Abstract
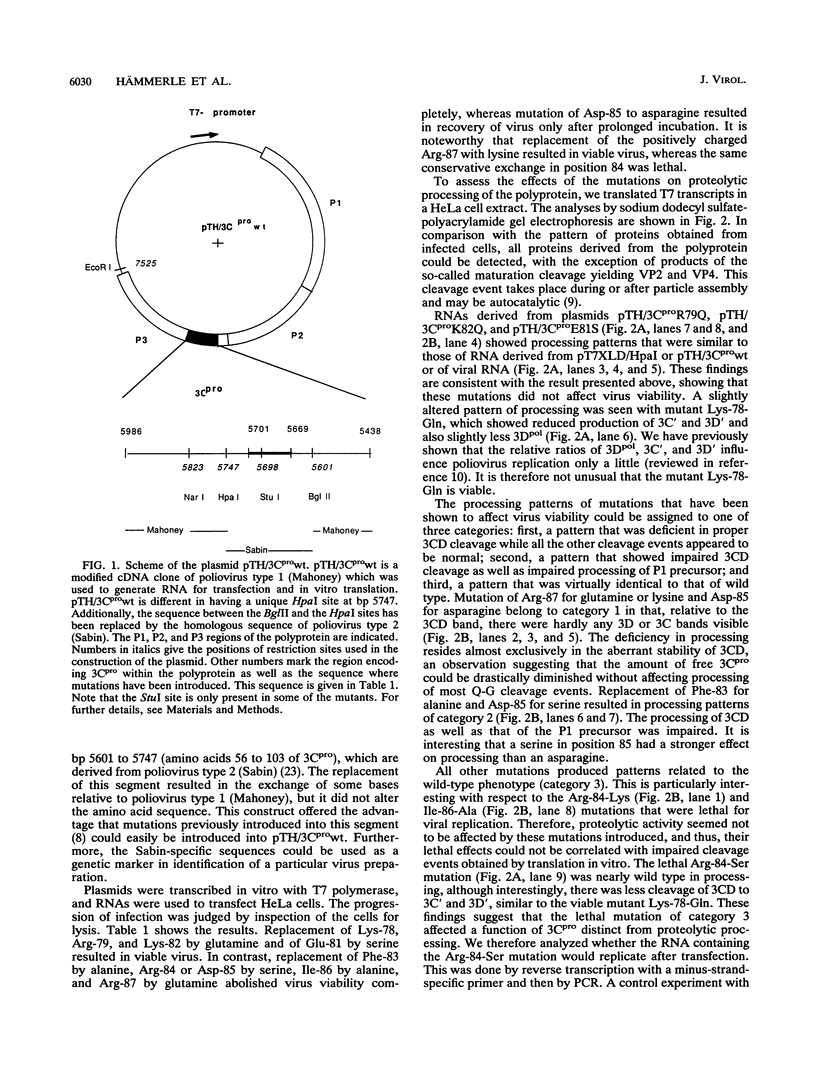
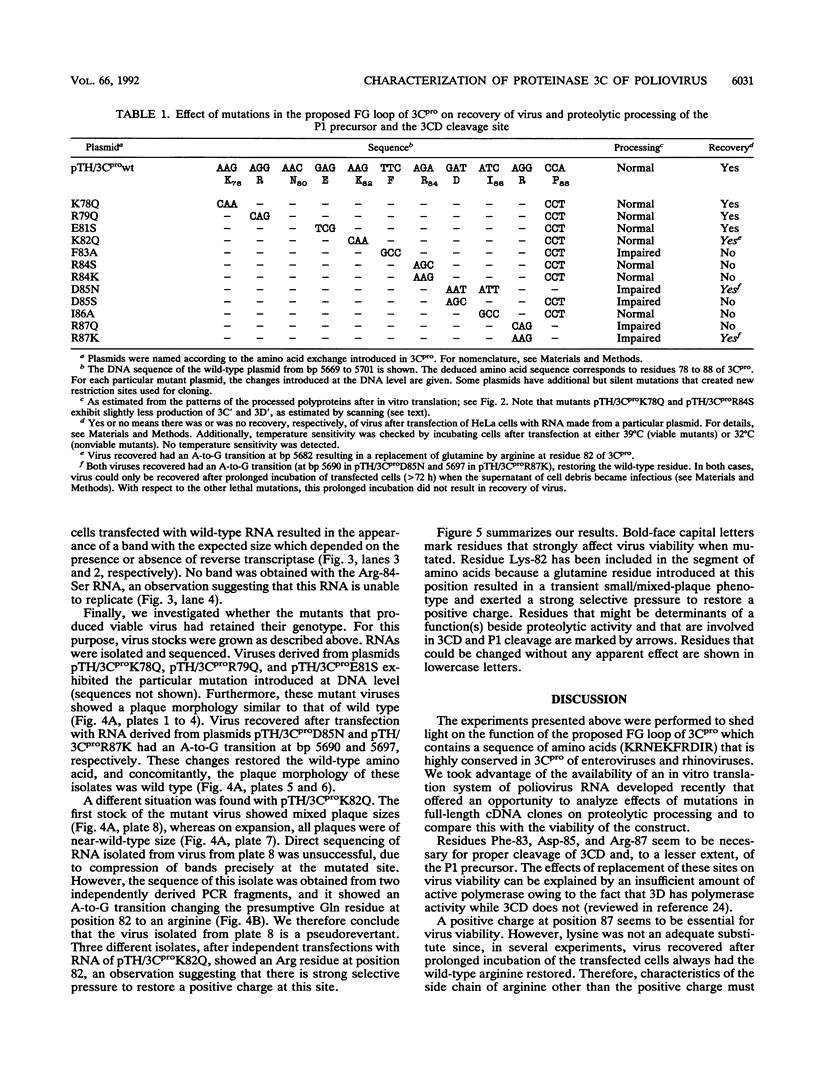
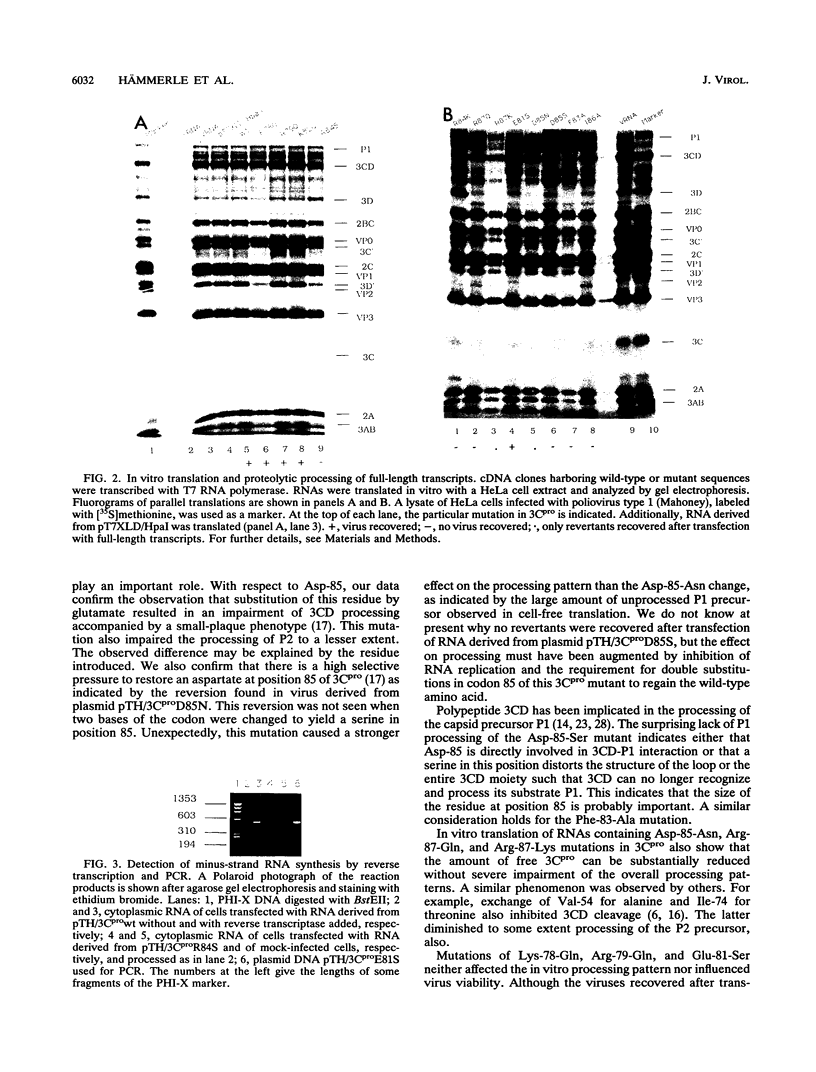
Mutations were introduced into a cDNA clone of poliovirus resulting in single-amino-acid substitutions within the region of the proposed FG loop of proteinase 3C. RNAs were made by in vitro transcription with T7 RNA polymerase and used to transfect HeLa cells. Virus viability was assessed as indicated by cell lysis. In parallel, RNAs were translated in vitro by using a HeLa cell lysate, and the patterns of the processed poly-proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Replacement of Lys-78, Arg-79, and Glu-81 had apparently no effect on virus viability and on proteolytic processing. In contrast, virus viability was abolished by mutation of Phe-83, Arg-84, Asp-85, Ile-86, and Arg-87. With respect to substitution of Phe-83, Asp-85, and Arg-87, these effects correlated with impaired processing of the 3CD cleavage site, separating 3C and 3D, and, to a lesser extent, of the P1 precursor. Replacement of Arg-84 and Ile-86, on the other hand, did not alter the processing pattern. Thus, the lethal effects in these mutant genomes may not have been caused by impaired processing. A special case was the mutant of Lys-82-Gln. Virus recovered from cells transfected with RNA carrying this mutation always contained an A-to-G transition which resulted in the replacement of glutamine for arginine. Our data suggest that residues in the proposed FG loop of proteinase 3C influence 3CD cleavage and that they are determinants of a function unrelated to proteolytic processing.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andino R., Rieckhof G. E., Baltimore D. A functional ribonucleoprotein complex forms around the 5' end of poliovirus RNA. Cell. 1990 Oct 19;63(2):369–380. doi: 10.1016/0092-8674(90)90170-j. [DOI] [PubMed] [Google Scholar]
