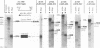Abstract
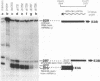
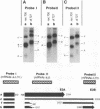
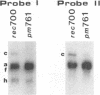
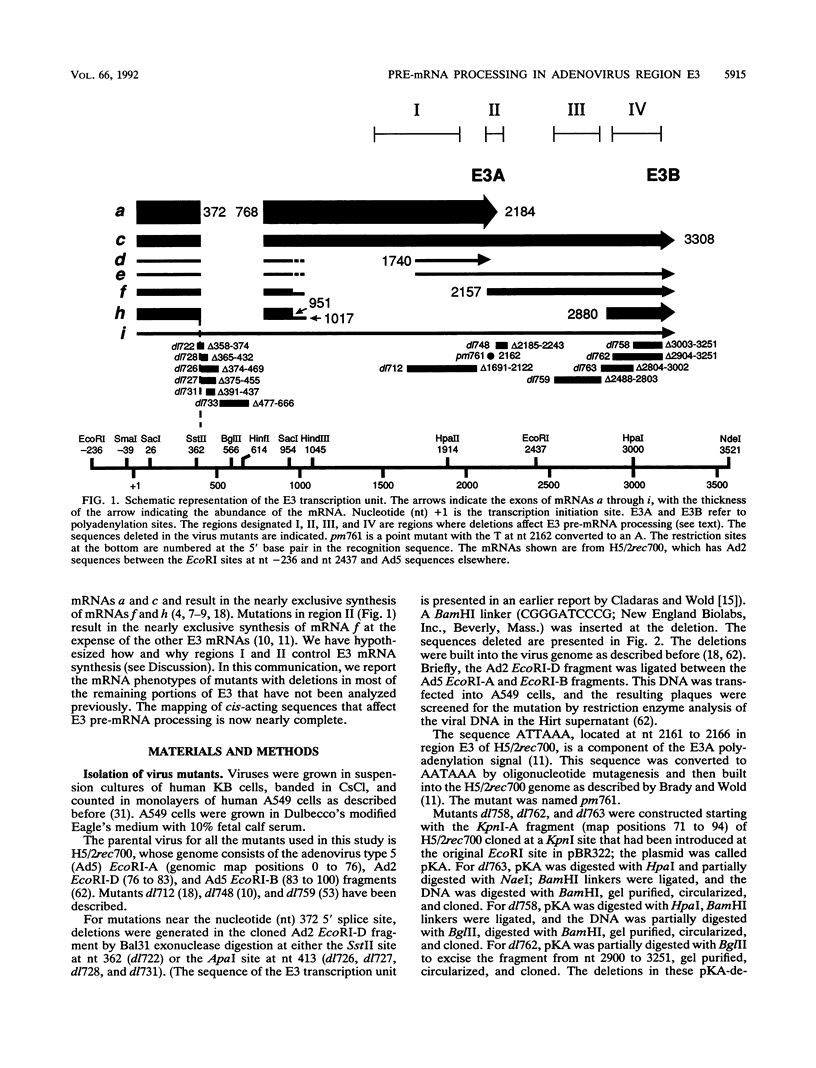
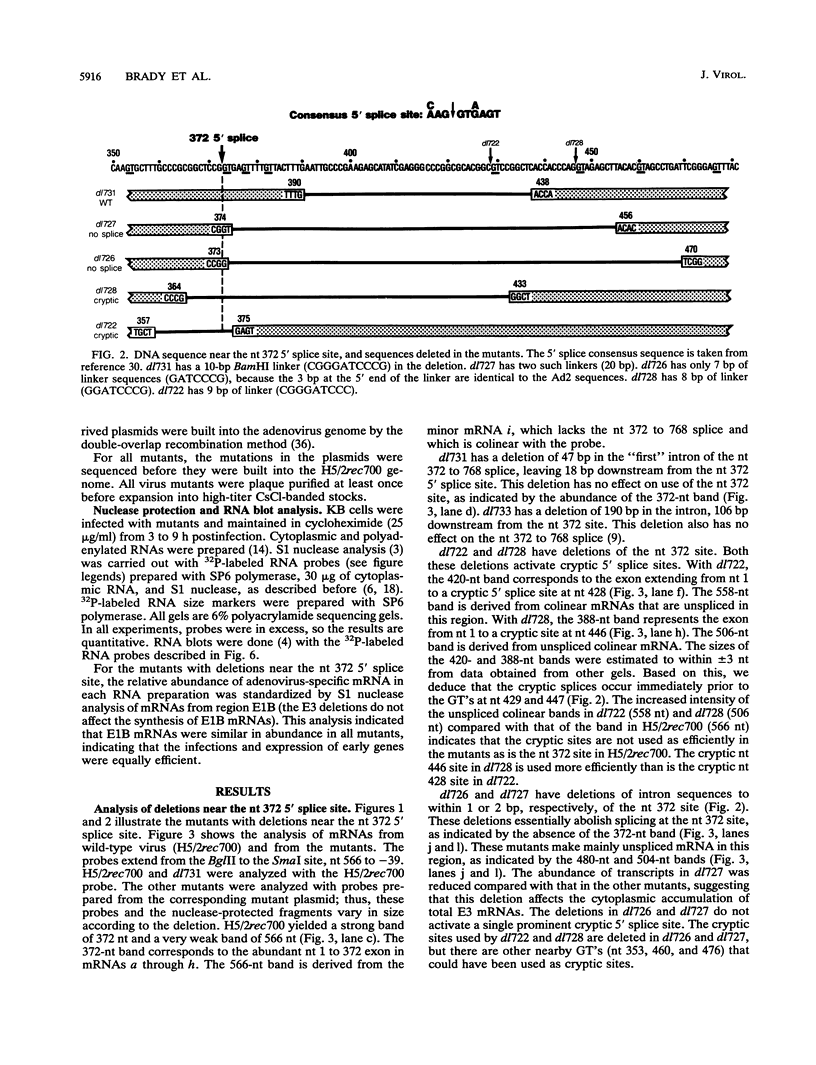
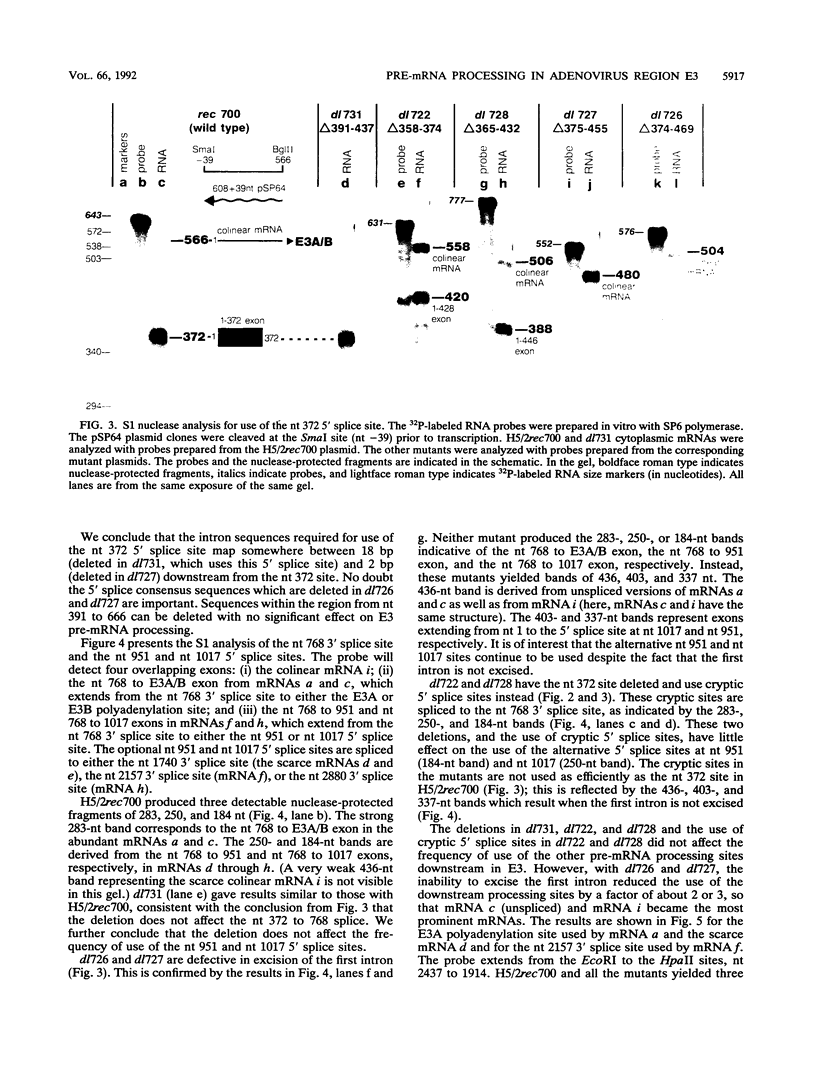
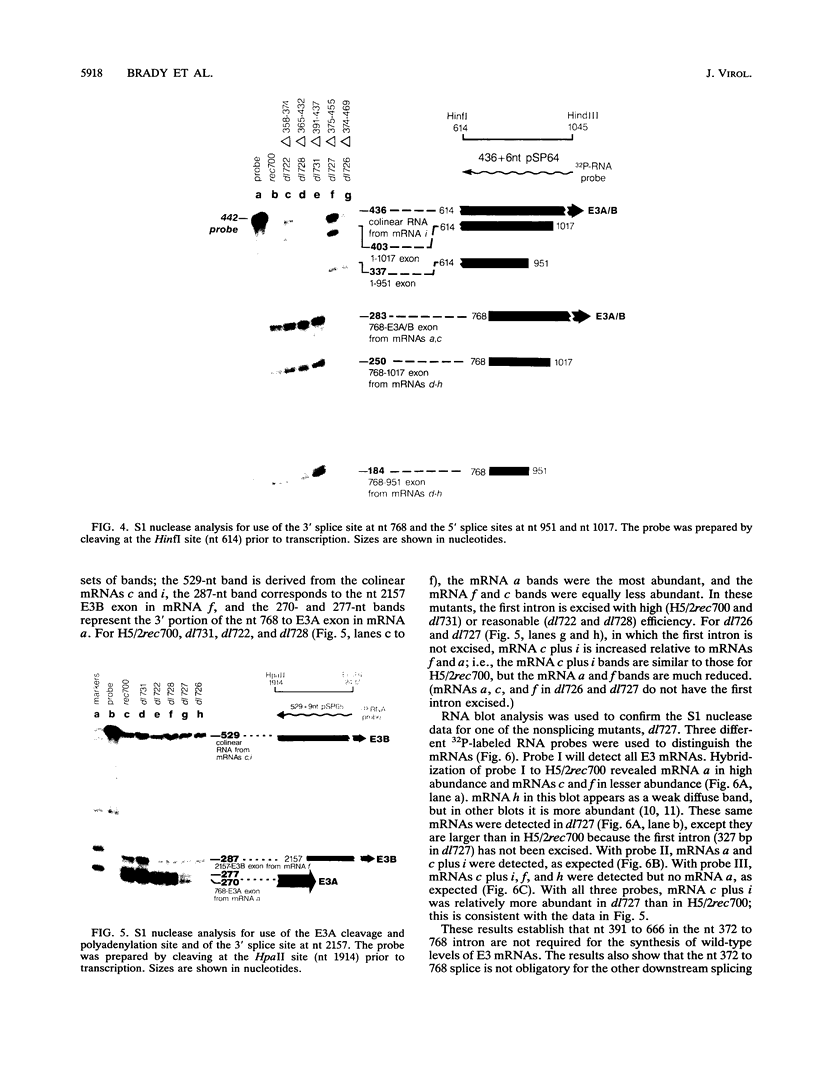
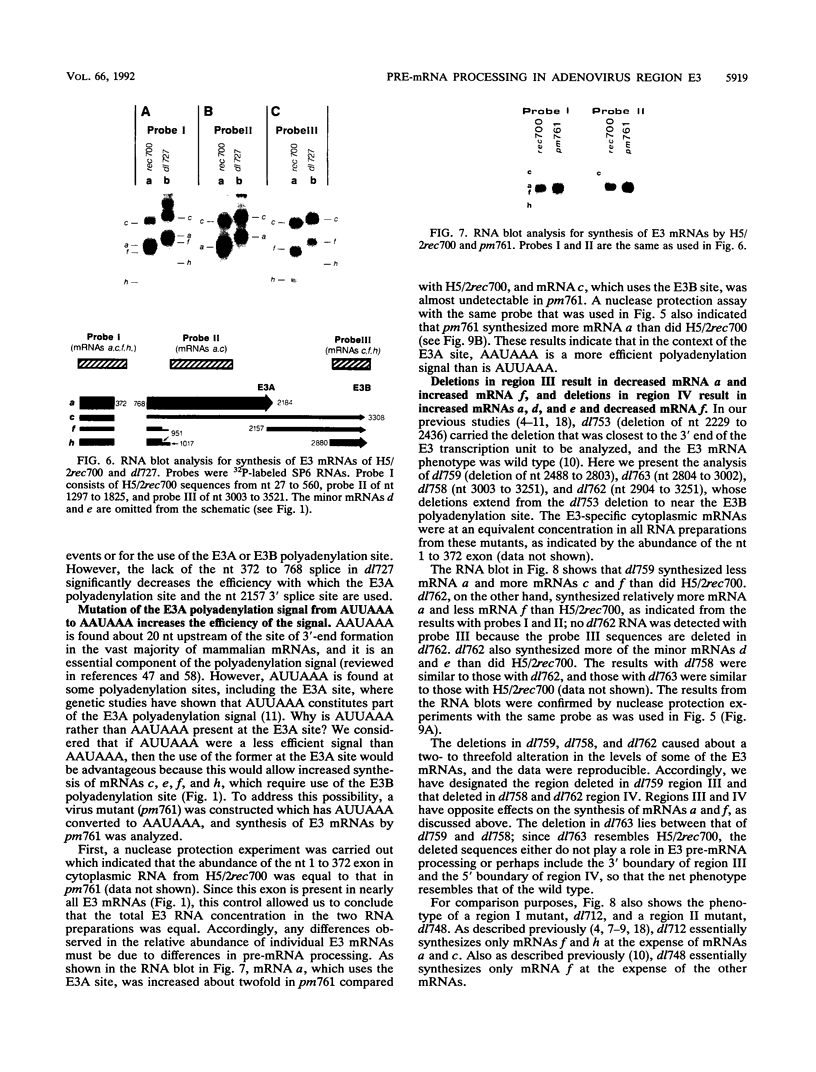
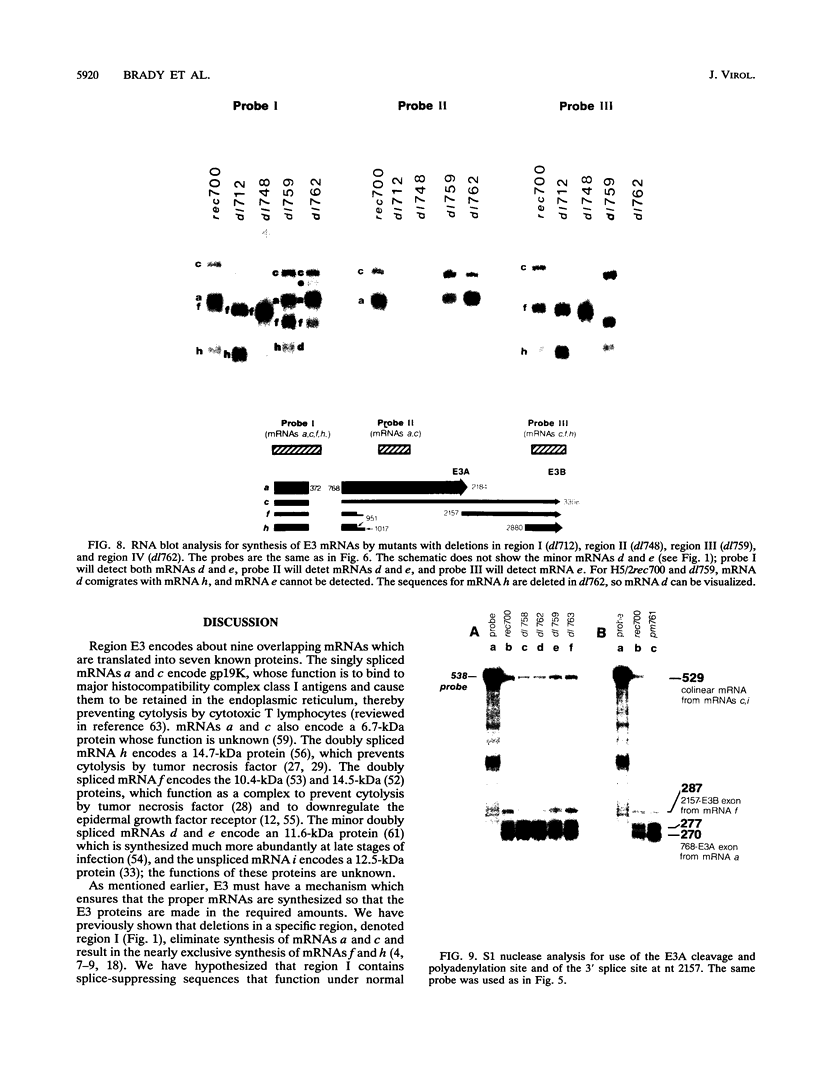
The E3 complex transcription unit of adenovirus encodes four major mRNAs (a, c, f, and h) and two minor (d and e) mRNAs with overlapping exons, alternative splice sites, and two polyadenylation sites, termed E3A (upstream) and E3B (downstream). mRNAs a and d use the E3A polyadenylation site, and mRNAs c, e, f, and h use the E3B site. We have analyzed virus mutants with deletions throughout the E3 region in order to identify cis-acting sequences that function in E3 pre-mRNA processing. The results presented in this report as well as previous results are summarized as follows. (i) Deletions in the first (5') intron at nucleotides (nt) 372 to 768 in E3 had no effect unless they removed the consensus sequence for the nt 372 5' splice site; however, the overall pattern of E3 mRNAs did not change significantly. (ii) Deletions in region I (nt 1441 to 2044) eliminated mRNAs a and c and resulted in corresponding increases in mRNAs f and h; we propose that region I contains sequences that suppress splicing. (iii) Mutations in region II (nt 2161 to 2243) resulted in nearly exclusive synthesis of mRNA f; this phenotype is understood and is discussed. (iv) Changing the AUUAAA component of the E3A poly(A) addition signal to AAUAAA resulted in increased mRNA a levels, suggesting that the E3A poly(A) addition signal is intrinsically inefficient. (v) Deletions in region III (nt 2488 to 3002) decreased mRNA a levels about two- to threefold and specifically increased mRNA f levels; we suggest that region III facilitates use of the E3A polyadenylation site. (vi) Deletions in region IV (nt 2904 to 3251) increased mRNA a levels about two- to threefold; we suggest that region IV may contain sequences that facilitate use of the E3B polyadenylation site. A map of sequences that determine alternative pre-mRNA processing in region E3 is now nearly complete.