Abstract
The conserved CDC5 family of Myb-related proteins performs an essential function in cell cycle control at G2/M. Although c-Myb and many Myb-related proteins act as transcription factors, herein, we implicate CDC5 proteins in pre-mRNA splicing. Mammalian CDC5 colocalizes with pre-mRNA splicing factors in the nuclei of mammalian cells, associates with core components of the splicing machinery in nuclear extracts, and interacts with the spliceosome throughout the splicing reaction in vitro. Furthermore, genetic depletion of the homolog of CDC5 in Saccharomyces cerevisiae, CEF1, blocks the first step of pre-mRNA processing in vivo. These data provide evidence that eukaryotic cells require CDC5 proteins for pre-mRNA splicing.
The Schizosaccharomyces pombe cdc5-120 mutant was isolated in a screen for mutants defective in cell cycle progression (1). At the restrictive temperature, cdc5-120 cells arrest growth in G2 (1, 2), indicating that cdc5+ function is required for G2/M progression. CDC5 has been conserved throughout evolution, and related genes have been cloned from Saccharomyces cerevisiae (termed CEF1; ref. 3), Arabidopsis thaliana (4), Drosophila melanogaster (3), Caenorhabditis elegans (3), Xenopus laevis (5), and Homo sapiens (3, 6, 7). We conclude that these proteins are conserved functionally, because D. melanogaster and human CDC5 (hCDC5) complement the cdc5-120 mutant, S. cerevisiae CEF1 is essential during G2/M in this evolutionarily distinct yeast (3), and overexpression of dominant negative forms of hCDC5 slows G2 progression in mammalian cells (8).
In their N termini, CDC5 proteins are highly related to the DNA-binding domain of human c-Myb (2, 3, 9). Whereas human c-Myb contains three Myb repeats, ≈50-amino acid motifs with characteristic spacing of tryptophan residues (9), CDC5 proteins contain two Myb repeats (R1 and R2) followed by a Myb-like-repeat (MLR3) that contains some, but not all, of the hallmarks of a typical Myb repeat (3). Based on their homologies to c-Myb, CDC5 proteins were hypothesized to carry out their essential function in cell cycle control through transcriptional regulation, a notion supported by the following observations: (i) the Myb repeats of S. pombe cdc5p fused to glutathione S-transferase-bound DNA cellulose (2); (ii) the Myb repeats of A. thaliana cdc5p selected a specific DNA sequence in a cyclic amplification and selection of targets protocol (4); and (iii) the C terminus of hCDC5 fused to the GAL4-DNA binding domain activated transcription in a reporter assay (8). To date, however, no downstream transcriptional targets for any of the CDC5 proteins have been identified.
hCDC5 was identified recently in a biochemical purification of the mammalian spliceosome assembled in vitro (10), indicating that CDC5 proteins may be involved in pre-mRNA splicing rather than transcriptional regulation. Herein, we extend this observation by showing that mammalian CDC5 colocalizes with splicing factors in the nuclei of mammalian cells, coimmunoprecipitates with core components of the splicing machinery from nuclear extracts, and interacts with the spliceosome throughout the splicing reaction in vitro. Finally, we present evidence that eukaryotic cells require the CDC5 family of proteins for pre-mRNA splicing in vivo.
Materials and Methods
DNA manipulations were performed by using standard techniques (11). Details of plasmid construction and oligonucleotide sequences are available on request.
Mammalian Cell Culture Conditions and Flow Cytometric Analysis.
NIH 3T3 and HeLa cells were cultured in DMEM and 10% (vol/vol) bovine calf serum or 10% (vol/vol) FBS, respectively. TsBN2 cells (12) were cultured and shifted to the restrictive temperature for 10 h as described (13). Asynchronously growing subconfluent NIH 3T3 cells were rendered quiescent by incubation for 24 h in DMEM containing 0.1% bovine calf serum. For flow cytometric analysis, cells were trypsinized, collected by centrifugation, and fixed overnight at 4°C in 70% (vol/vol) ethyl alcohol. The next day, cells were incubated for 30 min with 0.1 mg/ml RNase and 0.05 mg/ml propidium iodide in 50 mM sodium citrate. Flow cytometric analysis was conducted with a Becton Dickinson fluorescence-activated cell sorter (FACScan).
Generation of hCDC5 Polyclonal Antisera.
The C-terminal 440 and the N-terminal 363 amino acids of hCDC5 were produced separately in Escherichia coli as hexahistidine fusion proteins and used to generate polyclonal antisera in rabbits. Antisera were affinity-purified against hCDC5C or hCDC5N as described (14).
Protein Lysates, Subcellular Fractionation, Production, and Immunoprecipitation of hCDC5 in Vitro.
Subconfluent NIH 3T3 and HeLa cells were lysed in 1× SDS sample buffer [0.125 M Tris⋅HCl, pH 6.8/10% (vol/vol) glycerol/2% (vol/vol) SDS/10% (vol/vol) β-mercaptoethanol Bromophenol blue]. Lysates were heated to 95°C for 5 min and clarified before loading. Nuclear and cytoplasmic fractions of cultured cells were prepared as described (15). hCDC5 and 6XHis/Myc-tagged hCDC5 were produced in vitro in the presence of TRAN35S-LABEL (ICN) by using rabbit reticulocyte lysate (Promega). Nonidet P-40 buffer (500 μl; ref. 16) was added to each 50-μl reaction, and the lysates were clarified for 30 min at 4°C. Proteins were immunoprecipitated with 3 μl of preimmune or immune hCDC5C antiserum or 5 μg of the anti-Myc (9E10) monoclonal antibody. For immunoprecipitations with 9E10, affinity-purified rabbit anti-mouse secondary antibodies were added before addition of protein A Sepharose (Amersham Pharmacia).
