Abstract
Widespread interest in producing transgenic organisms is balanced by concern over ecological hazards, such as species extinction if such organisms were to be released into nature. An ecological risk associated with the introduction of a transgenic organism is that the transgene, though rare, can spread in a natural population. An increase in transgene frequency is often assumed to be unlikely because transgenic organisms typically have some viability disadvantage. Reduced viability is assumed to be common because transgenic individuals are best viewed as macromutants that lack any history of selection that could reduce negative fitness effects. However, these arguments ignore the potential advantageous effects of transgenes on some aspect of fitness such as mating success. Here, we examine the risk to a natural population after release of a few transgenic individuals when the transgene trait simultaneously increases transgenic male mating success and lowers the viability of transgenic offspring. We obtained relevant life history data by using the small cyprinodont fish, Japanese medaka (Oryzias latipes) as a model. Our deterministic equations predict that a transgene introduced into a natural population by a small number of transgenic fish will spread as a result of enhanced mating advantage, but the reduced viability of offspring will cause eventual local extinction of both populations. Such risks should be evaluated with each new transgenic animal before release.
Although production of transgenic organisms offers great agricultural potential, introduction of genetically modified organisms into natural populations could result in ecological hazards, such as species extinction (1–3). Such risk has been suggested to pose little environmental threat because transgenic organisms are evolutionary novelties that would have reduced viability (4, 5). However, transgenic organisms may also possess an advantage in some aspect of reproduction that may increase their success in nature. Although a variety of transgene traits have been incorporated into various species (6, 7), a commonly desired characteristic in transgenic fish species (important in aquaculture and sport fishing) is accelerated growth rate and larger adult body size (8). DNA sequences for growth hormone (GH) genes and cDNAs have been well characterized in fish, and transgenic fish of several species have now been produced (9, 10). Growth enhancements of up to several times that of wild type have been obtained, with growth advantages persisting throughout adulthood in some fish species (8, 11). In many animal species, including fish, body size is an important determinant of differential mating success (sexual selection) through advantages in competing for mates against members of the same sex (mate competition) and/or being preferred as a mate by the opposite sex (mate choice) (12). A recent review found that large body size conferred mating advantages in 40% of the 186 animal taxa surveyed (12). The potential for sexual selection to produce a rapid evolution of sexual traits has long been appreciated (12); here we consider its potential to increase transgene frequency and to eliminate populations, specifically when a sexual trait is affected by transgenes.
Materials and Methods
Study Organism.
As a model organism, we studied Japanese medaka (Oryzias latipes) (13) to explore the ecological consequences of transgene release into natural populations. Medaka were convenient study organisms for obtaining data on fitness components. Individuals were readily bred in the lab, were easily cultured, and attained sexual maturity in about two months. We produced a stock of transgenic medaka by inserting the human growth hormone gene (hGH), with a salmon promoter, sGH (14). We then conducted several experiments to document survival and reproductive differences between transgenic and wild-type medaka (15). We categorized these differences into four fitness components: (i) viability (offspring survival to sexual maturity), (ii) developmental (age at sexual maturation), (iii) fecundity (clutch size), and (iv) sexual selection (mating advantages). We modeled the introduction of a small number of transgenic individuals into a large wild-type population using recurrence equations (described below) to predict the consequences of the model, i.e., of increased male mating success but reduced offspring viability. Elsewhere, we examined the results of model predictions in which GH transgenes influenced developmental and fecundity fitness components as well as offspring viability (unpublished data). Different transgene lines are likely to vary in fitness even when the same transgene construct is used, because of differences in copy number and sites of transgene insertion. To take such variation into account as well as to make our model generally applicable to other organisms and transgene constructs, we used a range of parameter values for male mating success and offspring viability in our models. The range of values also encompassed the particular fitness component estimates that we obtained.
We conducted a 2 × 2 factorial experiment to assess the early viability of offspring produced from crosses involving transgenic and wild-type medaka parents (15). Each pairing combination consisted of 10 males and 10 females; eggs were obtained from each pair for a period of 10 days, producing a total of 1,910 fertile eggs. Viability was estimated as the percentage of 3-day-old fry that emerged. Results showed that early survival of transgenic young was 70% of that of the wild type (15).
Mating experiments using wild-type medaka were performed to measure the mating advantage that large males obtained over small males (16). We found that, regardless of protocol, large males obtained a 4-fold mating advantage (16). Such size-related mating advantages have been demonstrated in a variety of fish species; they can result from mate competition or mate choice or both (12). We do not expect transgenic male medaka to have a mating advantage over wild-type males, because the hGH transgene we inserted increased only juvenile growth rate, not final adult body size (14); that is, the size difference between transgenic and wild-type males disappeared by sexual maturity. Nonetheless, we modeled the possible effect of transgene release into wild-type populations when transgenes accelerate growth throughout adulthood, thus increasing transgenic male mating success, because these effects could occur with other transgene constructs in other fish species. For example, continued growth enhancements from GH genes occurs in adult salmonids (8), and the mating advantages of large males has been reported in several salmonid species (17–19).