- Andino R., Rieckhof G. E., Trono D., Baltimore D. Substitutions in the protease (3Cpro) gene of poliovirus can suppress a mutation in the 5' noncoding region. J Virol. 1990 Feb;64(2):607–612. doi: 10.1128/jvi.64.2.607-612.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazan J. F., Fletterick R. J. Viral cysteine proteases are homologous to the trypsin-like family of serine proteases: structural and functional implications. Proc Natl Acad Sci U S A. 1988 Nov;85(21):7872–7876. doi: 10.1073/pnas.85.21.7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calnan B. J., Tidor B., Biancalana S., Hudson D., Frankel A. D. Arginine-mediated RNA recognition: the arginine fork. Science. 1991 May 24;252(5009):1167–1171. doi: 10.1126/science.252.5009.1167. [DOI] [PubMed] [Google Scholar]
- Campos R., Villarreal L. P. An SV40 deletion mutant accumulates late transcripts in a paranuclear extract. Virology. 1982 May;119(1):1–11. doi: 10.1016/0042-6822(82)90059-9. [DOI] [PubMed] [Google Scholar]
- Dewalt P. G., Semler B. L. Site-directed mutagenesis of proteinase 3C results in a poliovirus deficient in synthesis of viral RNA polymerase. J Virol. 1987 Jul;61(7):2162–2170. doi: 10.1128/jvi.61.7.2162-2170.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A. E., Donchenko A. P., Blinov V. M., Koonin E. V. Cysteine proteases of positive strand RNA viruses and chymotrypsin-like serine proteases. A distinct protein superfamily with a common structural fold. FEBS Lett. 1989 Jan 30;243(2):103–114. doi: 10.1016/0014-5793(89)80109-7. [DOI] [PubMed] [Google Scholar]
