Abstract
Several scaffold proteins for neurotransmitter receptors have been identified as candidates for receptor targeting. However, the molecular mechanism underlying such receptor clustering and targeting to postsynaptic specializations remains unknown. PSD-Zip45 (also named Homer 1c/vesl-1L) consists of the NH2 terminus containing the enabled/VASP homology 1 domain and the COOH terminus containing the leucine zipper. Here, we demonstrate immunohistochemically that metabotropic glutamate receptor 1α (mGluR1α) and PSD-Zip45/Homer 1c are colocalized to synapses in the cerebellar molecular layer but not in the hippocampus. In cultured hippocampal neurons, PSD-Zip45/Homer1c and N-methyl-d-aspartate receptors are preferentially colocalized to dendritic spines. Cotransfection of mGluR1α or mGluR5 and PSD-Zip45/Homer 1c into COS-7 cells results in mGluR clustering induced by PSD-Zip45/Homer 1c. An in vitro multimerization assay shows that the extreme COOH-terminal leucine zipper is involved in self-multimerization of PSD-Zip45/Homer 1c. A clustering assay of mGluRs in COS-7 cells also reveals a critical role of this leucine-zipper motif of PSD-Zip45/Homer 1c in mGluR clustering. These results suggest that the leucine zipper of subsynaptic scaffold protein is a candidate motif involved in neurotransmitter receptor clustering at the central synapse.
Keywords: dendritic spine, EVH1 domain, NMDA receptors, receptor targeting, PSD-95
Recent studies have focused on the molecular bases of neurotransmitter receptor clustering and targeting into the central synapse. A series of PDZ domain-containing proteins have been identified as candidate molecules for such receptor clustering and/or targeting (reviewed in ref. 1). Yeast two-hybrid analyses have revealed interactions between an S/TXV sequence in the COOH terminus of the NR2 subunits of N-methyl-d-aspartate (NMDA) receptor or of Shaker’s K+ channels and the PDZ domain-containing protein PSD-95/SAP90 (2, 3). In heterologous cells, PSD-95 (or chapsyn-110) can cluster NMDA receptors and Shaker’s K+ channels (4). A disulfide-bridge formation between a pair of cysteine residues located near the NH2 terminus of PSD-95 has been proposed to be responsible for this clustering (5). Although this bridge can form only a dimer or tetramer of PSD-95, the disulfide-bridge formation is insufficient to explain the clustering. A conflicting result shows that this pair of cysteine residues is the palmitoylation site involved in the membrane sorting and in the synaptic targeting (6, 7). Therefore, the molecular mechanism underlying receptor clustering mediated by this protein remains unknown. A further analysis of PSD-95 mutant mice has revealed that, whereas NMDA receptors are targeted into excitatory synapses, the receptor-mediated signals appear to be perturbed (8). A family of PSD-95 binding proteins has also been isolated, although their functions are as yet unclear (9–11). The COOH termini of the GluR2 and GluR3 subunits of α-amino-3-hydroxy-5-methyl-4-isoxazole propionate (AMPA) receptors interact with the PDZ domain-containing protein GRIP (12), its related protein ABP (13), and a protein known to interact with protein kinase C, PICK1 (14). Of these, PICK1 induces AMPA receptor clustering in heterologous expression system (14). In spite of these accumulating evidences, there have been no insights into particular molecular motifs of these scaffold proteins in receptor clustering.
Homer 1a/vesl-1S, an immediate-response protein for a variety of neuronal stimuli (15, 16), binds to group 1 metabotropic glutamate receptors (mGluRs) (15). Because of the presence of a GLGF motif, this protein was initially considered to be a single PDZ domain-containing protein. Recent crystal structure analysis has revealed that the NH2–terminal region of this protein contains an enabled/VASP homology 1 (EVH1) domain, which binds to the COOH-terminal sequence, TPPSPFR, of mGluR1, but not to a S/TXV motif at the COOH terminus of the NR2 subunits of NMDA receptors or of Shaker’s K+ channels (17). We have recently isolated a member of the Homer 1a/vesl-1S protein family, named PSD-Zip45 (18). The same protein has been independently identified as Homer 1c (19) and vesl-1L (20). In this paper, we called this protein PSD-Zip45 for short. The NH2 terminus of this protein is identical to Homer 1a/vesl-1S, and its COOH terminus is composed of a coiled-coil structure and two leucine-zipper motifs (18–20). We have also demonstrated the self-multimerizing ability of the COOH terminus in this protein (18). These features suggest that this protein is a potent candidate for subsynaptic scaffold proteins.
Here, we demonstrated immunohistochemically that PSD-Zip45 and mGluR1α are largely colocalized to synapses of the cerebellar molecular layer. In contrast, the localizations of PSD-Zip45 and group 1 mGluRs in the hippocampus were segregated. In cultured hippocampal neurons, PSD-Zip45 and NMDA receptors were preferentially colocalized to dendritic spines. We further found that a unique leucine-zipper motif at the extreme COOH terminus of PSD-Zip45 is critically involved in self-multimerization and in PSD-Zip45-induced mGluR clustering. Thus, we provide the first insight in which a unique leucine-zipper-containing protein plays a critical role in receptor clustering at the central synapse.
