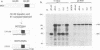Abstract
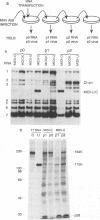
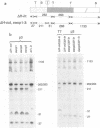
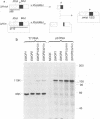
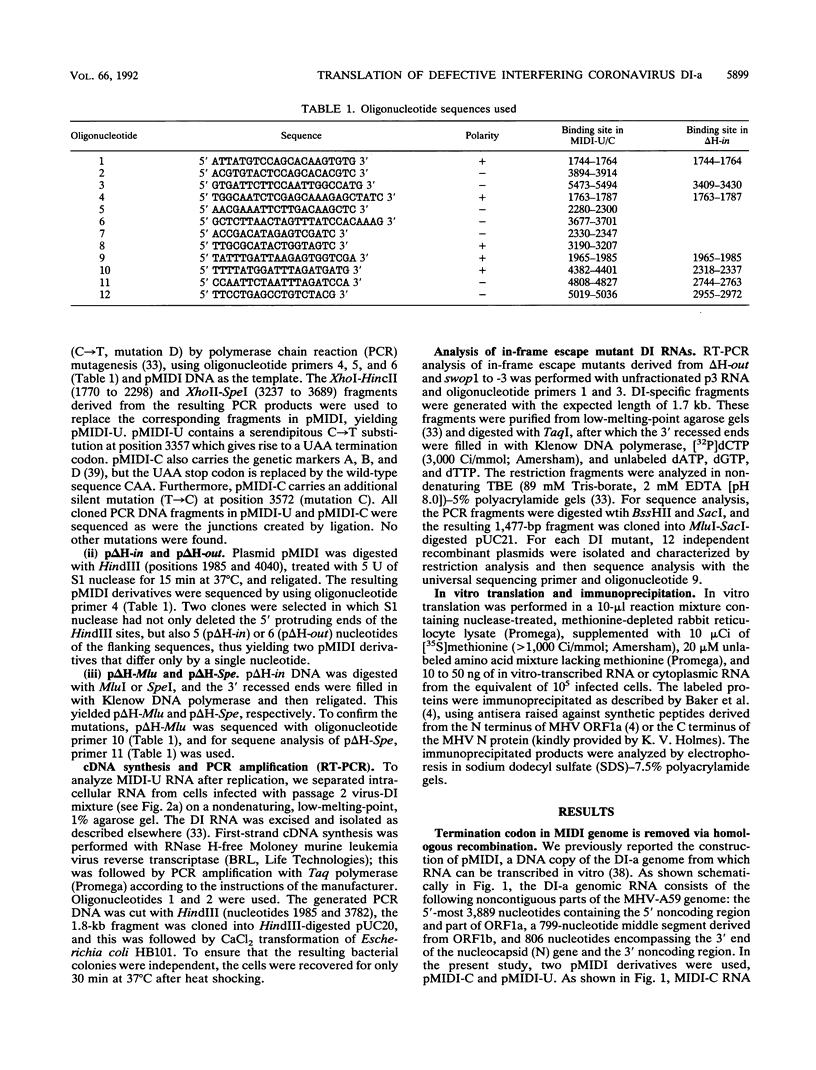
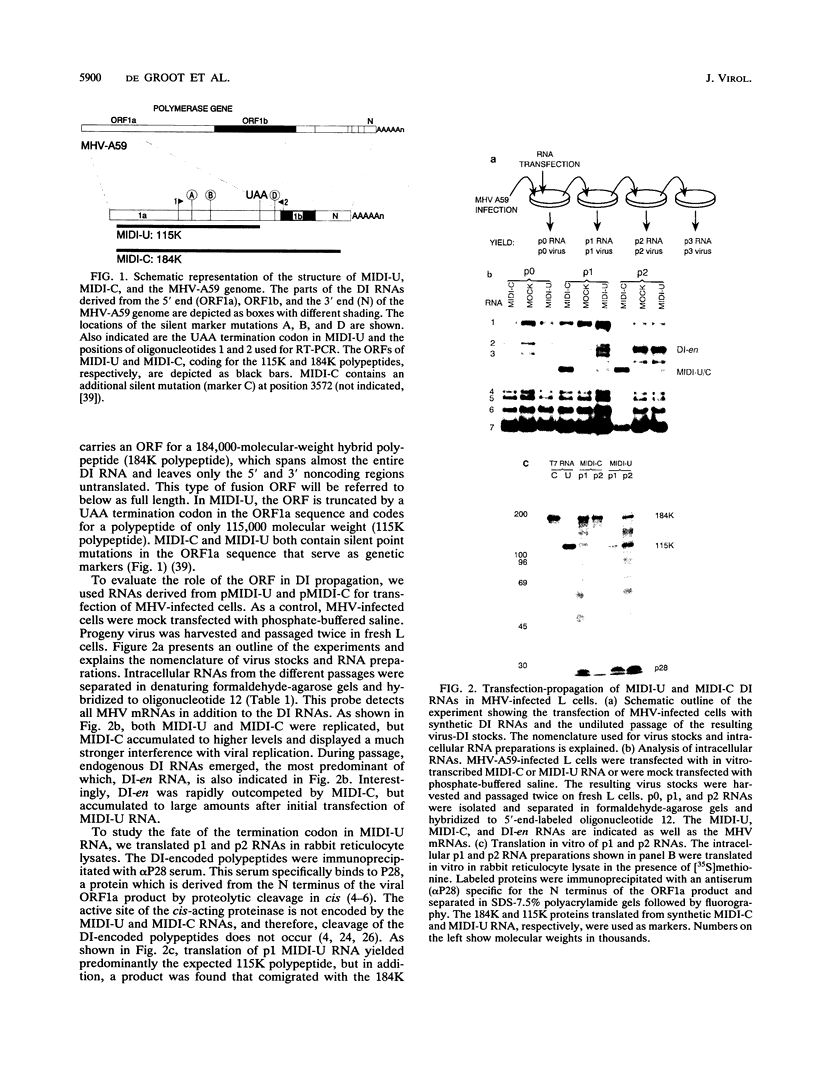
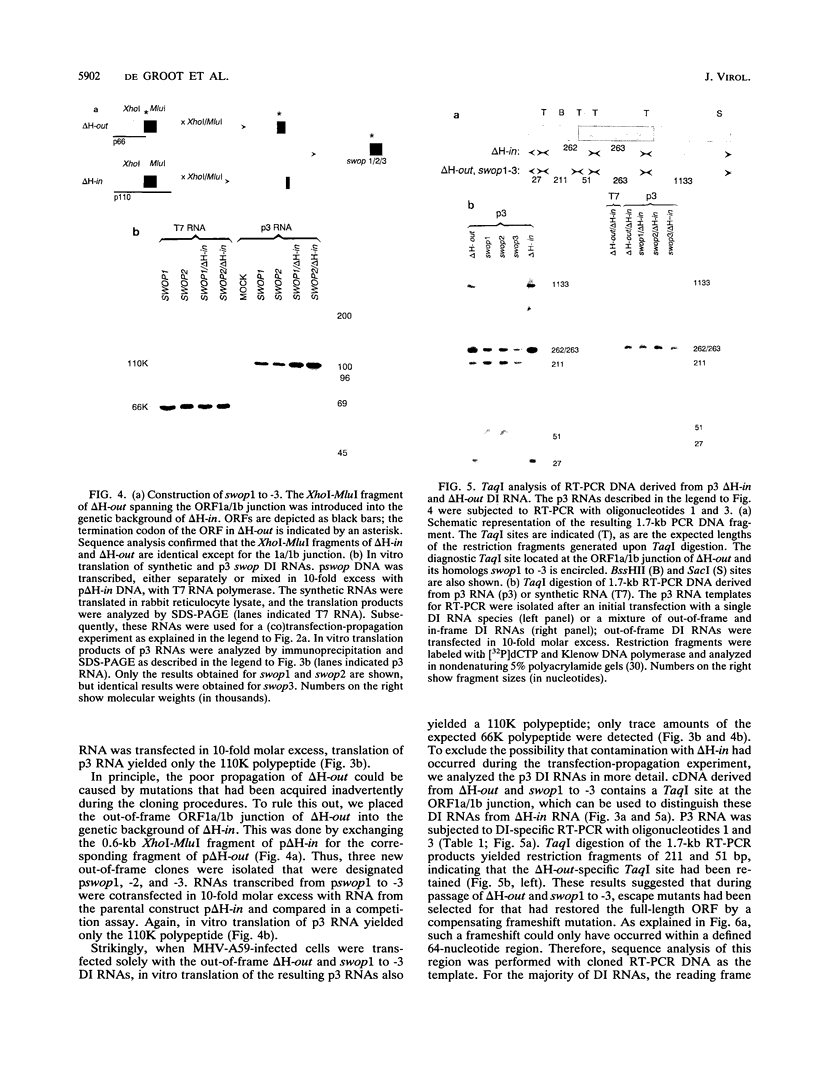
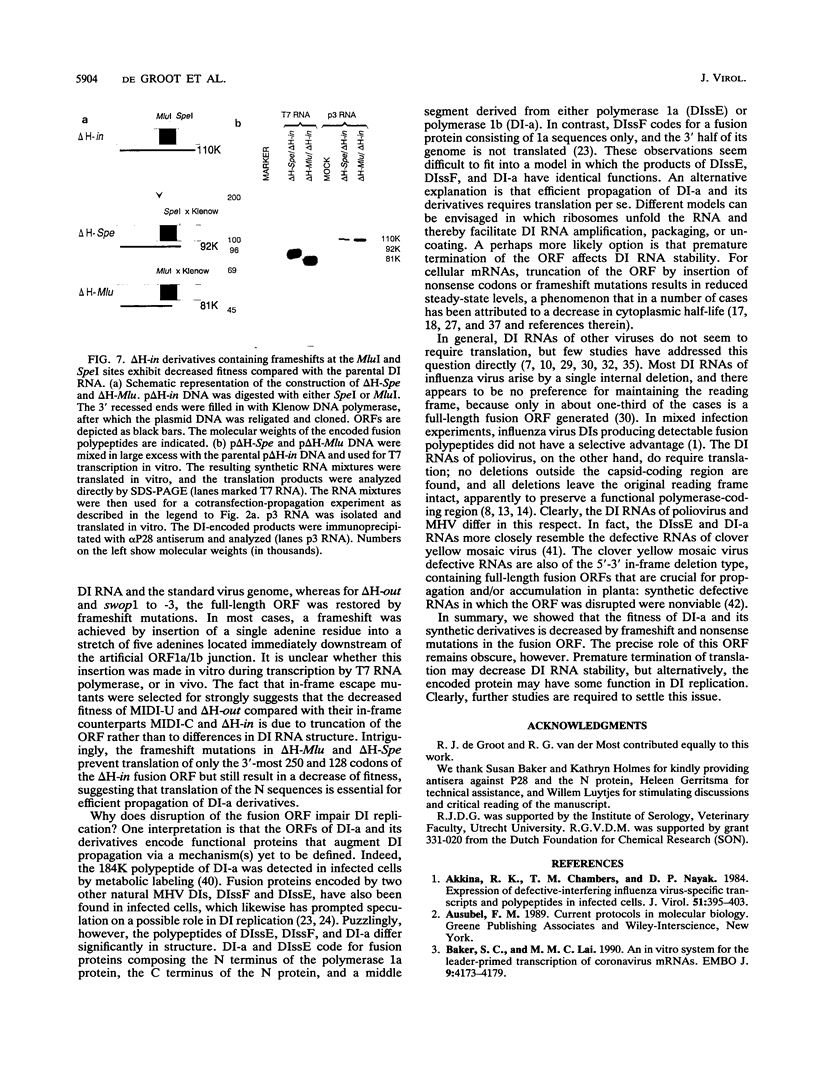
The genome of the defective interfering (DI) mouse hepatitis virus DI-a carries a large open reading frame (ORF) consisting of ORF1a, ORF1b, and nucleocapsid sequences. To test whether this fusion ORF is important for DI virus replication, we constructed derivatives of the DI-a genome in which the reading frame was truncated by a nonsense codon or a frameshift mutation. In vitro-transcribed DI RNAs were transfected into mouse hepatitis virus-infected cells followed by undiluted passage of the resulting virus-DI virus stocks. The following observations were made. (i) Truncation of the fusion ORF was not lethal but led to reduced accumulation of DI RNA. (ii) When pairs of nearly identical in-frame and out-of-frame DI RNAs were directly compared by cotransfection, DI viruses containing in-frame genomic RNAs prevailed within three successive passage even when the out-of-frame RNAs were transfected in 10-fold molar excess. (iii) When DI viruses containing out-of-frame genomic RNAs were passaged, mutants emerged and were selected for that had restored the reading frame. We conclude that translation of the fusion ORF is indeed required for efficient propagation of DI-a and its derivatives.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akkina R. K., Chambers T. M., Nayak D. P. Expression of defective-interfering influenza virus-specific transcripts and polypeptides in infected cells. J Virol. 1984 Aug;51(2):395–403. doi: 10.1128/jvi.51.2.395-403.