Immunoblotting.
Proteins were separated by SDS/PAGE and transferred to poly(vinylidene difluoride) membranes. Primary antibodies were used at dilutions of 1:1,000 or 1:2,000 for preimmune and immune hCDC5C antisera, respectively; 1:300 for affinity-purified hCDC5C antiserum; and at 0.5 μg/ml for preimmune IgG or affinity-purified hCDC5N antiserum. Monoclonal anti-actin antibodies (Amersham Pharmacia) were used at 1:2,000. Peroxidase-conjugated goat anti-rabbit or sheep anti-mouse secondary antibodies were used at 10,000-fold dilutions. Reactive proteins were detected by using enhanced chemiluminescence detection (Amersham Pharmacia).
Indirect Immunofluorescence.
Subconfluent cells attached to coverslips were fixed with 1:1 (vol/vol) methanol:acetone for 5 min at room temperature. Cells were blocked at 37°C with 3% (vol/vol) BSA, followed by incubation with 1:25 or 1:50 affinity-purified hCDC5C antiserum. For colocalization studies, cells were simultaneously incubated with 0.05 mg/ml anti-SC35 monoclonal antibody (PharMingen). Fluorescein goat anti-rabbit IgG conjugate (1:100 dilution of 2 mg/ml stock; Molecular Probes) and Texas Red goat anti-mouse IgG conjugate (1:100 dilution of 2 mg/ml; Molecular Probes) were used to detect the primary antibodies. DNA was visualized by incubating the cells in 0.5 μg/ml Hoechst 33258 (Sigma). Cells were photographed through a Zeiss Axioskop microscope with ×40 or ×100 objectives and appropriate filters. Images were captured with a cooled CCD camera (Optronics ZVS-47DEC, Goleta, CA). Confocal microscopy and overlay analysis were performed on a Zeiss LSM410 Confocal Laser Scanning Microscope with a ×63 objective.
Transient Transfection of Clk/Sty Kinase in NIH 3T3 Cells.
NIH 3T3 cells were transfected with a plasmid that constitutively expresses N-terminally Myc-tagged mouse Clk/Sty (pECE-M-STY; ref. 17). Transfections were performed with Lipofectamine (Life Technologies). After 24 h, cells were processed for indirect immunofluorescence with 9E10 monoclonal antibody and affinity-purified hCDC5C antiserum.
Immunoprecipitation of Small Nuclear RNAs (snRNAs), Small Nuclear Ribonucleoproteins (snRNPs), and Spliceosomes.
Immunoprecipitations were carried out in IPP150 (10 mM Tris⋅HCl, pH 8.0/150 mM KCl/0.1% NP-40) buffer essentially as described (18). Briefly, 20 μl of packed protein A Sepharose 4B fast flow resin (Sigma) was precoated with 20 μl of hybridoma ascites containing monoclonal antibodies against maltose-binding protein (AK105; ref. 48) or 40 μg of Sm Ab-1 (Clone Y12; Neomarkers, Fremont, CA). Beads (20 μl) were also precoated with 2 μl of either preimmune or immune hCDC5C antiserum. The beads were then incubated for 1 h at 4°C with 25 μl of HeLa-cell nuclear extract prepared as described below. snRNAs were immunoprecipitated with 100 μg of agarose-linked 2,2,7-trimethylguanosine (Ab-1) monoclonal antibodies (Oncogene Research Products, Cambridge, MA).
In vitro splicing assays were carried out in HeLa-cell nuclear extracts as described (19, 20). Capped and uniformly 32P-labeled β-globin pre-mRNA substrate was produced in vitro from pSP64-HβΔ6 (20, 21). β-Globin RNA species were visualized on 9% polyacrylamide (29:1, acrylamide/bisacrylamide)/8 M urea denaturing gels (20). Immunoprecipitation of spliceosomes was carried out after 45 or 90 min of the reaction in IP100 (50 mM Tris⋅HCl, pH 7.6/100 mM KCl/2 mM MgCl2/0.5 mM DTT/0.05% NP-40) buffer essentially as described, with 15 μl of protein A beads precoated with preimmune or immune anti-hCDC5C or 30 μl of Y12 hybridoma ascites (monoclonal anti-Sm; refs. 22 and 23).
S. cerevisiae Strains, Preparation of Total RNA, and Northern Blot Analysis.
KGY1120 and KGY1140 strains were grown under permissive conditions and shifted to restrictive conditions exactly as described (3). prp3–1(ts125; refs. 24 and 25), prp18 (ts503; ref. 24), and cdc28–1N (26) temperature-sensitive strains were grown in yeast extract/peptone/dextrose (27).
Total RNA was prepared from cells by extraction with hot acidic phenol as described (28). Total RNA (20 μg per sample) was electrophoresed on formaldehyde-agarose gels and capillary blotted to a Duralon-UV membrane (Stratagene). After UV crosslinking, blots were hybridized with labeled probes from RP51a, DYN2, DBP2, or GLC7 ORFs or PCR fragments representing the ACT1 or TUB3 intron sequences. Blots were exposed to PhosphorImager screens and visualized by using md image quant (version 3.3) or image quant for Macintosh 1.1 (Molecular Dynamics).
Results
Characterization of hCDC5 Antisera.