Experimental Procedures
Antibodies.
The following primary antibodies were used for immunostaining, immunoprecipitation, and Western blotting: mouse anti-FLAG M2 monoclonal antibody (IBI–Kodak); mouse anti-PSD-Zip45 monoclonal antibody (mAb 126H) (18); rabbit anti-mGluR1α polyclonal antibodies (Chemicon); rabbit polyclonal antibodies raised against the SH3 and GK domains of PSD-95 (18); anti-NR1 antibodies (Chemicon); anti-synaptophysin antibodies (Progen, Heidelberg); and rabbit anti-mGluR5 polyclonal antibodies (Upstate Biotechnology, Lake Placid, NY).
Plasmid Construction.
PSD-Zip45, its deletion mutants, and Homer 1a were amplified by PCR, and the products were subcloned into pGEX-6P-1 (Amersham Pharmacia Biotech). Wild-type and deletion mutants of PSD-Zip45 and Homer 1a were subcloned into mammalian expression vector pAct. Homer 1a, PSD45ΔZipA+B, and PSD45ΔZipB were tagged with FLAG in their C termini. The cDNA clones encoding mGluR1α (21) and mGluR5 (22) were subcloned into pcDNA3.1 (+) and pEF-BOS (23), respectively. Deletion mutants of mGluR1α lacking residues 1150–1158 (mGluR1αΔ1) and residues 1159–1179 (mGluR1αΔ2) were prepared. All constructs were confirmed by sequencing.
Expression and Purification of Recombinant Proteins.
Recombinant proteins expressed in Escherichia coli were purified by using glutathione-Sepharose 4B and PreScission Protease (Amersham Pharmacia Biotech) according to the manufacturer’s recommendations.
Multimerization Assay.
Recombinant proteins were diluted in 20 mM Tris⋅HCl, pH 7.0/50 mM NaCl (0.2 mg/ml) and treated with or without 0.05% glutaraldehyde at 30°C for 30 min, separated by SDS/PAGE (14%), and stained with Coomassie brilliant blue R.
Immunohistochemistry.
The brains of Sprague–Dawley rats (7 wk old) were fixed with 4% paraformaldehyde and cryoprotected. Fresh-frozen sections (10 μm thickness) were prepared, permeabilized with TBS (20 mM Tris⋅HCl, pH 7.5/0.9% NaCl) containing 0.1% TritonX-100 for 3 hr, and blocked with 1% BSA in TBS for 1 hr. The sections were then incubated with primary antibodies and secondary antibodies conjugated with Alexa 488 or 546 (Molecular Probes). Images were obtained with a Zeiss LSM 410 confocal microscope.
Neuronal Cell Culture and Immunocytochemistry.
Hippocampal neurons were prepared from rat embryonic brains at 18.5 d. The cells dispersed with papain were cultured in DMEM supplemented with B27 nutrient mixture (GIBCO). After 3 wk, cultured neurons were fixed in methanol at −20°C for 15 min, blocked with 3% skim milk in PBS, and incubated with primary and fluorescence-conjugated secondary antibodies.
Transfection by Using Microinjection Method and Quantification of Membrane-Targeted mGluR1α.
COS-7 cells were cultured in DMEM supplemented with 10% fetal calf serum. By using a micromanipulator (Narishige, Tokyo), plasmid DNAs were microinjected through a glass capillary into the nuclei. After 3 hr, the cells were fixed with 3.7% formaldehyde, and the expressed proteins were immunolabeled and then visualized by using Axiophot microscope (Zeiss). To quantify mGluR1α targeted into the plasma membrane of COS-7 cells, a myc epitope was introduced in the extracellular domain of mGluR1α. Fluorescence for membrane-targeted mGluR1α, which were labeled with anti-myc antibody without Triton X-100 permeabilization, was measured.
Competition Assay.
The cytoplasmic domain (residues 1042–1199) of mGluR1α was in vitro translated by using TNT T7 System (Promega) and labeled with [35S]methionine. Translated products were diluted with 19 vol of dilution buffer (20 mM Tris⋅HCl, pH 7.5/100 mM NaCl/1% Triton X-100/0.5 mg/ml BSA) and incubated overnight with purified PSD-Zip45 (3 μg) and various concentrations of Homer 1a. The complexes were brought down by using mAb 126H and protein A-coupled Sepharose beads (Amersham Pharmacia Biotech) at 4°C. After being washed, the immunoprecipitates were separated by SDS/PAGE (14%), and visualized by BAS5000 PhosphorImager (Fujifilm).
Results
Localization of PSD-Zip45 and Group 1 mGluRs in the Brain.