We used a range of mating and viability fitness parameters, including the values we obtained in experiments with a recurrence model that predicts changes in gene frequencies and population sizes when transgenic individuals invade a wild-type population (15).
Deterministic Model.
Our model assumes one locus with two alleles (three possible genotypes) and predicts changes in population number and gene frequency. We assume an arbitrary population with a maximum life span of d ages composed of Njaf females of genotype j in the ath age class and Nkam males of genotype k in the ath age class. The frequency of matings and offspring production determines the number of ages that need to be monitored. For a species such as medaka that mate and produce offspring every day after attaining sexual maturity, the ages were measured in days. Ages at sexual maturity were sjf and skm for females and males, respectively, with genotypes j and k. To determine gene frequency changes it was only necessary to determine the number of sexually mature individuals of each genotype and sex. The population size at time t would equal
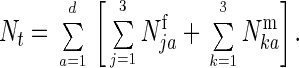 |
1 |
The relative frequency of sexually mature female and male genotypes at time t would equal
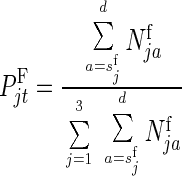 |
2 |
and
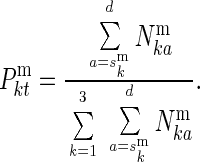 |
3 |
Let
fj = Relative mating advantage of jth female genotype, and
mk = Relative mating advantage of kth male genotype;
then the relative frequency with which genotypes j and k mate would equal
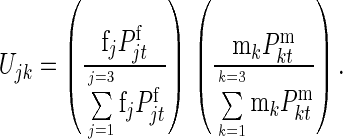 |
4 |
Let
cj = fecundity (clutch size) of the jth female genotype,
rk = Relative fertility of the kth male genotype,
Mijk = Expected frequency of genotype i at birth among offspring from a mating between genotypes j and k (Mendelian segregation ratio), and
vijk = Relative viability of a zygote of genotype i surviving from birth to sexual maturity, given that genotype i was produced from parents of genotypes j and k. We assumed, without loss of generality, that all deaths occurred in the first age class.
Assuming an equal sex ratio at birth, the expected number of offspring of genotype i of each sex surviving to the first age class from all matings would equal
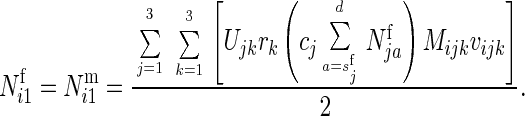 |
5 |
These individuals made up the first age class. When this age class entered the population, all other age classes advanced by one, and the oldest age class died off.
The population size at time t + 1 would equal
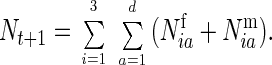 |
6 |
The relative frequency of the ith genotype at time t + 1 would equal
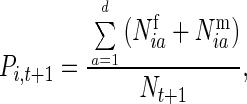 |
7 |
with gene frequencies of the two alleles equaling F1 = P1,t+1 + ½P2,t+1 and F2 = 1 − F1.
Results and Discussion
In the model, the initial population was structured with a stable age distribution giving a constant size (60,000), composed of wild-type fish with an equal sex ratio in each class. Based on experimental data (15), and adjusted by trial and error to achieve a stable age distribution, juvenile and adult mortality rates were set to 9.8% and 0.765% per day, respectively, for both genotypes, which resulted in an expected maximum life span of 150 days. Sixty homozygous transgenic fish of equal sex ratio were then introduced at sexual maturity. We assumed that transgenic and wild-type individuals were similar in age (at sexual maturity), fecundity, fertility, susceptibility to predation, and longevity; the only differential effects caused by the GH transgene were male mating success and offspring viability. We also assumed that the probability of mating was not frequency-dependent. For this model, population size was always assumed to be less than the carrying capacity; i.e., no density-dependent effects occurred. This assumption is known to be incorrect for some species. But for species that are declining in number because of heavy fishing pressure or other sources of mortality, the assumption is likely to be true. The above parameters were specified in the model, and genotype frequency, gene frequency, and population size were assessed each day. We expressed time to extinction in terms of the generation interval, the average age when all offspring were produced, which, in our laboratory experiments on medaka, equaled 96.9 days.
Predictions of the model were straightforward when transgenes affected only one fitness component. If transgenes reduced only juvenile survival, transgenic individuals would be quickly eliminated from any wild-type population. Our model predicted that if transgenic medaka suffered a 30% reduction in viability relative to the wild type, the transgene would be eliminated after about 10 generations (15). In contrast, if the GH transgene increased only the mating success of transgenic males relative to wild-type males, the gene would spread quickly. If adult transgenic males were 24% larger than adult wild-type males and thereby achieved the 4-fold mating advantage that we had observed in our mating experiments (16), the frequency of the transgene would exceed 50% in about five generations, and become fixed in the population in about 20 generations. In both of these situations, population size would remain essentially unchanged across generations, and the transgene would either be eliminated or go to fixation.