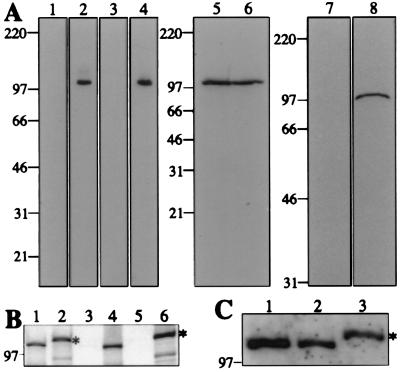
To facilitate studies of hCDC5, rabbit polyclonal antibodies were raised against the C-terminal 440 amino acids (hCDC5C) or N-terminal 363 amino acids (hCDC5N) of the protein. Crude and affinity-purified hCDC5C and hCDC5N sera recognized specifically a single protein species of 105 kDa from HeLa and NIH 3T3 whole-cell lysates (Fig. 1A; data not shown). To confirm that the 105-kDa proteins were indeed hCDC5 and mouse CDC5 (mCDC5), two full-length variants of hCDC5 differing only by the presence of a C-terminally located 6XHis/Myc epitope were produced by in vitro transcription/translation. Both proteins were specifically immunoprecipitated by the hCDC5C antiserum (Fig. 1B), and hCDC5 produced in vitro comigrated with mCDC5 from NIH 3T3 cells (Fig. 1C). Collectively, these data confirm that mammalian CDC5 migrates at 105 kDa and that our polyclonal antibodies recognize specifically mammalian CDC5 proteins.
Figure 1.
Characterization of hCDC5 antisera. (A) Immunoblots of whole-cell protein extracts from NIH 3T3 (lanes 1, 2, and 5) and HeLa (lanes 3, 4, and 6–8) cells probed with preimmune (lanes 1 and 3), immune (lanes 2 and 4), and affinity-purified (lanes 5 and 6) antisera raised against the C terminus of hCDC5 (hCDC5C), and preimmune IgG (lane 7) and affinity-purified antisera (lane 8) raised against the N terminus of hCDC5 (hCDC5N). (B and C) Analysis of hCDC5 produced in vitro. hCDC5 or 6XHis/Myc-tagged hCDC5 proteins were produced in vitro in the presence (B) or absence (C) of TRAN35S-LABEL. In B, total products of the reactions (lanes 1 and 2), preimmune (lane 3) or anti-hCDC5C (lane 4) immunoprecipitates, and immunoprecipitates of the 6XHis/Myc-tagged hCDC5 with no primary antibody (lane 5) or 9E10 antibody (lane 6) were resolved by SDS/PAGE. Proteins were detected by fluorography. In C, an NIH 3T3 cell lysate (lane 1), hCDC5 produced in vitro (lane 2), and 6XHis/Myc-tagged hCDC5 produced in vitro (lane 3) were resolved by SDS/PAGE and immunoblotted with anti-hCDC5C serum. In both B and C, asterisks denote the 6XHis/Myc-tagged species of hCDC5. For all panels, numbers to the left indicate molecular mass (in kDa).
Subcellular Localization of CDC5.
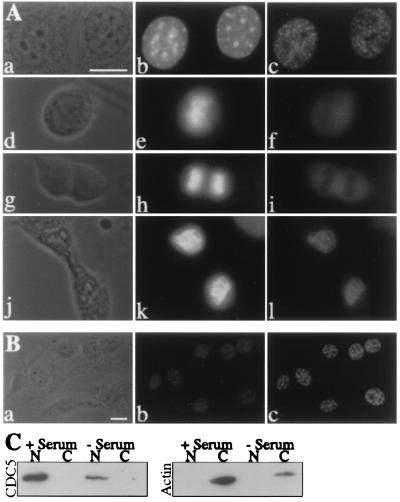
By indirect immunofluorescence, endogenous mCDC5 protein was localized to the nuclei of NIH 3T3 cells in a punctate pattern, except during metaphase and anaphase when mCDC5 became localized diffusely throughout the cell (Fig. 2A). mCDC5 was excluded from phase-dark regions of the nucleus presumed to be nucleoli. Cells in metaphase and anaphase, identified by cellular and chromosomal morphology, displayed a uniform distribution of mCDC5 with exclusion from segregating chromosomes. In telophase cells, mCDC5 localized exclusively to the reforming nucleus in a speckled pattern (Fig. 2A). Identical staining patterns were observed with affinity-purified hCDC5N antibodies, by using formaldehyde rather than methanol as fixative, in HeLa, CV-1, and COS-7 cell lines (data not shown). Cells stained with preimmune serum or secondary antibodies alone failed to produce fluorescent signals (data not shown).
Figure 2.
Subcellular localization of CDC5 throughout the cell cycle and in serum-deprived cells. (A) Phase contrast (a, d, g, and j) and fluorescence micrographs of NIH 3T3 cells stained with Hoechst (b, e, h, and k) and affinity-purified hCDC5C antiserum (c, f, i, and l). Cells in interphase (a–c), metaphase (d–f), anaphase (g–h), and telophase (j–l) are shown. (B) Localization of CDC5 in serum-deprived NIH 3T3 cells. Phase contrast (a) and fluorescence micrographs of NIH 3T3 cells deprived of serum for 24 h and stained with Hoechst (b) and affinity-purified hCDC5C antiserum (c). (C) Localization of CDC5 in nuclear and cytoplasmic fractions of asynchronously growing and serum-deprived NIH 3T3 cells. Equivalent amounts of nuclear and cytoplasmic fractions prepared from asynchronously growing NIH 3T3 cells or cells that were deprived of serum for 24 h were analyzed by immunoblotting by using either hCDC5C antiserum or a monoclonal anti-actin antibody. (Bars = 10 μm.)
To rule out the possibility that a cytoplasmic pool of mCDC5 had gone undetected by indirect immunofluorescence, NIH 3T3 cells were fractionated biochemically into nuclear and cytoplasmic pools. In agreement with the indirect immunofluorescence results, mCDC5 was detected in the nuclear but not the cytoplasmic fraction (Fig. 2C). Conversely, actin was detected in the cytoplasmic but not the nuclear fraction, indicating that the fractionation procedure had been successful (Fig. 2C).