- Harber J. J., Bradley J., Anderson C. W., Wimmer E. Catalysis of poliovirus VP0 maturation cleavage is not mediated by serine 10 of VP2. J Virol. 1991 Jan;65(1):326–334. doi: 10.1128/jvi.65.1.326-334.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellen C. U., Kräusslich H. G., Wimmer E. Proteolytic processing of polyproteins in the replication of RNA viruses. Biochemistry. 1989 Dec 26;28(26):9881–9890. doi: 10.1021/bi00452a001. [DOI] [PubMed] [Google Scholar]
- Higuchi R., Krummel B., Saiki R. K. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988 Aug 11;16(15):7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hämmerle T., Hellen C. U., Wimmer E. Site-directed mutagenesis of the putative catalytic triad of poliovirus 3C proteinase. J Biol Chem. 1991 Mar 25;266(9):5412–5416. [PubMed] [Google Scholar]
- Jang S. K., Kräusslich H. G., Nicklin M. J., Duke G. M., Palmenberg A. C., Wimmer E. A segment of the 5' nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988 Aug;62(8):2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jore J., De Geus B., Jackson R. J., Pouwels P. H., Enger-Valk B. E. Poliovirus protein 3CD is the active protease for processing of the precursor protein P1 in vitro. J Gen Virol. 1988 Jul;69(Pt 7):1627–1636. doi: 10.1099/0022-1317-69-7-1627. [DOI] [PubMed] [Google Scholar]
- Kean K. M., Agut H., Fichot O., Wimmer E., Girard M. A poliovirus mutant defective for self-cleavage at the COOH-terminus of the 3C protease exhibits secondary processing defects. Virology. 1988 Apr;163(2):330–340. doi: 10.1016/0042-6822(88)90273-5. [DOI] [PubMed] [Google Scholar]
- Kean K. M., Teterina N. L., Marc D., Girard M. Analysis of putative active site residues of the poliovirus 3C protease. Virology. 1991 Apr;181(2):609–619. doi: 10.1016/0042-6822(91)90894-h. [DOI] [PubMed] [Google Scholar]