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S. C., Lai M. M. An in vitro system for the leader-primed transcription of coronavirus mRNAs. EMBO J. 1990 Dec;9(12):4173–4179. doi: 10.1002/j.1460-2075.1990.tb07641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S. C., Shieh C. K., Soe L. H., Chang M. F., Vannier D. M., Lai M. M. Identification of a domain required for autoproteolytic cleavage of murine coronavirus gene A polyprotein. J Virol. 1989 Sep;63(9):3693–3699. doi: 10.1128/jvi.63.9.3693-3699.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison M. R., Perlman S. Translation and processing of mouse hepatitis virus virion RNA in a cell-free system. J Virol. 1986 Oct;60(1):12–18. doi: 10.1128/jvi.60.1.12-18.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison M., Perlman S. Identification of putative polymerase gene product in cells infected with murine coronavirus A59. Virology. 1987 Apr;157(2):565–568. doi: 10.1016/0042-6822(87)90303-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagino-Yamagishi K., Nomoto A. In vitro construction of poliovirus defective interfering particles. J Virol. 1989 Dec;63(12):5386–5392. doi: 10.1128/jvi.63.12.5386-5392.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann M. A., Sethna P. B., Brian D. A. Bovine coronavirus mRNA replication continues throughout persistent infection in cell culture. J Virol. 1990 Sep;64(9):4108–4114. doi: 10.1128/jvi.64.9.4108-4114.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A. S., Baltimore D. Defective viral particles and viral disease processes. Nature. 1970 Apr 25;226(5243):325–327. doi: 10.1038/226325a0. [DOI] [PubMed] [Google Scholar]
- Kaplan G., Racaniello V. R. Construction and characterization of poliovirus subgenomic replicons. J Virol. 1988 May;62(5):1687–1696. doi: 10.1128/jvi.62.5.1687-1696.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge S., Saito I., Nomoto A. Primary structure of poliovirus defective-interfering particle genomes and possible generation mechanisms of the particles. J Mol Biol. 1986 Dec 5;192(3):473–487. doi: 10.1016/0022-2836(86)90270-6. [DOI] [PubMed] [Google Scholar]