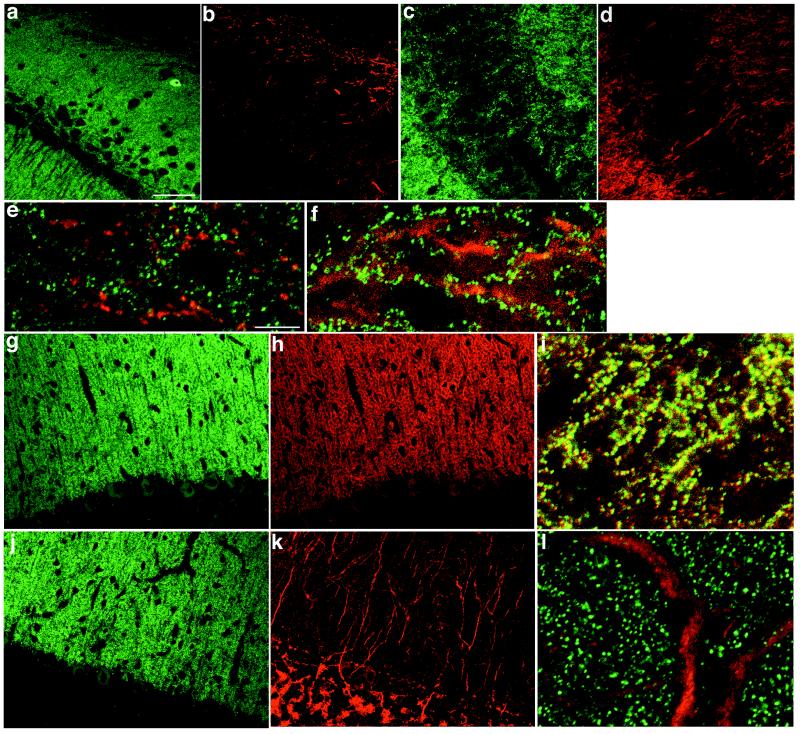
Intense immunoreactivity for PSD-Zip45 was observed in the cerebral cortex, the olfactory bulb, the hippocampus, and the cerebellar cortex, whereas faint reactivity was seen in the striatum and the brain stem (data not shown). Confocal microscopy showed that in hippocampal CA1 and CA3 areas, immunofluorescence was distributed in the stratum oriens and the stratum radiatum, but was less intense in the stratum pyramidale (Fig. 1 a and c). Immunofluorescence for mGluR1α was evident at the border between the stratum oriens and alveus of CA1 area (Fig. 1b). High-resolution images showed that punctate staining of PSD-Zip45 (green) and dendritic staining of mGluR1α (red) did not overlap (Fig. 1e). In CA1 and CA3 areas, mGluR5 staining was localized to the stratum oriens and the stratum radiatum and was less significant in the stratum pyramidale (Fig. 1d). High magnification of superimposed images revealed that punctate staining of PSD-Zip45 (green) was distributed along dendrites and cell bodies of hippocampal CA3 neurons, whereas mGluR5 staining (red) was diffusely distributed in dendrites and cell bodies of nonprincipal neurons (Fig. 1f).
Figure 1.
Distribution of PSD-Zip45 and group 1 mGluRs in the rat brain. Sagittal sections of CA1 (a, b, and e) and CA3 (c, d, and f) regions of the hippocampus and of the cerebellum (g–l) were double labeled with a mixture containing mAb 126H (green; a, c, e–g, i, j, and l) and rabbit polyclonal antibodies for mGluR1 (red; b, e, h, and i) or mGluR5 (red; d, f, k, and l). e, f, i, and l are superimposed images. Scale bars = 50 μm (a–d, g, h, j, and k), 10 μm (e, f, i, and l).
The PSD-Zip45 and mGluR1α immunoreactivities were observed mainly in the cerebellar molecular layer and were faint in the Purkinje cell bodies (Fig. 1 g and h). Double labeling of high-resolution images for PSD-Zip45 (green) and mGluR1α (red) yielded fused yellow punctate stainings along dendrites and neuronal cell bodies (Fig. 1i), indicating a colocalization of both proteins. The mGluR5 immunoreactivity was prominent in dendrites and cell bodies of Golgi cells (Fig. 1k). When superimposed, PSD-Zip45 (green) and mGluR5 (red) stainings in the cerebellar molecular layer were segregated (Fig. 1l).
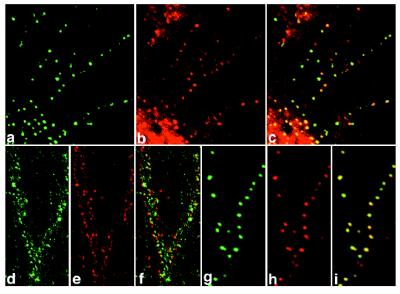
In cultured hippocampal neurons, punctate stainings of PSD-Zip45 and PSD-95, a postsynaptic protein, were colocalized well to dendrites and neuronal cell bodies (Fig. 2 a–c). By double labeling of PSD-Zip45 and synaptophysin, a specific presynaptic marker, the two immunostainings partially overlapped (Fig. 2 d–f). Punctate stainings of PSD-Zip45 and the NR1 subunit completely overlapped (Fig. 2 g–i). These results suggest that PSD-Zip45 and NMDA receptors are rather colocalized to dendritic spines.
