Abstract
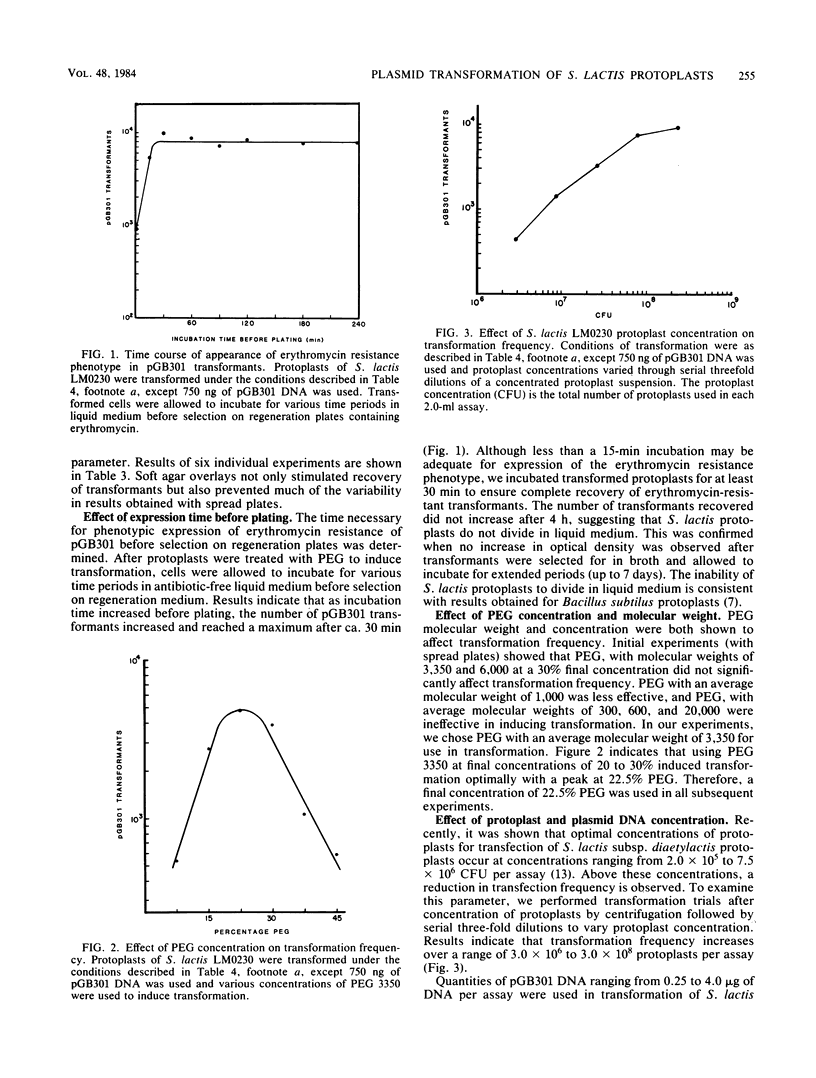
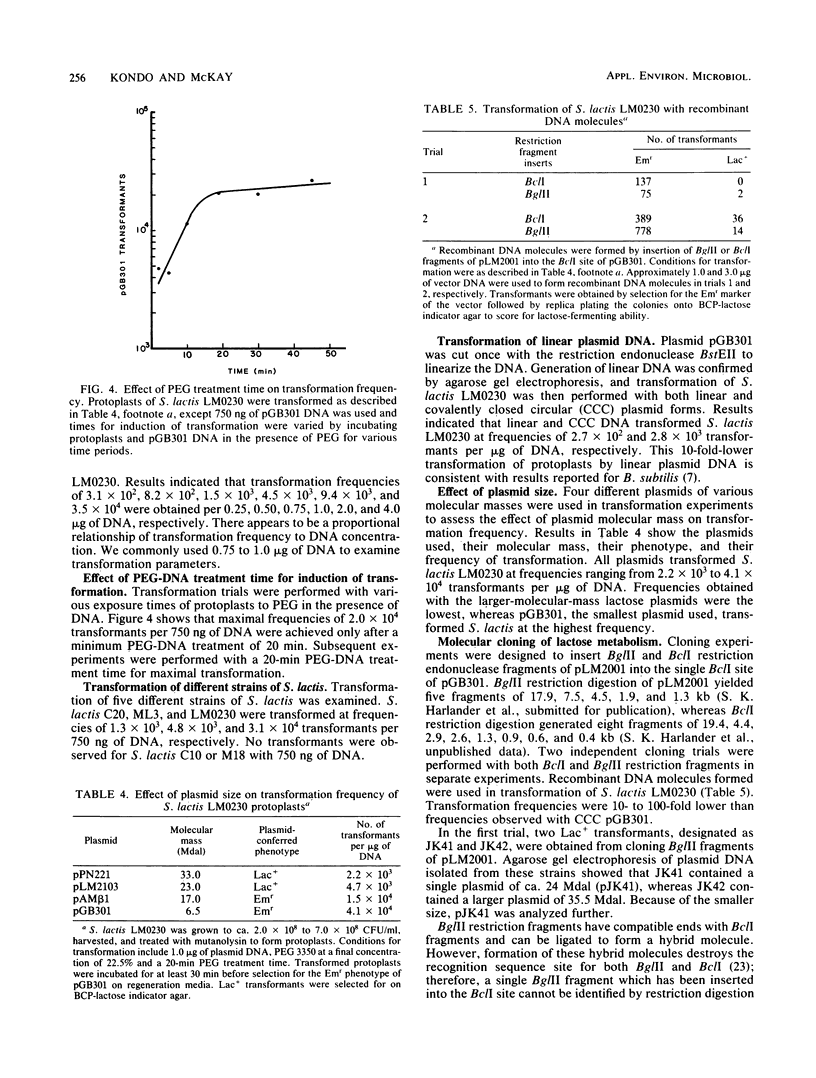
The parameters affecting polyethylene glycol-induced plasmid transformation of Streptococcus lactis LM0230 protoplasts were examined to increase the transformation frequency. In contrast to spreading protoplasts over the surface of an agar medium, their incorporation into soft agar overlays enhanced regeneration of protoplasts and eliminated variability in transformation frequencies. Polyethylene glycol with a molecular weight of 3,350 at a final concentration of 22.5% yielded optimal transformation. A 20-min polyethylene glycol treatment of protoplasts in the presence of DNA was necessary for maximal transformation. The number of transformants recovered increased as the protoplast and DNA concentration increased over a range of 3.0 X 10(6) to 3.0 X 10(8) protoplasts and 0.25 to 4.0 micrograms of DNA per assay, respectively. With these parameters, transformation was increased to 5 X 10(3) to 4 X 10(4) transformants per microgram of DNA. Linear and recombinant plasmid DNA transformed, but at frequencies 10- to 100-fold lower than that of covalently closed circular DNA. Transformation of recombinant DNA molecules enabled the cloning of restriction endonuclease fragments coding for lactose metabolism into S. lactis LM0230 with the Streptococcus sanguis cloning vector, pGB301. These results demonstrated that the transformation frequency is sufficient to clone plasmid-coded genes which should prove useful for strain improvement of dairy starter cultures.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson D. G., McKay L. L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983 Sep;46(3):549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke D., Gilmore M. S., Ferretti J. J. Plasmid pGB301, a new multiple resistance streptococcal cloning vehicle and its use in cloning of a gentamicin/kanamycin resistance determinant. Mol Gen Genet. 1981;182(3):414–421. doi: 10.1007/BF00293929. [DOI] [PubMed] [Google Scholar]
- Behnke D., Gilmore M. S. Location of antibiotic resistance determinants, copy control, and replication functions on the double-selective streptococcal cloning vector pGB301. Mol Gen Genet. 1981;184(1):115–120. doi: 10.1007/BF00271206. [DOI] [PubMed] [Google Scholar]
- Behnke D. Plasmid transformation of Streptococcus sanguis (Challis) occurs by circular and linear molecules. Mol Gen Genet. 1981;182(3):490–497. doi: 10.1007/BF00293940. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Ward J. M., Hopwood D. A. Transformation of plasmid DNA into Streptomyces at high frequency. Nature. 1978 Jul 27;274(5669):398–400. doi: 10.1038/274398a0. [DOI] [PubMed] [Google Scholar]
