradation of anthranilate negatively affects the production of PQS. Because of this relationship, it was of interest to determine whether PQS in turn regulates expression of the genes for anthranilate degradation. To test this idea, PqsR, the cognate regulator for PQS, was overexpressed in P. aeruginosa strain PAO1 from an arabinose-inducible promoter, and PantA::lacZ and PpqsA::lacZ reporter fusions were used to monitor promoter activity of the antABC and pqsABCDE operons, respectively, by β-galactosidase assays. As expected, activity of the pqsA promoter was induced greatly upon overexpression of PqsR. In contrast, that of antA was practically abolished (Fig. 5A), demonstrating that PqsR can lead to repression of the genes for anthranilate degradation.
FIGURE 5.
The ant genes are regulated by quorum sensing. A, strain PAO1, carrying an inducible copy of pqsR and either the PantA-lacZ or the PpqsA-lacZ fusion, was grown at 37 °C in MOPS-buffered LB supplemented with 0.4% l-arabinose for induction of PqsR. Each culture was then assayed for β-galactosidase activity. Shown are representative data from a single experiment. B–D. RT-PCR was used to measure expression of antA (B), antR (C), and pqsR (D) mRNA from the indicated strains grown at 37 °C in MOPS-buffered LB. Error bars show the range of two independent experiments. wt, wild type.
The PQS system is itself regulated by other quorum-sensing systems, making it likely that these other systems also regulate expression of the ant genes. In fact, previous microarray studies showed that RhlR and its cognate sensor molecule, C4-HSL, can exert a positive regulatory effect on antABC (36). Consistent with this observation, real-time PCR revealed a substantial decrease in antA expression upon deletion of rhlR (Fig. 5B). Additionally, the expression of antR was greatly reduced in ΔrhlR (Fig. 5C), suggesting that RhlR activates antA expression via activation of antR. RhlR was also shown to repress expression of the pqsABCDE genes for PQS production (37), but it is not yet clear whether RhlR regulated the ant and pqs genes directly or via repression of PqsR. Deletion of rhlR did result in a small increase in pqsR expression (Fig. 5D), but the induction seemed to be very sensitive to changes in growth phase. Further studies will therefore be required to determine the role of PqsR in the regulation of antA by RhlR.
Previous reports have also noted that LasR activates expression of the pqsABCDE genes (19, 37). Consistent with these data, deletion of lasR led to a significant decrease in pqsR expression (Fig. 5D). It seemed likely from these data that LasR may conversely repress expression of the ant genes; deletion of lasR, however, only resulted in a small increase in antA or antR expression and only at certain time points tested (Fig. 5, B and C). Alternatively, LasR has also been shown to activate rhlR expression (38, 39), in which case the deletion of lasR would lead to decreased rhlR expression and subsequent derepression of antA. Combined with the data presented here (Fig. 5, B–D), these studies suggest that the duality of LasR regulation leads to competition between RhlR- and PqsR-mediated regulation of antA.
We also examined the role of the orphaned quorum-sensing regulator QscR, which represses expression of both the lasR/lasI and the rhlR/rhlI quorum-sensing systems (40), in the regulation of antA. Deletion of qscR resulted in a dramatic increase in both antA and antR expression (Fig. 5, B and C), as well as a significant increase in pqsR expression (Fig. 5D). These data suggest that the activation of antA and antR by RhlR may out-compete PqsR-mediated repression of antA and antR (Fig. 6B) and further obscure the role of LasR in antA regulation. Overall, these data suggest the presence of a complex hierarchy of quorum-sensing regulation over the ant and pqs genes, in which the competition between various quorum signals contributes to the determination of how anthranilate is utilized.