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adami G., Nevins J. R. Splice site selection dominates over poly(A) site choice in RNA production from complex adenovirus transcription units. EMBO J. 1988 Jul;7(7):2107–2116. doi: 10.1002/j.1460-2075.1988.tb03050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrigo S., Beemon K. Regulation of Rous sarcoma virus RNA splicing and stability. Mol Cell Biol. 1988 Nov;8(11):4858–4867. doi: 10.1128/mcb.8.11.4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Bhat B. M., Brady H. A., Pursley M. H., Wold W. S. Deletion mutants that alter differential RNA processing in the E3 complex transcription unit of adenovirus. J Mol Biol. 1986 Aug 20;190(4):543–557. doi: 10.1016/0022-2836(86)90240-8. [DOI] [PubMed] [Google Scholar]
- Bhat B. M., Brady H. A., Wold W. S. Virus deletion mutants that affect a 3' splice site in the E3 transcription unit of adenovirus 2. Mol Cell Biol. 1985 Sep;5(9):2405–2413. doi: 10.1128/mcb.5.9.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat B. M., Wold W. S. A small deletion distant from a splice or polyadenylation site dramatically alters pre-mRNA processing in region E3 of adenovirus. J Virol. 1987 Dec;61(12):3938–3945. doi: 10.1128/jvi.61.12.3938-3945.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat B. M., Wold W. S. ATTAAA as well as downstream sequences are required for RNA 3'-end formation in the E3 complex transcription unit of adenovirus. Mol Cell Biol. 1985 Nov;5(11):3183–3193. doi: 10.1128/mcb.5.11.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat B. M., Wold W. S. Adenovirus mutants with splice-enhancing mutations in the E3 complex transcription unit are also defective in E3A RNA 3'-end formation. J Virol. 1986 Mar;57(3):1155–1158. doi: 10.1128/jvi.57.3.1155-1158.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat B. M., Wold W. S. Genetic analysis of mRNA synthesis in adenovirus region E3 at different stages of productive infection by RNA-processing mutants. J Virol. 1986 Oct;60(1):54–63. doi: 10.1128/jvi.60.1.54-63.