- Koonin E. V. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J Gen Virol. 1991 Sep;72(Pt 9):2197–2206. doi: 10.1099/0022-1317-72-9-2197. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lawson M. A., Semler B. L. Picornavirus protein processing--enzymes, substrates, and genetic regulation. Curr Top Microbiol Immunol. 1990;161:49–87. [PubMed] [Google Scholar]
- Molla A., Paul A. V., Wimmer E. Cell-free, de novo synthesis of poliovirus. Science. 1991 Dec 13;254(5038):1647–1651. doi: 10.1126/science.1661029. [DOI] [PubMed] [Google Scholar]
- Murdin A. D., Wimmer E. Construction of a poliovirus type 1/type 2 antigenic hybrid by manipulation of neutralization antigenic site II. J Virol. 1989 Dec;63(12):5251–5257. doi: 10.1128/jvi.63.12.5251-5257.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklin M. J., Harris K. S., Pallai P. V., Wimmer E. Poliovirus proteinase 3C: large-scale expression, purification, and specific cleavage activity on natural and synthetic substrates in vitro. J Virol. 1988 Dec;62(12):4586–4593. doi: 10.1128/jvi.62.12.4586-4593.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards O. C., Ehrenfeld E. Poliovirus RNA replication. Curr Top Microbiol Immunol. 1990;161:89–119. doi: 10.1007/978-3-642-75602-3_4. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ypma-Wong M. F., Dewalt P. G., Johnson V. H., Lamb J. G., Semler B. L. Protein 3CD is the major poliovirus proteinase responsible for cleavage of the P1 capsid precursor. Virology. 1988 Sep;166(1):265–270. doi: 10.1016/0042-6822(88)90172-9. [DOI] [PubMed] [Google Scholar]
- van der Werf S., Bradley J., Wimmer E., Studier F. W., Dunn J. J. Synthesis of infectious poliovirus RNA by purified T7 RNA polymerase. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2330–2334. doi: 10.1073/pnas.83.8.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]