- Lai M. M., Baric R. S., Makino S., Keck J. G., Egbert J., Leibowitz J. L., Stohlman S. A. Recombination between nonsegmented RNA genomes of murine coronaviruses. J Virol. 1985 Nov;56(2):449–456. doi: 10.1128/jvi.56.2.449-456.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. J., Shieh C. K., Gorbalenya A. E., Koonin E. V., La Monica N., Tuler J., Bagdzhadzhyan A., Lai M. M. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991 Feb;180(2):567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S. K., Sigmund C. D., Gross K. W., Maquat L. E. Nonsense codons in human beta-globin mRNA result in the production of mRNA degradation products. Mol Cell Biol. 1992 Mar;12(3):1149–1161. doi: 10.1128/mcb.12.3.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losson R., Lacroute F. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5134–5137. doi: 10.1073/pnas.76.10.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Fujioka N., Fujiwara K. Structure of the intracellular defective viral RNAs of defective interfering particles of mouse hepatitis virus. J Virol. 1985 May;54(2):329–336. doi: 10.1128/jvi.54.2.329-336.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Joo M., Makino J. K. A system for study of coronavirus mRNA synthesis: a regulated, expressed subgenomic defective interfering RNA results from intergenic site insertion. J Virol. 1991 Nov;65(11):6031–6041. doi: 10.1128/jvi.65.11.6031-6041.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Keck J. G., Stohlman S. A., Lai M. M. High-frequency RNA recombination of murine coronaviruses. J Virol. 1986 Mar;57(3):729–737. doi: 10.1128/jvi.57.3.729-737.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Lai M. M. High-frequency leader sequence switching during coronavirus defective interfering RNA replication. J Virol. 1989 Dec;63(12):5285–5292. doi: 10.1128/jvi.63.12.5285-5292.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Shieh C. K., Keck J. G., Lai M. M. Defective-interfering particles of murine coronavirus: mechanism of synthesis of defective viral RNAs. Virology. 