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady H. A., Wold W. S. Competition between splicing and polyadenylation reactions determines which adenovirus region E3 mRNAs are synthesized. Mol Cell Biol. 1988 Aug;8(8):3291–3297. doi: 10.1128/mcb.8.8.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady H. A., Wold W. S. Identification of a novel sequence that governs both polyadenylation and alternative splicing in region E3 of adenovirus. Nucleic Acids Res. 1987 Nov 25;15(22):9397–9416. doi: 10.1093/nar/15.22.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlin C. R., Tollefson A. E., Brady H. A., Hoffman B. L., Wold W. S. Epidermal growth factor receptor is down-regulated by a 10,400 MW protein encoded by the E3 region of adenovirus. Cell. 1989 Apr 7;57(1):135–144. doi: 10.1016/0092-8674(89)90179-7. [DOI] [PubMed] [Google Scholar]
- Carswell S., Alwine J. C. Efficiency of utilization of the simian virus 40 late polyadenylation site: effects of upstream sequences. Mol Cell Biol. 1989 Oct;9(10):4248–4258. doi: 10.1128/mcb.9.10.4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cladaras C., Bhat B., Wold W. S. Mapping the 5' ends, 3' ends, and splice sites of mRNAs from the early E3 transcription unit of adenovirus 5. Virology. 1985 Jan 15;140(1):44–54. doi: 10.1016/0042-6822(85)90444-1. [DOI] [PubMed] [Google Scholar]
- Cladaras C., Wold W. S. DNA sequence of the early E3 transcription unit of adenovirus 5. Virology. 1985 Jan 15;140(1):28–43. doi: 10.1016/0042-6822(85)90443-x. [DOI] [PubMed] [Google Scholar]
- Connelly S., Manley J. L. A CCAAT box sequence in the adenovirus major late promoter functions as part of an RNA polymerase II termination signal. Cell. 1989 May 19;57(4):561–571. doi: 10.1016/0092-8674(89)90126-8. [DOI] [PubMed] [Google Scholar]
- Connelly S., Manley J. L. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes Dev. 1988 Apr;2(4):440–452. doi: 10.1101/gad.2.4.440. [DOI] [PubMed] [Google Scholar]