It has been reported that exogenously expressed hCDC5 is cytoplasmic after serum deprivation, and that, on serum stimulation, hCDC5 translocates into the nucleus (6). To determine whether the localization of endogenous CDC5 was altered by serum conditions, indirect immunofluorescence was performed on NIH 3T3 cells that had been deprived of serum for 24 h. A significantly greater proportion (95%) of serum-deprived cells contained a 1 N content of DNA when compared with asynchronously growing serum-fed cells (63%). Thus, these cells had been appropriately serum starved. Serum-deprived NIH 3T3 cells displayed a punctate, nuclear staining pattern of CDC5, indistinguishable from that seen in asynchronously growing cells (Fig. 2B). The nuclear localization of CDC5 was again confirmed by biochemical fractionation (Fig. 2C). CDC5 was also detected in the nucleus of serum-deprived COS-7 and CV-1 cells under the conditions used previously (data not shown; ref. 6). The pattern of punctate staining was not altered in these cells after serum addition (data not shown).
CDC5 Localizes to the Nuclear Speckles.
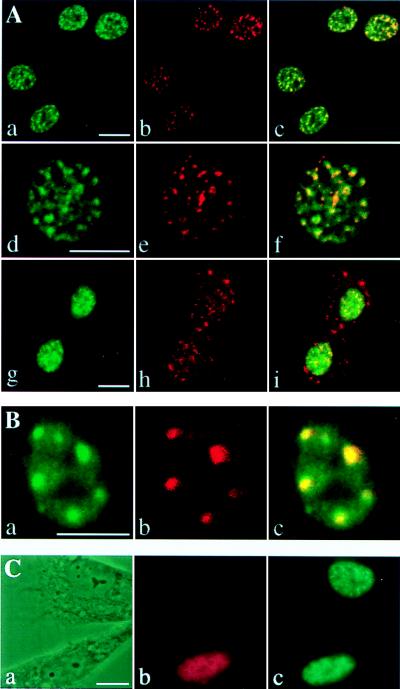
The nuclear, punctate distribution of CDC5 resembled that of factors involved in pre-mRNA splicing, which are concentrated into “nuclear speckles” (for review see ref. 29). To determine whether CDC5 localized to nuclear speckles, NIH 3T3 cells were costained for a non-snRNP component of the spliceosome and a marker of the nuclear speckle (SC35; refs. 30 and 31) by using an anti-SC35 monoclonal antibody and for mCDC5 by using affinity-purified hCDC5C antiserum. In all interphase nuclei observed, the distribution of brightly staining mCDC5 foci mirrored the SC35 staining pattern (Fig. 3A), although the more diffuse pool of mCDC5 did not colocalize with SC35. Colocalization of CDC5 with SC35 was also observed in asynchronously growing and serum-deprived NIH 3T3, CV-1, and COS-7 cells (data not shown). No staining was observed when the secondary antibodies used to detect CDC5 antibodies and the SC35 monoclonal antibody were interchanged (data not shown).
Figure 3.
CDC5 is a component of the nuclear speckles. (A) NIH 3T3 cells were costained with affinity-purified hCDC5C antiserum and a monoclonal antibody against SC35. Fluorescein and Texas Red-conjugated secondary antibodies were used to detect the distribution of CDC5 (a, d, and g) and SC35 (b, e, and h), respectively. Confocal images of cells in interphase (a–f) and telophase (g–i) were obtained and merged (c, f, and i). Regions of colocalization are yellow in merged images. (B) Asynchronously growing tsBN2 cells were shifted to the nonpermissive temperature and costained with affinity-purified hCDC5C antiserum (a) and a monoclonal antibody to SC35 (b). The confocal images were merged in c. (C) Myc-tagged Clk/Sty kinase was transfected into NIH 3T3 cells. Phase contrast images (a) of cells costained with anti-Myc epitope monoclonal antibodies (9E10; b) and affinity-purified hCDC5C antiserum (c) at 24 h after transfection. (Bars = 10 μm.)
During metaphase and early anaphase, SC35 staining is uniform throughout the cell body (32). In late anaphase and telophase, SC35 antigens reassociate into speckles that appear in both the cytoplasm and reforming telophase nuclei (Fig. 3A; 32). In contrast, during telophase, mCDC5 was localized completely to the reforming nuclei in foci that colocalized with the nuclear subset of SC35 speckles (Fig. 3A). These data indicate that CDC5 and SC35 translocate into the nucleus with different kinetics after chromosome segregation.
We reasoned that, if CDC5 proteins are indeed bona fide components of the nuclear speckles, then treatments that alter the spatial distribution of speckle proteins (for review see ref. 29) should similarly modulate the subnuclear localization of mammalian CDC5. In particular, tsBN2 cells (12) reorganize splicing factors into 4–10 large clusters at the restrictive temperature (13). To determine whether the CDC5 distribution is similarly reorganized, tsBN2 cells were shifted to 39.5°C for 10 h and costained for SC35 and CDC5. At 32.5°C, SC35 and CDC5 colocalized and displayed a normal speckled distribution (data not shown). In contrast, at 39.5°C, SC35 antigens and CDC5 reorganized into abnormally large clusters (Fig. 3B), indicating that CDC5 behaves like other speckle-associated proteins under these conditions.