1988 Mar;163(1):104–111. doi: 10.1016/0042-6822(88)90237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Taguchi F., Fujiwara K. Defective interfering particles of mouse hepatitis virus. Virology. 1984 Feb;133(1):9–17. doi: 10.1016/0042-6822(84)90420-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Yokomori K., Lai M. M. Analysis of efficiently packaged defective interfering RNAs of murine coronavirus: localization of a possible RNA-packaging signal. J Virol. 1990 Dec;64(12):6045–6053. doi: 10.1128/jvi.64.12.6045-6053.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat L. E., Kinniburgh A. J., Rachmilewitz E. A., Ross J. Unstable beta-globin mRNA in mRNA-deficient beta o thalassemia. Cell. 1981 Dec;27(3 Pt 2):543–553. doi: 10.1016/0092-8674(81)90396-2. [DOI] [PubMed] [Google Scholar]
- Meinkoth J., Wahl G. Hybridization of nucleic acids immobilized on solid supports. Anal Biochem. 1984 May 1;138(2):267–284. doi: 10.1016/0003-2697(84)90808-x. [DOI] [PubMed] [Google Scholar]
- Monroe S. S., Schlesinger S. Common and distinct regions of defective-interfering RNAs of Sindbis virus. J Virol. 1984 Mar;49(3):865–872. doi: 10.1128/jvi.49.3.865-872.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore L. L., Bostick D. A., Garry R. F. Sindbis virus infection decreases intracellular pH: alkaline medium inhibits processing of Sindbis virus polyproteins. Virology. 1988 Sep;166(1):1–9. doi: 10.1016/0042-6822(88)90139-0. [DOI] [PubMed] [Google Scholar]
- Pachuk C. J., Bredenbeek P. J., Zoltick P. W., Spaan W. J., Weiss S. R. Molecular cloning of the gene encoding the putative polymerase of mouse hepatitis coronavirus, strain A59. Virology. 1989 Jul;171(1):141–148. doi: 10.1016/0042-6822(89)90520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux L., Simon A. E., Holland J. J. Effects of defective interfering viruses on virus replication and pathogenesis in vitro and in vivo. Adv Virus Res. 1991;40:181–211. doi: 10.1016/S0065-3527(08)60279-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S. G., Sawicki D. L. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J Virol. 