- DeZazzo J. D., Imperiale M. J. Sequences upstream of AAUAAA influence poly(A) site selection in a complex transcription unit. Mol Cell Biol. 1989 Nov;9(11):4951–4961. doi: 10.1128/mcb.9.11.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZazzo J. D., Kilpatrick J. E., Imperiale M. J. Involvement of long terminal repeat U3 sequences overlapping the transcription control region in human immunodeficiency virus type 1 mRNA 3' end formation. Mol Cell Biol. 1991 Mar;11(3):1624–1630. doi: 10.1128/mcb.11.3.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher S. L., Bhat B. M., Pursley M. H., Cladaras C., Wold W. S. Novel deletion mutants that enhance a distant upstream 5' splice in the E3 transcription unit of adenovirus 2. Nucleic Acids Res. 1985 Aug 26;13(16):5771–5788. doi: 10.1093/nar/13.16.5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenjoud L., Gallinaro H., Kister L., Meyer S., Jacob M. Identification of a specific exon sequence that is a major determinant in the selection between a natural and a cryptic 5' splice site. Mol Cell Biol. 1991 Sep;11(9):4581–4590. doi: 10.1128/mcb.11.9.4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser N. W., Baker C. C., Moore M. A., Ziff E. B. Poly(A) sites of adenovirus serotype 2 transcription units. J Mol Biol. 1982 Mar 5;155(3):207–233. doi: 10.1016/0022-2836(82)90002-x. [DOI] [PubMed] [Google Scholar]
- Fu X. D., Katz R. A., Skalka A. M., Maniatis T. The role of branchpoint and 3'-exon sequences in the control of balanced splicing of avian retrovirus RNA. Genes Dev. 1991 Feb;5(2):211–220. doi: 10.1101/gad.5.2.211. [DOI] [PubMed] [Google Scholar]
- Gallinaro H., Sittler A., Jacob M. In vivo splicing of the premRNAs from early region 3 of adenovirus-2: association of precursors, intermediates and products with hnRNP. Nucleic Acids Res. 1986 May 27;14(10):4171–4185. doi: 10.1093/nar/14.10.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H., Manley J. L. A protein factor, ASF, controls cell-specific alternative splicing of SV40 early pre-mRNA in vitro. Cell. 1990 Jul 13;62(1):25–34. doi: 10.1016/0092-8674(90)90236-8. [DOI] [PubMed] [Google Scholar]
- Ge H., Zuo P., Manley J. L. Primary structure of the human splicing factor ASF reveals similarities with Drosophila regulators. Cell. 1991 Jul 26;66(2):373–382. doi: 10.1016/0092-8674(91)90626-a. [DOI] [PubMed] [Google Scholar]
- Gooding L. R., Elmore L. W., Tollefson A. E., Brady H. A., Wold W. S. A 14,700 MW protein from the E3 region of adenovirus inhibits cytolysis by tumor necrosis factor. Cell. 1988 May 6;53(3):341–346. doi: 10.1016/0092-8674(88)90154-7. [DOI] [PubMed] [Google Scholar]