To disperse nuclear speckles, we overexpressed the SR protein kinase Clk/Sty in NIH 3T3 cells (33). A plasmid expressing Myc-tagged Clk/Sty under control of the cytomegalovirus promoter was transfected into NIH 3T3 cells, and 24 h later, cells were fixed and costained for the Myc epitope, which allowed the identification of transfected cells and mCDC5. As described previously, transfected Clk/Sty localized diffusely throughout the nucleus (33). In transfected cells, mCDC5 was significantly more dispersed compared with untransfected cells (Fig. 3C). These data indicate that CDC5 is a bona fide component of the nuclear speckles.
hCDC5 Associates with Core Components of the Splicing Machinery and the Spliceosome in Vitro.
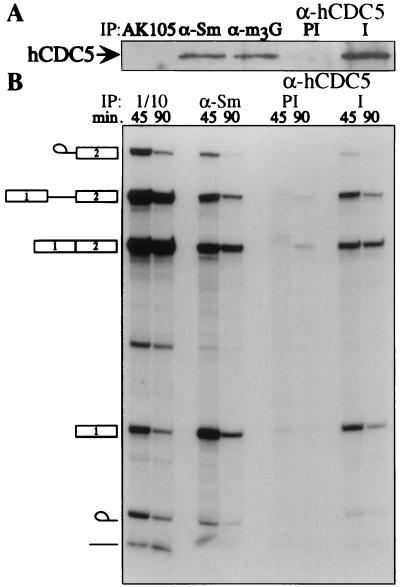
The observation that CDC5 colocalized with pre-mRNA splicing factors raised the possibility that CDC5 associates in vivo with components of the pre-mRNA splicing machinery. Indeed, hCDC5 was found in both anti-Sm and anti-m3G immunoprecipitates prepared from HeLa-cell nuclear extracts (Fig. 4A), and approximately 20% of total hCDC5 in the extract could be immunoprecipitated by using anti-m3G antibodies (data not shown). We next investigated whether hCDC5 antiserum could immunoprecipitate spliceosomes from in vitro splicing reactions. After incubation of a nuclear splicing extract containing 32P-labeled β-globin pre-mRNA for 45 or 90 min, both anti-Sm (Y12 hybridoma ascites; ref. 23) and anti-CDC5 but not preimmune antibodies immunoprecipitated approximately 5% of β-globin pre-mRNA, the lariat intermediate, and mature β-globin mRNA (Fig. 4B). By quantitating the maximal amount of β-globin immunoprecipitated by anti-hCDC5 serum relative to that of anti-Sm antibodies, we determined that hCDC5 was associated with approximately one-third of the spliceosomes in vitro (data not shown).
Figure 4.
hCDC5 interacts with core components of the splicing machinery and the spliceosome in vitro. (A) Immunoprecipitations (IP) from a nuclear splicing extract were performed with three monoclonal antibodies [irrelevant monoclonal antibody (AK105), anti-Sm (Y12), and anti-snRNA cap (anti-trimethylguanosine; α-m3G)] and preimmune (PI) or immune (I) hCDC5C antiserum. Immunoprecipitates were blotted with hCDC5C antiserum. (B) hCDC5 or Sm proteins were immunoprecipitated from an in vitro splicing reaction containing labeled β-globin pre-mRNA at 45 or 90 min after addition of pre-mRNA, and 1/10 of the total reactions were run as standards. The identities of the RNA species are indicated to the left.
S. cerevisiae Cells Lacking the Ortholog of S. pombe cdc5+, CEF1, Are Defective in pre-mRNA Splicing.
Finally, we wanted to test whether eukaryotic cells require Cdc5 proteins for pre-mRNA splicing in vivo. We decided to analyze S. cerevisiae cells genetically depleted of CEF1 for pre-mRNA splicing defects. A S. cerevisiae strain (KGY1120) lacking endogenous CEF1 and harboring plasmid-borne CEF1 cDNA under the control of the GAL1 promoter arrests growth during G2/M after 10 h of CEF1 repression (3). We looked for accumulation of immature forms of several intron-containing genes by Northern blot analysis over a 10-h time course of CEF1 repression. Cef1p is detectable until 8 h of repression (data not shown). KGY1120 mRNA was compared with that isolated from four control strains: (i) KGY1140 (3), a strain isogenic to KGY1120 but containing endogenous CEF1 as a control for growth conditions; (ii) prp3–1, a positive control for a defect in the first step of splicing (24); (iii) prp18 (ts503), a positive control for a defect in the second step of splicing (24); and (iv) cdc28-1N, a G2 arrest (26) control to ensure that any observed splicing defects were not secondary to cell cycle arrest.
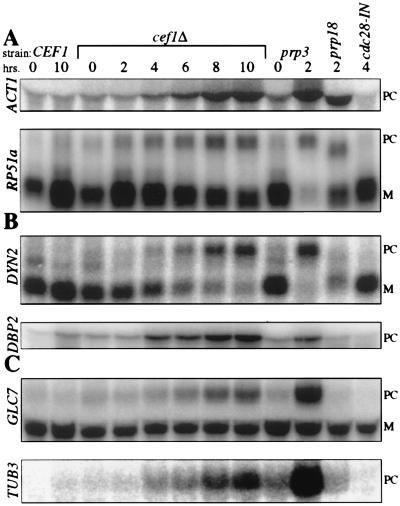
Initially, we assayed two intron-containing transcripts that are routinely used to analyze splicing defects in prp mutants ACT1 and RP51a (24). In KGY1120 cells under CEF1 repression, intron-containing forms of both transcripts steadily accumulated and peaked by 10 h (Fig. 5A). At this time, the abundance of each unspliced transcript was comparable to that observed in prp3–1 cells (Fig. 5A). Next, we assayed two transcripts with unusual intron properties, DYN2, one of two transcripts in S. cerevisiae that contains two introns, and DBP2, the transcript with the largest intron in the S. cerevisiae genome (34). Similar to ACT1 and RP51a, DYN2 and DBP2 unspliced transcripts accumulated progressively after CEF1 repression (Fig. 5B). At 10 h, DYN2 and DBP2 pre-mRNAs had roughly equivalent or greater abundance, respectively, than the same pre-mRNAs in the prp3–1 mutant. Furthermore, the levels of mature DYN2 decreased dramatically throughout the time course. Finally, we analyzed two intron-containing transcripts, GLC7 and TUB3, that encode proteins required for mitotic progression (35–38). Unspliced GLC7 accumulated throughout the time course of CEF1 repression (Fig. 5C). Similarly, intron-containing transcripts of the α-tubulin encoding TUB3 (36) increased throughout the time course (Fig. 5C). Taken together, these data indicate that CEF1 is essential for pre-mRNA splicing in vivo.