Figure 2.
Immunocytochemical localization of PSD-Zip45 in cultured hippocampal neurons. At 21 days in culture, hippocampal neurons were double labeled with antibodies for PSD-Zip45 (green; a, d, and g) and PSD-95 (red; b), synaptophysin (red; e), or the NR1 subunit of NMDA receptors (red; h). The images were examined by confocal microscopy. c, f, and i are superimposed images.
Cluster Formation of Group 1 mGluRs Induced by PSD-Zip45.
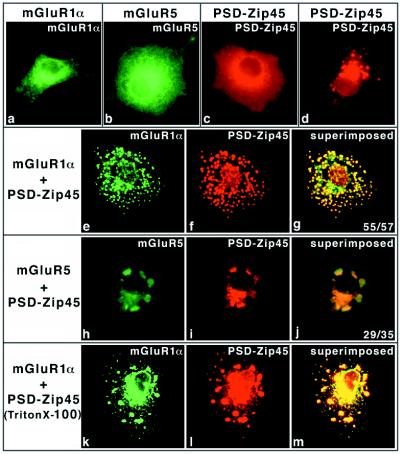
Using immunoprecipitation and Western blotting, we confirmed interaction between PSD-Zip45 and mGluR1α or mGluR5 in COS-7 cells (data not shown). To investigate the cellular localization of the proteins, plasmid DNAs encoding PSD-Zip45, mGluR1α, or mGluR5 alone or in combination were microinjected into COS-7 cells. The proteins were immunocytochemically observed at 3 hr after microinjection. When mGluR1α or mGluR5 was expressed alone, the stainings were diffuse from the plasma membrane to the cytoplasm (Fig. 3 a and b). In PSD-Zip45-microinjected cells, the staining was distributed throughout the cytoplasm and the plasma membrane. Speckle-like labelings were faintly observed (Fig. 3c), and their labelings were increased with increasing incubation time (9 hr) after microinjection (Fig. 3d). mGluR1α alone did not show such speckle-like labelings even after a 9-hr incubation (data not shown). When they were coexpressed, mGluR1α and PSD-Zip45 stainings with multiple clusters were strikingly colocalized to the plasma membrane and the perinuclear region (Fig. 3 e–g). The efficiency of mGluR1α clustering induced by PSD-Zip45 was more than 90%. Careful focusing made it possible to distinguish the localization of immunostainings between at the plasma membrane and the cytoplasmic structures (data not shown). mGluR5 clustering was also induced by PSD-Zip45 (Fig. 3 h–j).
Figure 3.
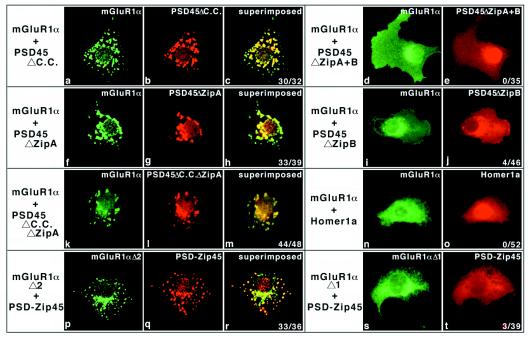
Group 1 mGluR clustering induced by PSD-Zip45. COS-7 cells were microinjected with plasmid DNAs listed at the top or left of each column. After microinjection (3 hr), the cells were fixed and double labeled with mAb 126H and/or anti-mGluR1α or anti-mGluR5 polyclonal antibodies. In the case of d, the cells were stained with mAb 126H at 9 hr after microinjection. The stained proteins are indicated (Top). In some experiments (k–m), the cells were permeabilized with 0.1% TritonX-100 before fixation. (g and j, Inset) Clustering scores (cell number showing clusters/examined cells). Final concentrations of plasmid DNAs are as follows; mGluR1α (0.1 mg/ml), mGluR5 (0.2 mg/ml), and PSD-Zip45 (0.2 mg/ml).
Even when coexpressed, cells were extracted with Triton X-100 before fixation, and mGluR1α and PSD-Zip45 coclusters were still evident (Fig. 3 k–m), indicating a cytoskeletal linkage. We also examined the effect of PSD-Zip45 on membrane targeting of mGluR1α. Fluorescence quantification assays revealed no significant difference with or without PSD-Zip45 (data not shown), suggesting that PSD-Zip45 has no effect on membrane targeting of mGluR1α.