- Gooding L. R., Ranheim T. S., Tollefson A. E., Aquino L., Duerksen-Hughes P., Horton T. M., Wold W. S. The 10,400- and 14,500-dalton proteins encoded by region E3 of adenovirus function together to protect many but not all mouse cell lines against lysis by tumor necrosis factor. J Virol. 1991 Aug;65(8):4114–4123. doi: 10.1128/jvi.65.8.4114-4123.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooding L. R., Sofola I. O., Tollefson A. E., Duerksen-Hughes P., Wold W. S. The adenovirus E3-14.7K protein is a general inhibitor of tumor necrosis factor-mediated cytolysis. J Immunol. 1990 Nov 1;145(9):3080–3086. [PubMed] [Google Scholar]
- Green M. R. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu Rev Cell Biol. 1991;7:559–599. doi: 10.1146/annurev.cb.07.110191.003015. [DOI] [PubMed] [Google Scholar]
- Green M., Wold W. S. Human adenoviruses: growth, purification, and transfection assay. Methods Enzymol. 1979;58:425–435. doi: 10.1016/s0076-6879(79)58157-9. [DOI] [PubMed] [Google Scholar]
- Harper J. E., Manley J. L. A novel protein factor is required for use of distal alternative 5' splice sites in vitro. Mol Cell Biol. 1991 Dec;11(12):5945–5953. doi: 10.1128/mcb.11.12.5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins L. K., Wold W. S. A 12,500 MW protein is coded by region E3 of adenovirus. Virology. 1992 Jun;188(2):486–494. doi: 10.1016/0042-6822(92)90502-g. [DOI] [PubMed] [Google Scholar]
- Hedley M. L., Maniatis T. Sex-specific splicing and polyadenylation of dsx pre-mRNA requires a sequence that binds specifically to tra-2 protein in vitro. Cell. 1991 May 17;65(4):579–586. doi: 10.1016/0092-8674(91)90090-l. [DOI] [PubMed] [Google Scholar]
- Inoue K., Hoshijima K., Sakamoto H., Shimura Y. Binding of the Drosophila sex-lethal gene product to the alternative splice site of transformer primary transcript. Nature. 1990 Mar 29;344(6265):461–463. doi: 10.1038/344461a0. [DOI] [PubMed] [Google Scholar]