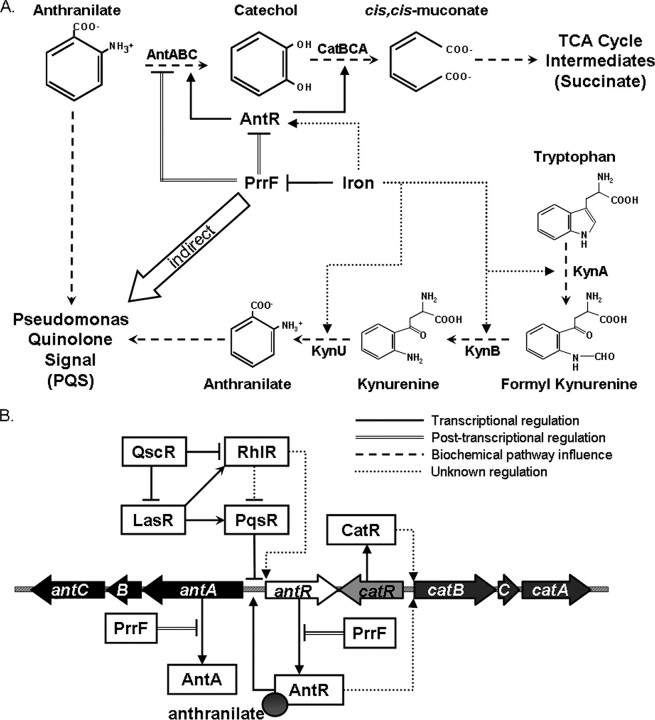
FIGURE 6.
Regulatory network of iron, PrrF, anthranilate, and quorum sensing in P. aeruginosa. A, under iron-depleted conditions, PrrF represses antABC and catABC, sparing anthranilate for PQS synthesis. The repression of antA by PrrF may occur by direct interaction with the antA mRNA and indirectly via degradation of the antR mRNA. Iron also activates expression of antR, leading to activation of antA, by an unknown, PrrF-independent, mechanism. Iron may also activate expression of genes encoding enzymes in the kynurenine pathway, providing anthranilate for PQS production under high iron conditions. TCA, tricarboxylic acid. B, AntR, an AraC-type regulator, activates expression of antR, antABC, and possibly catBCA. Anthranilate increases the ability of AntR to bind to the antA promoter and activates expression of antR and antABC. Quorum-sensing regulators (QscR, LasR, RhlR, and PqsR) also coordinate the expression of antR and antABC.
DISCUSSION
This study provides a detailed report of the regulatory mechanism by which iron affects quorum sensing in P. aeruginosa. The data presented in this study demonstrate the extensive control that iron exerts on P. aeruginosa physiology via the PrrF small regulatory RNAs, the roles of which extend beyond iron homeostasis and into control over metabolism and virulence. The extent of PrrF regulation is demonstrated by the marked effect that prrF1,2 deletion has on expression of the genes for anthranilate degradation and the result of this regulation with regard to PQS production. We observed some variation in the degree of derepression of antABC upon prrF1,2 deletion in our studies. This may be a result of varying concentrations of anthranilate, or other related metabolites, in the dialyzed tryptic soy broth that was used in our expression studies. The nature of this complex medium, as well as some inherent variability in the dialysis procedure, which allows us to remove a large proportion of iron, likely leads to variations in the concentrations of some metabolites from one batch of medium to another.
Fig. 6A shows our model of how the PrrF RNAs affect PQS production, in which PrrF controls PQS production not by a direct regulatory event, but instead, by means of a sparing effect that PrrF has on anthranilate, i.e. anthranilate is spared for PQS production from the repression of antABC by PrrF under iron-limiting conditions. This study highlights how the regulation of metabolic processes can strongly affect the production of virulence factors in P. aeruginosa. Furthermore, the role of the PrrF RNAs in this regulatory pathway provides another example of how iron and Fur contribute to the regulation of a number of virulence mechanisms in P. aeruginosa.