Figure 5.
S. cerevisiae cells lacking CEF1 are defective in pre-mRNA splicing. Strains containing (CEF1) or lacking the endogenous copy of CEF1 (cef1Δ) harboring plasmid-borne CEF1 cDNA under the control of the GAL1 promoter were maintained in synthetic medium containing raffinose and galactose. CEF1 expression was repressed by shifting the cells to synthetic medium containing glucose (SD). Aliquots of cells were collected at the number of hours indicated after the shift into SD. Total RNA was also purified from temperature-sensitive mutants prp3–1, prp18 (ts503), and cdc28–1N shifted to the restrictive temperature (35.5°C) for the number of hours indicated. Total RNA (20 μg) was electrophoresed and blotted. (A) Northern blots probed with the ACT1 intron sequence or the RP51a ORF. (B) Northern blots probed with the labeled ORFs for DYN2 and DBP2. (C) Northern blots probed with the GLC7 ORF or TUB3 intron sequence. PC, precursor mRNA; M, mature mRNA.
Discussion
CDC5 proteins are essential for G2/M progression in S. pombe (2), S. cerevisiae (3), and likely in higher eukaryotic cells (8). Despite a well documented role for these proteins in cell cycle progression, their biochemical function or functions are unknown. Consistent with CDC5 proteins containing homology to the well characterized DNA-binding factor and transcriptional activator c-Myb, we and others have demonstrated that the Myb domains of CDC5 proteins display affinity for double-stranded DNA (2, 4). Furthermore, the C terminus of hCDC5 fused to the GAL4-DNA-binding domain activated transcription in a transcriptional reporter assay (8). Beyond these properties, however, there is little evidence to suggest that CDC5 proteins function as transcription factors in vivo.
In fact, a recent report that hCDC5 associates with the spliceosome assembled in vitro (10) raised the possibility that CDC5 proteins are involved in pre-mRNA splicing. Our data confirm and extend this idea and show that mammalian CDC5 colocalizes with pre-mRNA splicing factors in mammalian cells, associates with core components of the splicing machinery in nuclear extracts, and interacts with the spliceosome throughout the splicing reaction in vitro. We have also shown that genetic depletion of S. cerevisiae CEF1 blocks pre-mRNA splicing in vivo. These data provide evidence that eukaryotic cells require CDC5 proteins for pre-mRNA splicing. While an earlier version of this report was under review, similar conclusions were reached regarding the requirement for Cef1p in pre-mRNA splicing in vivo (39).
Although our data, as well as those of others (10, 39), are consistent with the notion that CDC5 proteins function directly in pre-mRNA splicing, we are unable to draw such a strong conclusion. To do so, it would have to be shown that immunodepletion of hCDC5 abrogated pre-mRNA splicing in vitro. Subsequent readdition of hCDC5 should then restore pre-mRNA splicing to the depleted extract. Although we were able to remove the majority of hCDC5 from a HeLa-cell S100 extract by immunodepletion, ≈5% of the protein remained in the extract, and the depleted extract spliced β-globin (data not shown). We did observe, however, that preincubation of S100 extract with affinity-purified hCDC5N antibodies inhibited the splicing reaction, whereas equivalent amounts of control antibodies had no effect on the reaction (data not shown). That antibodies to hCDC5 inhibited pre-mRNA splicing in vitro corroborates the finding that antibodies to Cef1p inhibit pre-mRNA splicing in S. cerevisiae extracts (39). Thus, further investigations into hCDC5/Cef1p function are likely to reveal that these proteins play a direct role in the process of pre-mRNA splicing.
We have observed that mammalian CDC5 localized to the nucleus regardless of serum conditions. This result contrasts with a previous report indicating that epitope-tagged and overexpressed hCDC5 localized exclusively to the cytoplasm in serum-deprived cells (6, 8). There are at least two simple explanations for this discrepancy. First, whereas the previous study examined the localization of epitope-tagged cytomegalovirus-expressed hCDC5, we have examined the localization of endogenous CDC5. Second, both of these studies observed the localization of CDC5 at steady state. Because some splicing factors shuttle continuously between the cytoplasm and the nucleus (40), the difference between our observations may result from nucleocytoplasmic shuttling of mammalian CDC5. Perhaps serum starvation causes cytoplasmic retention of overexpressed hCDC5, thereby explaining the observations reported in the previous study.
Recent data call attention to the idea that pre-mRNA splicing and cell cycle control are linked (reviewed in ref. 41). As examples, most S. pombe prp mutants display cell cycle phenotypes (42, 43); S. pombe cdc28+, a putative RNA helicase, is required for both pre-mRNA splicing and G2/M progression (44); and cyclin E associates with and phosphorylates protein components of the U2 snRNP (45). Several hypotheses have been formulated to explain how a protein would function simultaneously in cell cycle control and pre-mRNA splicing (for review see ref. 41). One model predicts that a block in pre-mRNA splicing depletes a transcript or transcripts necessary for cell cycle progression. Indeed, it has been suggested that the mitotic arrest of S. cerevisiae prp22 mutants results from a failure to splice the mRNAs for TUB1 and TUB3 (46). Because we have found that unprocessed TUB3 transcripts accumulate in cells lacking CEF1, these cells might also arrest during G2/M (3) because of a deficiency in α-tubulin protein.