- Kapoor Q. S., Chinnadurai G. Method for introducing site-specific mutations into adenovirus 2 genome: construction of a small deletion mutant in VA-RNAI gene. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2184–2188. doi: 10.1073/pnas.78.4.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjems J., Frankel A. D., Sharp P. A. Specific regulation of mRNA splicing in vitro by a peptide from HIV-1 Rev. Cell. 1991 Oct 4;67(1):169–178. doi: 10.1016/0092-8674(91)90580-r. [DOI] [PubMed] [Google Scholar]
- Krainer A. R., Conway G. C., Kozak D. Purification and characterization of pre-mRNA splicing factor SF2 from HeLa cells. Genes Dev. 1990 Jul;4(7):1158–1171. doi: 10.1101/gad.4.7.1158. [DOI] [PubMed] [Google Scholar]
- Krainer A. R., Conway G. C., Kozak D. The essential pre-mRNA splicing factor SF2 influences 5' splice site selection by activating proximal sites. Cell. 1990 Jul 13;62(1):35–42. doi: 10.1016/0092-8674(90)90237-9. [DOI] [PubMed] [Google Scholar]
- Krainer A. R., Mayeda A., Kozak D., Binns G. Functional expression of cloned human splicing factor SF2: homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell. 1991 Jul 26;66(2):383–394. doi: 10.1016/0092-8674(91)90627-b. [DOI] [PubMed] [Google Scholar]
- Le Moullec J. M., Akusjärvi G., Stålhandske P., Pettersson U., Chambraud B., Gilardi P., Nasri M., Perricaudet M. Polyadenylic acid addition sites in the adenovirus type 2 major late transcription unit. J Virol. 1983 Oct;48(1):127–134. doi: 10.1128/jvi.48.1.127-134.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T. Mechanisms of alternative pre-mRNA splicing. Science. 1991 Jan 4;251(4989):33–34. doi: 10.1126/science.1824726. [DOI] [PubMed] [Google Scholar]
- Maquat L. E. Nuclear mRNA export. Curr Opin Cell Biol. 1991 Dec;3(6):1004–1012. doi: 10.1016/0955-0674(91)90121-e. [DOI] [PubMed] [Google Scholar]
- McNally M. T., Beemon K. Intronic sequences and 3' splice sites control Rous sarcoma virus RNA splicing. J Virol. 1992 Jan;66(1):6–11. doi: 10.1128/jvi.66.1.6-11.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally M. T., Gontarek R. R., Beemon K. Characterization of Rous sarcoma virus intronic sequences that negatively regulate splicing. Virology. 1991 Nov;185(1):99–108. doi: 10.1016/0042-6822(91)90758-4. [DOI] [PubMed] [Google Scholar]