An interesting aspect of our data is the observation that high iron, which increases the expression levels of the genes for anthranilate degradation by both PrrF-dependent and PrrF-independent mechanisms, did not diminish the ability of PAO1 to synthesize PQS in our experiments. In fact, the addition of iron increased PQS production in the wild type strain and restored PQS synthesis to the ΔprrF12 mutant (Fig. 2). The observation that high iron induces the expression of genes encoding enzymes in the kynurenine pathway (Table 1), which can supply anthranilate for PQS synthesis (21), may explain this apparent inconsistency in our model. These data suggest that pathways for both anthranilate biosynthesis and degradation are turned on in iron-replete conditions. The induction of anthranilate biosynthesis pathways may also explain the ability of PAO1 overexpressing antR to maintain levels of PQS production. The rationale for the increased expression of the genes for anthranilate degradation by high iron is not clear but may allow for increased energy production via respiration by providing a tricarboxylic acid cycle intermediate (i.e. succinate). Therefore, the activation of both anthranilate degradation and biosynthesis pathways during high iron may allow for both energy production and PQS synthesis.
We also reveal the extensive control that P. aeruginosa places on the degradation of anthranilate, underlining the importance of this metabolic branch point. Fig. 6B shows a model of how quorum sensing and anthranilate together coordinate the expression of the ant genes, emphasizing the numerous regulatory factors that compete for control over ant gene expression. PqsR, which activates the expression of the PQS biosynthetic genes, also contributes to repression of antA and antR, allowing for increased availability of the anthranilate precursor for PQS synthesis. Although LasR clearly activates expression of pqsR, it appears to play a competing role in the regulation of antA via activation of rhlR. This sort of quorum-sensing antagonism has been demonstrated for the pqsABCDE operon (37) and may be obscuring the effects of LasR on antA expression in our experiments. Although many unknowns still exist in the regulatory cascade shown in Fig. 6B, this study provides a starting point for studies on the intricate relationship between quorum sensing and the metabolism of anthranilate.
Our model also postulates that the repression of antABC could occur via direct interaction of PrrF with the mRNAs encoding both AntA and its activator, AntR (Fig. 6A). The region of complementarity to PrrF in antA does not overlap the translation initiation site, as is standard for most RyhB-regulated genes (41, 42). Instead, the complementarity to the antA mRNA occurs ∼400 bases into a 2-kb open reading frame, and the primer-probe binding site used for real-time PCR is located downstream from the putative PrrF binding sites. Three other sets of genes identified by our microarray analysis also showed complementarity to PrrF in regions that were not near the translation initiation site (supplemental Table S2). Interestingly, all but one of these base-pairing regions were identified in polycistronic transcripts, suggesting a slightly different mechanism for PrrF-mediated degradation of these messages. It remains unclear whether small RNA binding or the resulting block in translation is more important for target mRNA degradation (42). Thus, it is unknown whether or not binding of PrrF anywhere within a target RNA could lead to complete degradation of the mRNA. It is important to note that the base-pairing regions identified in supplemental Table S2 have not been experimentally tested and only provide a starting point for studying the mechanism of PrrF-mediated mRNA degradation. It is also important to recognize that the presence of complementarity does not always translate to regulation; for bfrB, a region of complementarity was identified, but other factors seem to be involved in the iron-mediated regulation of this gene. Several of the regions listed in supplemental Table S2 are currently under investigation for their roles in PrrF-mediated degradation.
The impact of PrrF on PQS synthesis presents a novel role for iron regulation in virulence. The P. aeruginosa-infected CF lung is a dynamic environment, with environmental indicators, including iron availability, varying drastically at different stages of infection. Although P. aeruginosa may encounter differing iron concentrations depending on time and location, the importance of iron acquisition and iron-dependent regulation for a successful infection is well established. Previous studies have shown that iron acquisition systems, specifically the uptake of pyoverdine, allow for the activation of several virulence genes (43–45). Other studies have suggested that the CF lung can be an iron-depleted environment; Palmer et al. (46) observed that P. aeruginosa grown in the presence of CF sputum greatly induce genes for iron acquisition. The genes for both anthranilate and PQS synthesis are also strongly induced under these conditions, suggesting that this signal is an important aspect in CF lung infection (46). Although the PrrF RNAs do not seem to be required for PQS synthesis under iron-replete conditions, our data suggest that the PrrF RNAs can protect anthranilate stores during iron-limiting stages of infection, ensuring that this precursor is available for production of the PQS signal regardless of iron availability.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants R37-NIH AI15940 (to M. L. V.) and R01-AI46682 and 5R01-GM059026-09 (to E. P. G.). This work was also supported by the Korea Research Foundation Grant KRF-2007-331-C00222 funded by the Korean Government (to J.-H. L.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains two supplemental tables.