Because precursor mRNAs were the most abundant species to accumulate in cells lacking Cef1p, it is likely that CDC5 proteins are required for the first biochemical step of pre-mRNA splicing. Notably, genetic depletion of CEF1 did not affect the abundance of several intronless transcripts, indicating that CEF1 function is not globally required for mRNA production (3). Because the function of S. pombe cdc5+ is structurally and functionally conserved in eukaryotes (3), these data are consistent with the notion that all eukaryotic cells require CDC5 proteins for pre-mRNA processing. Consistent with this hypothesis, we have shown recently that S. pombe cdc5p is also required for pre-mRNA splicing in vivo and associates with a snRNP-containing complex (47). The CDC5 family of proteins represents a conserved subfamily of Myb-related proteins necessary for pre-mRNA splicing.
Acknowledgments
We are grateful to all members of the Gould laboratory, past and present, especially A. Feoktistova for dedicated technical assistance and W. H. McDonald and M. Wright for valuable discussions. We thank M. Murray, S. Chew, A. Mayeda, and M. Hastings in the Krainer laboratory for valuable discussions, reagents, and help with in vitro splicing assays. We thank M. Rush for the tsBN2 cells and S. Hanks and S. Hann for NIH 3T3, COS-7, and CV-1 cells. B. Rymond, J. Patton, and S. Reed kindly provided yeast strains. We thank J. Bell for the Clk/Sty kinase. Confocal microscopy was performed in part through the use of the Vanderbilt University Medical Center Cell Imaging Resource supported by National Cancer Institute Grant CA68485 and National Institute of Diabetes, Digestive, and Kidney Disease Grant DK 20593. This work was supported by National Institutes of Health Grant GM47728 (to K.L.G.) and National Cancer Institute Grant CA13106 (to A.R.K.). C.G.B. was supported by National Institutes of Health Medical Scientist Training Program Grant GM07347. K.L.G. is an Associate Investigator of the Howard Hughes Medical Institute.
Abbreviations
- hCDC5
human CDC5
- snRNA
small nuclear RNA
- snRNP
small nuclear ribonucleoprotein
- mCDC5
mouse CDC5
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Nurse P, Thuriaux P, Nasmyth K. Mol Gen Genet. 1976;146:167–178. doi: 10.1007/BF00268085. [DOI] [PubMed] [Google Scholar]
- 2.Ohi R, McCollum D, Hirani B, Den Haese G J, Zhang X, Burke J D, Turner K, Gould K L. EMBO J. 1994;13:471–483. doi: 10.1002/j.1460-2075.1994.tb06282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohi R, Feoktistova A, McCann S, Valentine V, Look A T, Lipsick J S, Gould K L. Mol Cell Biol. 1998;18:4097–4108. doi: 10.1128/mcb.18.7.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirayama T, Shinozaki K. Proc Natl Acad Sci USA. 1996;93:13371–13376. doi: 10.1073/pnas.93.23.13371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stukenberg P T, Lustig K D, McGarry T J, King R W, Kuang J, Kirschner M W. Curr Biol. 1997;7:338–348. doi: 10.1016/s0960-9822(06)00157-6. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein H S, Coughlin S R. J Biol Chem. 1997;272:5833–5837. doi: 10.1074/jbc.272.9.5833. [DOI] [PubMed] [Google Scholar]
- 7.Groenen P M, Vanderlinden G, Devriendt K, Fryns J P, Van de Ven W J. Genomics. 1998;49:218–229. doi: 10.1006/geno.1998.5254. [DOI] [PubMed] [Google Scholar]
- 8.Bernstein H S, Coughlin S R. J Biol Chem. 1998;273:4666–4671. doi: 10.1074/jbc.273.8.4666. [DOI] [PubMed] [Google Scholar]
- 9.Lipsick J S. Oncogene. 1996;13:223–235. [PubMed] [Google Scholar]
- 10.Neubauer G, King A, Rappsilber J, Calvio C, Watson M, Ajuh P, Sleeman J, Lamond A, Mann M. Nat Genet. 1998;20:46–50. doi: 10.1038/1700. [DOI] [PubMed] [Google Scholar]
- 11.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley; 1997. [Google Scholar]
- 12.Nishimoto T, Eilen E, Basilico C. Cell. 1978;15:475–483. doi: 10.1016/0092-8674(78)90017-x. [DOI] [PubMed] [Google Scholar]
- 13.Huang S, Mayeda A, Krainer A R, Spector D L. Mol Biol Cell. 1997;8:1143–1157. doi: 10.1091/mbc.8.6.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olmsted J B. J Biol Chem. 1981;256:11955–11957. [PubMed] [Google Scholar]
- 15.Krude T, Jackman M, Pines J, Laskey R A. Cell. 1997;88:109–119. doi: 10.1016/s0092-8674(00)81863-2. [DOI] [PubMed] [Google Scholar]
- 16.Gould K L, Moreno S, Owen D J, Sazer S, Nurse P. EMBO J. 1991;10:3297–3309. doi: 10.1002/j.1460-2075.1991.tb04894.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colwill K, Feng L L, Yeakley J M, Gish G D, Cáceres J F, Pawson T, Fu X D. J Biol Chem. 1996;271:24569–24575. doi: 10.1074/jbc.271.40.24569. [DOI] [PubMed] [Google Scholar]