- Padgett R. A., Grabowski P. J., Konarska M. M., Seiler S., Sharp P. A. Splicing of messenger RNA precursors. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. Poly(A) signals. Cell. 1991 Feb 22;64(4):671–674. doi: 10.1016/0092-8674(91)90495-k. [DOI] [PubMed] [Google Scholar]
- Russnak R., Ganem D. Sequences 5' to the polyadenylation signal mediate differential poly(A) site use in hepatitis B viruses. Genes Dev. 1990 May;4(5):764–776. doi: 10.1101/gad.4.5.764. [DOI] [PubMed] [Google Scholar]
- Sheets M. D., Ogg S. C., Wickens M. P. Point mutations in AAUAAA and the poly (A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res. 1990 Oct 11;18(19):5799–5805. doi: 10.1093/nar/18.19.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebel C. W., Rio D. C. Regulated splicing of the Drosophila P transposable element third intron in vitro: somatic repression. Science. 1990 Jun 8;248(4960):1200–1208. doi: 10.1126/science.2161558. [DOI] [PubMed] [Google Scholar]
- Stoltzfus C. M., Fogarty S. J. Multiple regions in the Rous sarcoma virus src gene intron act in cis to affect the accumulation of unspliced RNA. J Virol. 1989 Apr;63(4):1669–1676. doi: 10.1128/jvi.63.4.1669-1676.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollefson A. E., Krajcsi P., Pursley M. H., Gooding L. R., Wold W. S. A 14,500 MW protein is coded by region E3 of group C human adenoviruses. Virology. 1990 Mar;175(1):19–29. doi: 10.1016/0042-6822(90)90182-q. [DOI] [PubMed] [Google Scholar]
- Tollefson A. E., Krajcsi P., Yei S. P., Carlin C. R., Wold W. S. A 10,400-molecular-weight membrane protein is coded by region E3 of adenovirus. J Virol. 1990 Feb;64(2):794–801. doi: 10.1128/jvi.64.2.794-801.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollefson A. E., Scaria A., Saha S. K., Wold W. S. The 11,600-MW protein encoded by region E3 of adenovirus is expressed early but is greatly amplified at late stages of infection. J Virol. 1992 Jun;66(6):3633–3642. doi: 10.1128/jvi.66.6.3633-3642.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollefson A. E., Stewart A. R., Yei S. P., Saha S. K., Wold W. S. The 10,400- and 14,500-dalton proteins encoded by region E3 of adenovirus form a complex and function together to down-regulate the epidermal growth factor receptor. J Virol. 