Self-Multimerization and mGluR Clustering Mediated by a Unique Leucine-Zipper Motif of PSD-Zip45.
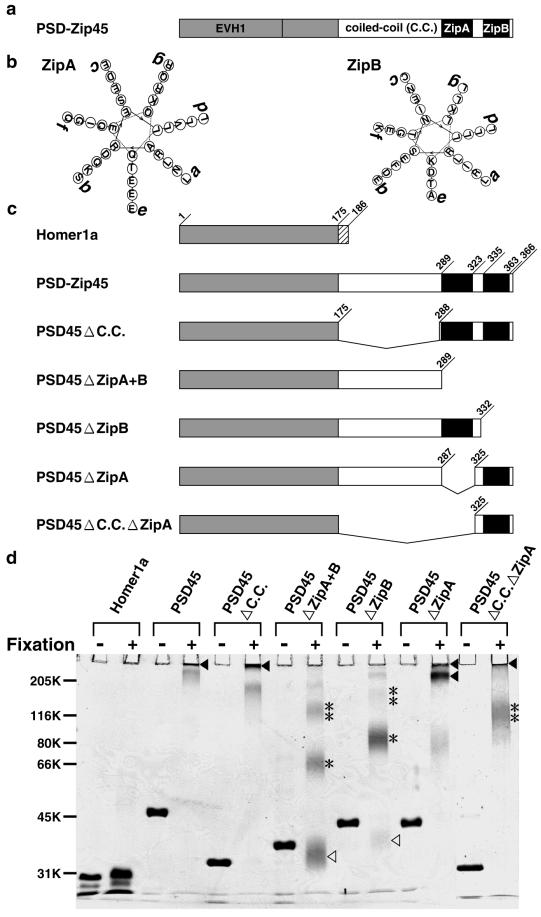
We previously demonstrated the self-multimerizing ability of PSD-Zip45 mediated through its COOH terminus, using in vitro translation assay (18). To further identify a critical motif(s) contributing to self-multimerization, we used purified PSD-Zip45 and its deleted proteins (Fig. 4c). The proteins treated with or without glutaraldehyde were subjected to SDS/PAGE, followed by Coomassie staining (Fig. 4d). Without crosslinking, each protein migrated as a monomer on SDS/PAGE. When crosslinked, PSD-Zip45, PSD45ΔC.C., PSD45ΔZipA, and PSD45ΔC.C.ΔZipA were retained on or near the gel top. Even with 5% SDS/PAGE, the crosslinked proteins did not enter the gels or formed high molecular mass complexes (>300 kDa), indicating self-multimerization. Of these, PSD45ΔC.C.ΔZipA, which consists of the NH2 terminus of PSD-Zip45 and ZipB domain, formed multimers in addition tetramers. The PSD45ΔZipA+B and PSD45ΔZipB migrated as monomers, dimers, and tetramers. Because of intramolecular crosslinking, migrated bands were broad. Homer 1a showed neither dimerization nor multimerization.
Figure 4.
Self-multimerization of PSD-Zip45 mediated through the extreme COOH-terminal leucine zipper motif. (a) Schematic diagram of structural domains of PSD-Zip45. (b) Helical wheel projection of ZipA and ZipB, respectively. (c) Schematic diagram of PSD-Zip45 and its deletion mutants. (d) Self-multimerization assay. All recombinant proteins were treated with or without glutaraldehyde and were separated by SDS/PAGE (14%). Δ, monomer; *, dimer; **, tetramer; ▴, multimer.
We then investigated a critical motif(s) of PSD-Zip45 involved in mGluR clustering. In addition to PSD-Zip45, PSD45ΔC.C, PSD45ΔZipA, and PSD45ΔC.C.ΔZipA induced mGluR1α clustering (Fig. 5 a–c, f–h, and k–m), but PSD45ΔZipA + B, PSD45ΔZipB, and Homer 1a did not (Fig. 5 d and e, i and j, n and o). The same results were obtained by using mGluR5 (data not shown). Taken together, a multimer, but not dimer or tetramer, formation of PSD-Zip45 or its deletion mutants is essential for mGluR clustering. These results indicate that ZipB in PSD-Zip45 plays a critical role in self-multimerization and PSD-Zip45-induced mGluR clustering.
Figure 5.
Identification of critical motif(s) of PSD-Zip45 required for mGluR1α clustering. Plasmid DNAs listed at the left of each column were microinjected into COS-7 cells. After 3 hr, the cells were fixed and stained with appropriate antibodies. (c, e, h, r, o, r, and t Inset) Clustering scores. Final concentrations of plasmid DNAs are as follows: mGluR1α and its deletion mutants (0.1 mg/ml) and PSD-Zip45 and its deletion mutants (0.2 mg/ml). mGluR1αΔ1 and mGluR1αΔ2 are mutant forms lacking residues 1150–1158 (LTPPSPFRD) and 1159–1179 (SVASGSSVPSSPVSESVLCTP), respectively.