- Brown B. J., Carlton B. C. Plasmid-mediated transformation in Bacillus megaterium. J Bacteriol. 1980 May;142(2):508–512. doi: 10.1128/jb.142.2.508-512.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S., Cohen S. N. High frequency transformation of Bacillus subtilis protoplasts by plasmid DNA. Mol Gen Genet. 1979 Jan 5;168(1):111–115. doi: 10.1007/BF00267940. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Yagi Y., Dunny G. M., Schultz S. K. Characterization of three plasmid deoxyribonucleic acid molecules in a strain of Streptococcus faecalis: identification of a plasmid determining erythromycin resistance. J Bacteriol. 1974 Jan;117(1):283–289. doi: 10.1128/jb.117.1.283-289.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efstathiou J. D., McKay L. L. Inorganic salts resistance associated with a lactose-fermenting plasmid in Streptococcus lactis. J Bacteriol. 1977 Apr;130(1):257–265. doi: 10.1128/jb.130.1.257-265.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasson M. J. Genetic transfer systems in lactic acid bacteria. Antonie Van Leeuwenhoek. 1983 Sep;49(3):275–282. doi: 10.1007/BF00399503. [DOI] [PubMed] [Google Scholar]
- Hopwood D. A. Genetic studies with bacterial protoplasts. Annu Rev Microbiol. 1981;35:237–272. doi: 10.1146/annurev.mi.35.100181.001321. [DOI] [PubMed] [Google Scholar]
- Imanaka T., Fujii M., Aramori I., Aiba S. Transformation of Bacillus stearothermophilus with plasmid DNA and characterization of shuttle vector plasmids between Bacillus stearothermophilus and Bacillus subtilis. J Bacteriol. 1982 Mar;149(3):824–830. doi: 10.1128/jb.149.3.824-830.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob A. E., Hobbs S. J. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J Bacteriol. 1974 Feb;117(2):360–372. doi: 10.1128/jb.117.2.360-372.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaenhammer T. R., McKay L. L., Baldwin K. A. Improved lysis of group N streptococci for isolation and rapid characterization of plasmid deoxyribonucleic acid. Appl Environ Microbiol. 1978 Mar;35(3):592–600. doi: 10.1128/aem.35.3.592-600.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo J. K., McKay L. L. Transformation of Streptococcus lactis Protoplasts by Plasmid DNA. Appl Environ Microbiol. 1982 May;43(5):1213–1215. doi: 10.1128/aem.43.5.1213-1215.