1991 Jun;65(6):3095–3105. doi: 10.1128/jvi.65.6.3095-3105.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollefson A. E., Wold W. S. Identification and gene mapping of a 14,700-molecular-weight protein encoded by region E3 of group C adenoviruses. J Virol. 1988 Jan;62(1):33–39. doi: 10.1128/jvi.62.1.33-39.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valsamakis A., Zeichner S., Carswell S., Alwine J. C. The human immunodeficiency virus type 1 polyadenylylation signal: a 3' long terminal repeat element upstream of the AAUAAA necessary for efficient polyadenylylation. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2108–2112. doi: 10.1073/pnas.88.6.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M. How the messenger got its tail: addition of poly(A) in the nucleus. Trends Biochem Sci. 1990 Jul;15(7):277–281. doi: 10.1016/0968-0004(90)90054-f. [DOI] [PubMed] [Google Scholar]
- Wilson-Rawls J., Saha S. K., Krajcsi P., Tollefson A. E., Gooding L. R., Wold W. S. A 6700 MW membrane protein is encoded by region E3 of adenovirus type 2. Virology. 1990 Sep;178(1):204–212. doi: 10.1016/0042-6822(90)90395-8. [DOI] [PubMed] [Google Scholar]
- Wilusz J., Pettine S. M., Shenk T. Functional analysis of point mutations in the AAUAAA motif of the SV40 late polyadenylation signal. Nucleic Acids Res. 1989 May 25;17(10):3899–3908. doi: 10.1093/nar/17.10.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold W. S., Cladaras C., Magie S. C., Yacoub N. Mapping a new gene that encodes an 11,600-molecular-weight protein in the E3 transcription unit of adenovirus 2. J Virol. 1984 Nov;52(2):307–313. doi: 10.1128/jvi.52.2.307-313.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold W. S., Deutscher S. L., Takemori N., Bhat B. M., Magie S. C. Evidence that AGUAUAUGA and CCAAGAUGA initiate translation in the same mRNA region E3 of adenovirus. Virology. 1986 Jan 15;148(1):168–180. doi: 10.1016/0042-6822(86)90412-5. [DOI] [PubMed] [Google Scholar]
- Wold W. S., Gooding L. R. Region E3 of adenovirus: a cassette of genes involved in host immunosurveillance and virus-cell interactions. Virology. 1991 Sep;184(1):1–8. doi: 10.1016/0042-6822(91)90815-s. [DOI] [PubMed] [Google Scholar]