1990 Mar;64(3):1050–1056. doi: 10.1128/jvi.64.3.1050-1056.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E. J., den Boon J. A., Horzinek M. C., Spaan W. J. Characterization of defective interfering RNAs of Berne virus. J Gen Virol. 1991 Jul;72(Pt 7):1635–1643. doi: 10.1099/0022-1317-72-7-1635. [DOI] [PubMed] [Google Scholar]
- Spaan W. J., Rottier P. J., Horzinek M. C., van der Zeijst B. A. Isolation and identification of virus-specific mRNAs in cells infected with mouse hepatitis virus (MHV-A59). Virology. 1981 Jan 30;108(2):424–434. doi: 10.1016/0042-6822(81)90449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vancanneyt G., Rosahl S., Willmitzer L. Translatability of a plant-mRNA strongly influences its accumulation in transgenic plants. Nucleic Acids Res. 1990 May 25;18(10):2917–2921. doi: 10.1093/nar/18.10.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K. A., Bancroft J. B., Mackie G. A. Coding capacity determines in vivo accumulation of a defective RNA of clover yellow mosaic virus. J Virol. 1992 May;66(5):3069–3076. doi: 10.1128/jvi.66.5.3069-3076.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K. A., Bancroft J. B., Mackie G. A. Defective RNAs of clover yellow mosaic virus encode nonstructural/coat protein fusion products. Virology. 1991 Aug;183(2):479–486. doi: 10.1016/0042-6822(91)90977-J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Most R. G., Bredenbeek P. J., Spaan W. J. A domain at the 3' end of the polymerase gene is essential for encapsidation of coronavirus defective interfering RNAs. J Virol. 1991 Jun;65(6):3219–3226. doi: 10.1128/jvi.65.6.3219-3226.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Most R. G., Heijnen L., Spaan W. J., de Groot R. J. Homologous RNA recombination allows efficient introduction of site-specific mutations into the genome of coronavirus MHV-A59 via synthetic co-replicating RNAs. Nucleic Acids Res. 1992 Jul 11;20(13):3375–3381. doi: 10.1093/nar/20.13.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]