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhl S. A., Larsen L. D., McKay L. L. Plasmid Profiles of Lactose-Negative and Proteinase-Deficient Mutants of Streptococcus lactis C10, ML(3), and M18. Appl Environ Microbiol. 1979 Jun;37(6):1193–1195. doi: 10.1128/aem.37.6.1193-1195.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer P. R., Waldrop A. A., Ward D. C. Enzymatic synthesis of biotin-labeled polynucleotides: novel nucleic acid affinity probes. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6633–6637. doi: 10.1073/pnas.78.11.6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A., Walsh P. M. Conjugal transfer of genetic information in group N streptococci. Appl Environ Microbiol. 1980 Jul;40(1):84–89. doi: 10.1128/aem.40.1.84-91.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A., Zottola E. A. Loss of lactose metabolism in lactic streptococci. Appl Microbiol. 1972 Jun;23(6):1090–1096. doi: 10.1128/am.23.6.1090-1096.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. L. Functional properties of plasmids in lactic streptococci. Antonie Van Leeuwenhoek. 1983 Sep;49(3):259–274. doi: 10.1007/BF00399502. [DOI] [PubMed] [Google Scholar]
- Shoemaker N. B., Smith M. D., Guild W. R. Organization and transfer of heterologous chloramphenicol and tetracycline resistance genes in pneumococcus. J Bacteriol. 1979 Aug;139(2):432–441. doi: 10.1128/jb.139.2.432-441.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sladek T. L., Maniloff J. Polyethylene glycol-dependent transfection of Acholeplasma laidlawii with mycoplasma virus L2 DNA. J Bacteriol. 1983 Aug;155(2):734–741. doi: 10.1128/jb.155.2.734-741.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Suarez J. E., Chater K. F. Polyethylene glycol-assisted transfection of Streptomyces protoplasts. J Bacteriol. 1980 Apr;142(1):8–14. doi: 10.1128/jb.142.1.8-14.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi B. E., Sandine W. E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975 Jun;29(6):807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh P. M., McKay L. L. Recombinant plasmid associated cell aggregation and high-frequency conjugation of Streptococcus lactis ML3. J Bacteriol. 1981 Jun;146(3):937–944. doi: 10.1128/jb.146.3.937-944.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokogawa K., Kawata S., Nishimura S., Ikeda Y., Yoshimura Y. Mutanolysin, bacteriolytic agent for cariogenic Streptococci: partial purification and properties. Antimicrob Agents Chemother. 1974 Aug;6(2):156–165. doi: 10.1128/aac.6.2.156. [DOI] [PMC free article] [PubMed] [Google Scholar]