Tu et al. have identified a proline-rich sequence in the cytoplasmic domain of group 1 mGluRs as a binding motif for PSD-Zip45 (24). Consistent with their finding, mGluR1αΔ1, but not mGluR1αΔ2, lost an ability of cluster formation induced by PSD-Zip45 (Fig. 5 p–t), indicating a requirement of PPXPF motif for the PSD-Zip45 binding.
Inhibition of PSD-Zip45-Induced mGluR1α Clustering by Homer 1a.
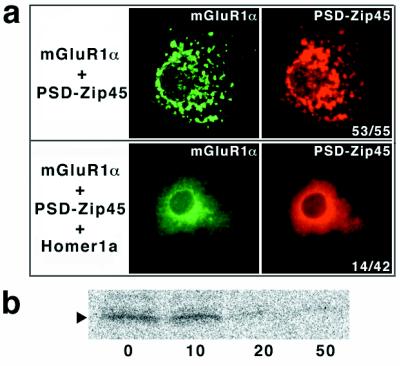
When PSD-Zip45 and Homer 1a plasmid DNAs were microinjected into COS-7 cells in a 1:10 ratio, PSD-Zip45-induced mGluR1α clustering was inhibited (Fig. 6a). Because of a difficulty in determining amounts of expressed proteins in COS-7 cells, we examined the effect of Homer1a on the PSD-Zip45 binding to the cytoplasmic domain of mGluR1α (Cyt mGluR1α). By using mAb 126H, the [35S]Cyt mGluR1α bound to PSD-Zip45 was detected in the immunoprecipitates. Interaction between PSD-Zip45 and [35S]Cyt mGluR1α was strongly suppressed by addition of more than a 10-fold molar excess of Homer 1a (Fig. 6b).
Figure 6.
Homer 1a inhibits interaction between mGluR1α and PSD-Zip45. (a) Inhibition of PSD-Zip45-induced mGluR1α clustering by Homer 1a. Plasmid DNAs for mGluR1α and PSD-Zip45 with (Lower) or without (Upper) Homer 1a were microinjected into COS7 cells. Final concentrations of plasmid DNAs are mGluR1α (0.1 mg/ml), PSD-Zip45 (0.2 mg/ml), and Homer 1a (2.0 mg/ml). After 3 hr, the cells were fixed and double labeled with appropriate antibodies. The stained proteins are indicated (Top). (b) Effect of Homer 1a on interaction between PSD-Zip45 and mGluR1α. The cytoplasmic domain of mGluR1α labeled with [35S]methionine (indicated by arrowheads) was incubated with PSD-Zip45 (3 μg/assay) and increasing amounts of Homer 1a. Molar ratios of PSD-Zip45 to Homer 1a were 0, 10, 20, and 50. The complex was immunoprecipitated with mAb 126H, and analyzed.
Discussion
Consistent with previous reports (27–29), mGluR1α and mGluR5 immunostainings in the hippocampus are observed mainly in dendrites and cell bodies of nonprincipal neurons (Fig. 1). In the cerebellum, punctate staining of mGluR1α is enriched in the molecular layer, and mGluR5 staining is restricted to the Golgi cells (Fig. 1). PSD-Zip45 shows a discrete punctate localization along dendrites and cell bodies of neurons in the hippocampus and the cerebellum (Fig. 1), indicating its localization to the synapse. In the cerebellar molecular layer, PSD-Zip45 and mGluR1α are entirely colocalized to synapses, suggesting that PSD-Zip45 may be involved in mGluR1α targeting to the cerebellar synapse. However, the distributions of PSD-Zip45 and group 1 mGluRs in the hippocampus and those of PSD-Zip45 and mGluR5 in the cerebellum are spatially segregated (Fig. 1). In cultured hippocampal neurons, punctate stainings for PSD-Zip45, PSD-95, and the NR1 subunit of NMDA receptors show complete overlapping patterns, whereas PSD-Zip45 and synaptophysin stainings partially overlap (Fig. 2). These results implicate the localization of PSD-Zip45 in hippocampal dendritic spines and suggest the possible linkage between PSD-Zip45 and NMDA receptors. Further study is required for PSD-Zip45 and NMDA receptor interaction mediated by some scaffold proteins, because their direct interaction has not been proven (15, 17).