Footnotes
The abbreviations used are: CF, cystic fibrosis; PQS, Pseudomonas quinolone signal; Fur, ferric uptake regulator; TSB, tryptic soy broth; DTSB, dialyzed TSB; MOPS, 4-morpholinepropanesulfonic acid; IPTG, isopropyl-1-thio-β-d-galactopyranoside; HSL, homoserine lactone; RT-PCR, reverse transcription-PCR.
M. L. Vasil, unpublished data.
A. Vasil and M. Vasil, unpublished data.
References
- 1.Prince, R. W., Cox, C. D., and Vasil, M. L. (1993) J. Bacteriol. 175 2589–2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Griggs, D. W., and Konisky, J. (1989) J. Bacteriol. 171 1048–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ochsner, U. A., Wilderman, P. J., Vasil, A. I., and Vasil, M. L. (2002) Mol. Microbiol. 45 1277–1287 [DOI] [PubMed] [Google Scholar]
- 4.Hunt, T. A., Peng, W. T., Loubens, I., and Storey, D. G. (2002) Microbiology (Read.) 148 3183–3193 [DOI] [PubMed] [Google Scholar]
- 5.Leoni, L., Orsi, N., de Lorenzo, V., and Visca, P. (2000) J. Bacteriol. 182 1481–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vasil, M. L., and Ochsner, U. A. (1999) Mol. Microbiol. 34 399–413 [DOI] [PubMed] [Google Scholar]
- 7.Masse, E., and Gottesman, S. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 4620–4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilderman, P. J., Sowa, N. A., FitzGerald, D. J., FitzGerald, P. C., Gottesman, S., Ochsner, U. A., and Vasil, M. L. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 9792–9797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vasil, M. (2007) BioMetals 20 587–601 [DOI] [PubMed] [Google Scholar]
- 10.Erickson, D. L., Endersby, R., Kirkham, A., Stuber, K., Vollman, D. D., Rabin, H. R., Mitchell, I., and Storey, D. G. (2002) Infect. Immun. 70 1783–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu, H., Song, Z., Givskov, M., Doring, G., Worlitzsch, D., Mathee, K., Rygaard, J., and Hoiby, N. (2001) Microbiology (Read.) 147 1105–1113 [DOI] [PubMed] [Google Scholar]
- 12.Rumbaugh, K. P., Griswold, J. A., Iglewski, B. H., and Hamood, A. N. (1999) Infect. Immun. 67 5854–5862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Latifi, A., Winson, M. K., Foglino, M., Bycroft, B. W., Stewart, G. S., Lazdunski, A., and Williams, P. (1995) Mol. Microbiol. 17 333–343 [DOI] [PubMed] [Google Scholar]
- 14.Pearson, J. P., Gray, K. M., Passador, L., Tucker, K. D., Eberhard, A., Iglewski, B. H., and Greenberg, E. P. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 197–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ochsner, U. A., and Reiser, J. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 6424–6428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brint, J. M., and Ohman, D. E. (1995) J. Bacteriol. 177 7155–7163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pesci, E. C., Milbank, J. B., Pearson, J. P., McKnight, S., Kende, A. S., Greenberg, E. P., and Iglewski, B. H. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 11229–11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallagher, L. A., McKnight, S. L., Kuznetsova, M. S., Pesci, E. C., and Manoil, C. (2002) J. Bacteriol. 184 6472–6480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wade, D. S., Calfee, M. W., Rocha, E. R., Ling, E. A., Engstrom, E., Coleman, J. P., and Pesci, E. C. (2005) J. Bacteriol. 187 4372–4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calfee, M. W., Coleman, J. P., and Pesci, E. C. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 11633–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farrow, J. M., III, and Pesci, E. C. (2007) J. Bacteriol. 189 3425–3433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Essar, D. W., Eberly, L., Hadero, A., and Crawford, I. P. (1990) J. Bacteriol. 172 884–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collier, D. N., Anderson, L., McKnight, S. L., Noah, T. L., Knowles, M., Boucher, R., Schwab, U., Gilligan, P., and Pesci, E. C. (2002) FEMS Microbiol. Lett. 215 41–46 [DOI] [PubMed] [Google Scholar]