- 18.Blencowe B J, Carmo-Fonseca M, Behrens S E, Lührmann R, Lamond A I. J Cell Sci. 1993;105:685–697. doi: 10.1242/jcs.105.3.685. [DOI] [PubMed] [Google Scholar]
- 19.Mayeda A, Krainer A R. Methods Mol Biol. 1999;118:309–314. doi: 10.1385/1-59259-676-2:309. [DOI] [PubMed] [Google Scholar]
- 20.Mayeda A, Krainer A R. Methods Mol Biol. 1999;118:315–322. doi: 10.1385/1-59259-676-2:315. [DOI] [PubMed] [Google Scholar]
- 21.Krainer A R, Maniatis T, Ruskin B, Green M R. Cell. 1984;36:993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- 22.Blencowe B J, Nickerson J A, Issner R, Penman S, Sharp P A. J Cell Biol. 1994;127:593–607. doi: 10.1083/jcb.127.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lerner E A, Lerner M R, Janeway C A, Jr, Steitz J A. Proc Natl Acad Sci USA. 1981;78:2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vijayraghavan U, Company M, Abelson J. Genes Dev. 1989;3:1206–1216. doi: 10.1101/gad.3.8.1206. [DOI] [PubMed] [Google Scholar]
- 25.Blanton S, Srinivasan A, Rymond B C. Mol Cell Biol. 1992;12:3939–3947. doi: 10.1128/mcb.12.9.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piggott J R, Rai R, Carter B L. Nature (London) 1982;298:391–393. doi: 10.1038/298391a0. [DOI] [PubMed] [Google Scholar]
- 27.Guthrie C, Fink G R, editors. Guide to Yeast Genetics and Molecular Biology. San Diego: Academic; 1991. [Google Scholar]
- 28.Collart M A, Oliviero S. In: Current Protocols in Molecular Biology. Ausubel F M, Brent R, Kinston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Vol. 2. New York: Wiley; 1993. pp. 13.12.1–13.12.50. [Google Scholar]
- 29.Misteli T, Spector D L. Curr Opin Cell Biol. 1998;10:323–331. doi: 10.1016/s0955-0674(98)80007-0. [DOI] [PubMed] [Google Scholar]
- 30.Fu X D, Maniatis T. Science. 1992;256:535–538. doi: 10.1126/science.1373910. [DOI] [PubMed] [Google Scholar]
- 31.Fu X D, Maniatis T. Nature (London) 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- 32.Spector D L, Fu X D, Maniatis T. EMBO J. 1991;10:3467–3481. doi: 10.1002/j.1460-2075.1991.tb04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Colwill K, Pawson T, Andrews B, Prasad J, Manley J L, Bell J C, Duncan P I. EMBO J. 1996;15:265–275. [PMC free article] [PubMed] [Google Scholar]
- 34.Spingola M, Grate L, Haussler D, Ares M., Jr RNA. 1999;5:221–234. doi: 10.1017/s1355838299981682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hisamoto N, Sugimoto K, Matsumoto K. Mol Cell Biol. 1994;14:3158–3165. doi: 10.1128/mcb.14.5.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schatz P J, Pillus L, Grisafi P, Solomon F, Botstein D. Mol Cell Biol. 1986;6:3711–3721. doi: 10.1128/mcb.6.11.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schatz P J, Solomon F, Botstein D. Genetics. 1988;120:681–695. doi: 10.1093/genetics/120.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jacobs C W, Adams A E, Szaniszlo P J, Pringle J R. J Cell Biol. 1988;107:1409–1426. doi: 10.1083/jcb.107.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai W Y, Chow Y T, Chen H R, Huang K T, Hong R I, Jan S P, Kuo N Y, Tsao T Y, Chen C H, Cheng S C. J Biol Chem. 1999;274:9455–9462. doi: 10.1074/jbc.274.14.9455. [DOI] [PubMed] [Google Scholar]
- 40.Cáceres J F, Screaton G R, Krainer A R. Genes Dev. 1998;12:55–66. doi: 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burns C G, Gould K L. In: Post-Transcriptional Regulation of Gene Expression and Its Importance to the Endocrine System. Chew S L, editor. Vol. 25. Basel: Karger; 1999. pp. 59–82. [Google Scholar]
- 42.Urushiyama S, Tani T, Ohshima Y. Mol Gen Genet. 1996;253:118–127. doi: 10.1007/s004380050304. [DOI] [PubMed] [Google Scholar]
- 43.Potashkin J, Kim D, Fons M, Humphrey T, Frendewey D. Curr Genet. 1998;34:153–163. doi: 10.1007/s002940050381. [DOI] [PubMed] [Google Scholar]
- 44.Lundgren K, Allan S, Urushiyama S, Tani T, Ohshima Y, Frendewey D, Beach D. Mol Biol Cell. 1996;7:1083–1094. doi: 10.1091/mbc.7.7.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seghezzi W, Chua K, Shanahan F, Gozani O, Reed R, Lees E. Mol Cell Biol. 1998;18:4526–4536. doi: 10.1128/mcb.18.8.4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hwang L H, Murray A W. Mol Biol Cell. 1997;8:1877–1887. doi: 10.1091/mbc.8.10.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McDonald W H, Ohi R, Smelkova N, Frendewey D, Gould K L. Mol Cell Biol. 1999;19:5352–5362. doi: 10.1128/mcb.19.8.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hanamura A, Cáceres J F, Mayeda A, Franza B R, Jr, Krainer A R. RNA. 1998;4:430–444. [PMC free article] [PubMed] [Google Scholar]