The leucine-zipper motif is important for dimerization of transcription factors, such as fos and jun. Heptad leucine repeats are also present in a variety of cytoplasmic and transmembrane proteins, including cytoskeletal proteins and voltage-gated ion channels (27). A few leucine-zipper motif-containing proteins in the neuromuscular junction have been reported. Rapsyn (43-K protein), a neuromuscular protein involved in acetylcholine receptor (AChR) clustering, contains one leucine-zipper motif. Mutants that lack this motif retain self clustering but are unable to cluster the AChR (28). In addition to rapsyn, there are two neuromuscular proteins that contain leucine-zipper motifs: dystrophin and dystrobrevin. The COOH termini of both proteins appear to bind each other through heterodimerization of two leucine-zipper motifs (29). As demonstrated here, in vitro self-multimerization (Fig. 4d) and mGluR clustering assays (Figs. 3 and 5) indicate that self-multimerization of PSD-Zip45 mediated through ZipB plays a critical role in the induction of mGluR clustering. This leucine-zipper motif is composed of four heptad repeats (abcdefg)4, in which leucine (or isoleucine) residues occur uniquely at positions a, d, and g. Charged residues (glutamic acid, aspartic acid, and lysine) are positioned to make interhelical ion pairs (Fig. 4b). This motif is very stable and is responsible for self-multimerization. Together, the former reports regarding the neuromuscular junction and our study suggest that the leucine zipper may be a common structural motif for protein–protein interactions at subsynaptic sites. In particular, to our knowledge, PSD-Zip45 is the first example of scaffold protein for receptor clustering mediated by a unique leucine-zipper motif at the central synapse.
Although the NH2-terminal 175 amino acids of PSD-Zip45 and those of Homer 1a are identical, PSD-Zip45 is constitutively expressed, and Homer 1a is an immediate-response protein for a variety of neuronal stimuli (15, 16). Because PSD-Zip45-induced mGluR1α clustering is inhibited by overexpression of Homer 1a (Fig. 6), Homer 1a is speculated to be a natural competitor for PSD-Zip45. In vitro binding assay reveals that more than a 10-fold molar excess of Homer 1a is required for the inhibition of PSD-Zip45 and mGluR1α interaction. However, the expression level of Homer 1a induced by electroconvulsive seizure is calculated to be approximately equivalent to that of PSD-Zip45. This elevated level of Homer 1a is not sufficient to exert a competitive action on PSD-Zip45 and mGluR1α interaction in vivo. Further study will be required to reveal the biological significance of Homer 1a.
Regarding mGluR1α–PSD-Zip45 interaction, it has been demonstrated that the COOH-terminal sequence, TPPSPFR, of mGluR1α binds to the EVH1 domain of PSD-Zip45 (17, 24). Consistently, PSD-Zip45 cannot induce a cluster formation of mGluR1αΔ1, which lacks the COOH-terminal LTPPSPFRD sequence, in COS-7 cells (Fig. 5). Crystal structure study has advanced our understanding of this interaction (17). Because the NH2-terminal 141 amino acids of PSD-Zip45 are homologous to the EVH1 domain, this protein is expected to interact with the subcortical cytoskeleton (30). In fact, mGluR1α clusters complexed by PSD-Zip45 in COS-7 cells are resistant to Triton X-100 extraction (Fig. 3), suggesting a cytoskeletal linkage. We are currently investigating such cytoskeletal proteins (or their associated proteins) that bind PSD-Zip45.
Acknowledgments
We thank Dr. S. Nakanishi (Kyoto University) for kindly providing cDNA clones encoding mGluR1α and mGluR5. This work was supported partly by Grants-in-Aid for Center of Excellence research (to K.S.) and partly by Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan.
Abbreviations
- NMDA
N-methyl-d-aspartate
- AMPA
α-amino-3-hydroxy-5-methyl-4-isoxazole propionate
- EVH1
enabled/VASP homology 1
- mGluR
metabotropic glutamate receptor
References
- 1.Craig A M. Neuron. 1998;21:459–462. doi: 10.1016/s0896-6273(00)80555-3. [DOI] [PubMed] [Google Scholar]
- 2.Kornau H-C, Schenker L T, Kennedy M B, Seeburg P H. Science. 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- 3.Kim E, Niethammer M, Rothschild A, Jan Y H, Sheng M. Nature (London) 1995;378:85–88. doi: 10.1038/378085a0. [DOI] [PubMed] [Google Scholar]