- 24.Guina, T., Purvine, S. O., Yi, E. C., Eng, J., Goodlett, D. R., Aebersold, R., and Miller, S. I. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 2771–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoang, T. T., Karkhoff-Schweizer, R. R., Kutchma, A. J., and Schweizer, H. P. (1998) Gene (Amst.) 212 77–86 [DOI] [PubMed] [Google Scholar]
- 26.Hoang, T. T., Kutchma, A. J., Becher, A., and Schweizer, H. P. (2000) Plasmid 43 59–72 [DOI] [PubMed] [Google Scholar]
- 27.Yanisch-Perron, C., Vieira, J., and Messing, J. (1985) Gene (Amst.) 33 103–119 [DOI] [PubMed] [Google Scholar]
- 28.Newman, J. R., and Fuqua, C. (1999) Gene (Amst.) 227 197–203 [DOI] [PubMed] [Google Scholar]
- 29.Farinha, M. A., and Kropinski, A. M. (1990) J. Bacteriol. 172 3496–3499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morrison, J. F. (1954) Biochem. J. 58 685–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, J. H., Lequette, Y., and Greenberg, E. P. (2006) Mol. Microbiol. 59 602–609 [DOI] [PubMed] [Google Scholar]
- 32.Becher, A., and Schweizer, H. P. (2000) BioTechniques 29 948–952 [DOI] [PubMed] [Google Scholar]
- 33.Masse, E., Vanderpool, C. K., and Gottesman, S. (2005) J. Bacteriol. 187 6962–6971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Somerville, G., Mikoryak, C. A., and Reitzer, L. (1999) J. Bacteriol. 181 1072–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Urata, M., Miyakoshi, M., Kai, S., Maeda, K., Habe, H., Omori, T., Yamane, H., and Nojiri, H. (2004) J. Bacteriol. 186 6815–6823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schuster, M., Lostroh, C. P., Ogi, T., and Greenberg, E. P. (2003) J. Bacteriol. 185 2066–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGrath, S., Wade, D. S., and Pesci, E. C. (2004) FEMS Microbiol. Lett. 230 27–34 [DOI] [PubMed] [Google Scholar]
- 38.Medina, G., Juarez, K., Diaz, R., and Soberon-Chavez, G. (2003) Microbiology (Read.) 149 3073–3081 [DOI] [PubMed] [Google Scholar]
- 39.Pesci, E. C., Pearson, J. P., Seed, P. C., and Iglewski, B. H. (1997) J. Bacteriol. 179 3127–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chugani, S. A., Whiteley, M., Lee, K. M., D'Argenio, D., Manoil, C., and Greenberg, E. P. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 2752–2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aiba, H. (2007) Curr. Opin. Microbiol. 10 134–139 [DOI] [PubMed] [Google Scholar]
- 42.Gottesman, S., McCullen, C. A., Guillier, M., Vanderpool, C. K., Majdalani, N., Benhammou, J., Thompson, K. M., FitzGerald, P. C., Sowa, N. A., and FitzGerald, D. J. (2006) Cold Spring Harbor Symp. Quant. Biol. 71 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beare, P. A., For, R. J., Martin, L. W., and Lamont, I. L. (2003) Mol. Microbiol. 47 195–207 [DOI] [PubMed] [Google Scholar]
- 44.Lamont, I. L., Beare, P. A., Ochsner, U., Vasil, A. I., and Vasil, M. L. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 7072–7077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meyer, J. M., Neely, A., Stintzi, A., Georges, C., and Holder, I. A. (1996) Infect. Immun. 64 518–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palmer, K. L., Mashburn, L. M., Singh, P. K., and Whiteley, M. (2005) J. Bacteriol. 187 5267–5277 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.