- 4.Kim E, Cho K-O. Neuron. 1996;17:103–113. doi: 10.1016/s0896-6273(00)80284-6. [DOI] [PubMed] [Google Scholar]
- 5.Hsueh Y-P, Kim E, Sheng M. Neuron. 1997;18:803–814. doi: 10.1016/s0896-6273(00)80319-0. [DOI] [PubMed] [Google Scholar]
- 6.Topinka J R, Bredt D S. Neuron. 1998;20:125–134. doi: 10.1016/s0896-6273(00)80440-7. [DOI] [PubMed] [Google Scholar]
- 7.Craven S E, El-Husseini A E. Neuron. 1999;22:497–509. doi: 10.1016/s0896-6273(00)80705-9. [DOI] [PubMed] [Google Scholar]
- 8.Migaud M, Charlesworth P, Dempster M, Webster L C, Watabe A M, Makhinson M, He Y, Ramsay M F, Morris R G M, Morrison J H, et al. Nature (London) 1998;396:433–439. doi: 10.1038/24790. [DOI] [PubMed] [Google Scholar]
- 9.Kim E, Naisbitt S, Hsueh Y-P, Rao A, Rothschild A, Craig A M, Sheng M. J Cell Biol. 1997;136:669–678. doi: 10.1083/jcb.136.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takeuchi M, Hata Y, Hirao K, Toyoda A, Irie M, Takai Y. J Biol Chem. 1997;272:11943–11951. doi: 10.1074/jbc.272.18.11943. [DOI] [PubMed] [Google Scholar]
- 11.Kawashima N, Takamiya K, Sun J, Kitabatake A, Sobue K. FEBS Lett. 1997;418:301–304. doi: 10.1016/s0014-5793(97)01399-9. [DOI] [PubMed] [Google Scholar]
- 12.Dong H, O’Brien R J, Fung E T, Lanahan A A, Worley P F, Huganir R L. Nature (London) 1997;386:279–284. doi: 10.1038/386279a0. [DOI] [PubMed] [Google Scholar]
- 13.Srivastava S, Osten P, Vilim F S, Khatri L, Inman G, States B, Daly C, DeSouza S, Abagyan R, Valtschanoff J G, et al. Neuron. 1998;21:581–591. doi: 10.1016/s0896-6273(00)80568-1. [DOI] [PubMed] [Google Scholar]
- 14.Xia J, Zhang X, Staudinger J, Huganir R L. Neuron. 1999;22:179–187. doi: 10.1016/s0896-6273(00)80689-3. [DOI] [PubMed] [Google Scholar]
- 15.Brakeman P R, Lanahan A A, O’Brien R, K, Roche K, Barnes C A, Huganir R L, Worley P F. Nature (London) 1997;386:284–288. doi: 10.1038/386284a0. [DOI] [PubMed] [Google Scholar]
- 16.Kato A, Ozawa F, Saitoh Y, Hirai K, Inokuchi K. FEBS Lett. 1997;412:183–189. doi: 10.1016/s0014-5793(97)00775-8. [DOI] [PubMed] [Google Scholar]
- 17.Prehoda K E, Lee D J, Lim W A. Cell. 1999;97:471–480. doi: 10.1016/s0092-8674(00)80757-6. [DOI] [PubMed] [Google Scholar]
- 18.Sun J, Tadokoro S, Imanaka T, Murakami S D, Nakamura M, Kashiwada K, Ko J, Nishida W, Sobue K. FEBS Lett. 1998;437:304–308. doi: 10.1016/s0014-5793(98)01256-3. [DOI] [PubMed] [Google Scholar]
- 19.Xiao B, Tu J C, Petralia R S, Yuan J P, Doan A, Breder C D, Ruggiero A, Lanahan A A, Wenthold R J, Worley P F. Neuron. 1998;21:707–716. doi: 10.1016/s0896-6273(00)80588-7. [DOI] [PubMed] [Google Scholar]
- 20.Kato A, Ozawa F, Saitoh Y, Fukazawa Y, Sugiyama H, Inokuchi K. J Biol Chem. 1998;273:23969–23975. doi: 10.1074/jbc.273.37.23969. [DOI] [PubMed] [Google Scholar]
- 21.Masu M, Tanabe Y, Tsuchida K, Shigemoto R, Nakanishi S. Nature (London) 1991;349:760–765. doi: 10.1038/349760a0. [DOI] [PubMed] [Google Scholar]
- 22.Abe T, Sugihara H, Nawa H, Shigemoto R, Mizuno N, Nakanishi S. J Biol Chem. 1992;267:13361–13368. [PubMed] [Google Scholar]
- 23.Mizushima S, Nagata S. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tu J C, Xiao B, Yuan J P, Lanahan A A, Leoffert K, Li M, Linden D J, Worley P F. Neuron. 1998;21:717–726. doi: 10.1016/s0896-6273(00)80589-9. [DOI] [PubMed] [Google Scholar]
- 25.Shigemoto R, Kinoshita A, Wada E, Nomura S, Ohishi H, Takada M, Flor P J, Neki A, Abe T, Nakanishi S, Mizuno N. J Neurosci. 1997;17:7503–7522. doi: 10.1523/JNEUROSCI.17-19-07503.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romano C, Sesma M A, McDonald C T, O’Malley K, Van den Pol A N, Olney J W. J Comp Neurol. 1995;355:455–469. doi: 10.1002/cne.903550310. [DOI] [PubMed] [Google Scholar]
- 27.Lupas A. Trends Biochem Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- 28.Phillips W D, Maimone M M, Merlie J P. J Cell Biol. 1991;115:1713–1723. doi: 10.1083/jcb.115.6.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sadoulet-Puccio H M, Rajala M, Kunkel L M. Proc Natl Acad Sci USA. 1997;94:12413–12418. doi: 10.1073/pnas.94.23.12413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beckerle M C. Cell. 1998;95:741–748. doi: 10.1016/s0092-8674(00)81697-9. [DOI] [PubMed] [Google Scholar]