In contrast, combining the effects of the transgene on mating success and offspring viability is predicted to result in the local extinction of any wild-type population invaded by transgenic organisms. The male mating advantage would act to increase the frequency of the transgene in the population; however, the viability disadvantage suffered by all offspring carrying the transgene would reduce the population size by 50% in less than six generations and completely eliminate the population in about 40 generations. These population projections result because the males that produce the least fit offspring obtain a disproportionate share of the matings. We refer to this type of extinction as the “Trojan gene effect,” because the mating advantage provides a mechanism for the transgene to enter and spread in a population, and the viability reduction eventually results in population extinction. Such a conflict between offspring viability and male mating advantage based on large body size has been theorized to be one of the processes that can cause species extinction (20, 21).
Both the advantageous and disadvantageous effects of such sexual traits are usually considered to be sex-limited; however, the transgene we considered has a sex-limited advantage (male mating success), but no sex limitation on viability reduction. As a result, population extinction should occur even more rapidly. In theory, counterselection against the transgene and thereby rescuing a population from extinction is possible. Such counterselection could take two forms. Modifying genes might be selected that mitigate the degree of viability reduction of the transgene. Alternatively, if the transgenic male mating advantage results mostly from female preference for large males, females with alternative mating preferences could be favored by selection, halting or reversing the spread of the transgene. If the mating advantage of transgenic males resulted mostly from success in mate competition, we would expect no such selection against the transgene. Our prediction of population extinction must, however, be interpreted cautiously. A critical assumption of our deterministic model is that the viability reduction of transgenic organisms remains constant, even with a lowering of population density.
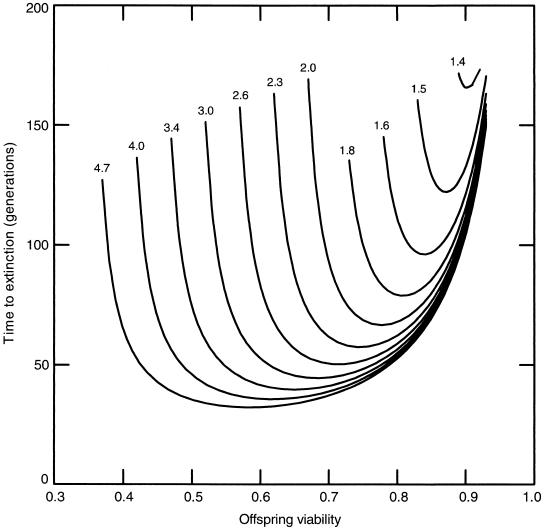
The predicted time course for extinction of a wild-type population after the release of transgenic individuals varies as a function of the rate of transgene spread, which is influenced by the relative mating advantage of transgenic males and by the severity of viability reduction in transgenic young (Fig. 1). For example, our model predicted that if the viability of transgenic young were 70% of that of wild-type young, as was the case with the hGH-sGH transgenic medaka we produced, population extinction would result only when transgenic males obtained a 2-fold or greater mating advantage over wild-type males.
Figure 1.
Predicted time to extinction of a wild-type medaka population as a function of the mating advantage (numbers above curves) of transgenic males relative to that of wild-type males and the relative viability of transgenic offspring.
Increasing the viability of transgenic offspring in the simulations produced a counterintuitive result, however. If the viability of transgenic young was increased to 85% of that of wild-type offspring, population extinction was predicted to occur over a wider range of male mating advantages, even though the time to extinction was greater. Thus, as the viability of transgenic offspring approaches that of wild type, risk of extinction may actually increase. Two situations resulted in the highest risk: a high mating advantage and a moderate viability reduction (Fig. 1). A mating advantage of at least 4-fold produced a risk over a range of viabilities from about 0.45 to 0.9; a viability reduction in the range of 0.7 to 0.9 resulted in the risk of extinction over the widest range of mating advantages. These trends were predicted because, at one extreme, a transgene that greatly reduced offspring viability would be quickly eliminated unless it were counterbalanced by a very high male mating advantage. At the other extreme, in the case of a transgene that produced high viability of transgenic young, a lower male mating advantage could drive the gene to high frequency in the population, resulting in a lower genetic load and requiring more generations for population extinction.
Local extinction of a wild-type population from a release of transgenic individuals could also have cascading negative effects on the community. In contrast, if transgenic males were created intentionally to drive to extinction a wild-type population of, for example, a species of pests, it could serve as a mechanism for biological control.
Acknowledgments
We thank J. Lucas, P. Waser, Anne Kapuscinski, and an anonymous reviewer for helpful comments. This research was supported by U.S. Department of Agriculture National Biological Impact Assessment Program grants ( 93-33120-9468 and 97-39210-4997).
Abbreviation
- GH
growth hormone
References
- 1.Tiedje J M, Colwell R K, Grossman Y L, Hodson R E, Lenski R E, Mack R N, Regal P J. Ecology. 1989;70:298–315. [Google Scholar]
- 2.Kapuscinski A R, Hallerman E M. Can J Fish Aquat Sci. 1991;48:99–107. [Google Scholar]
- 3.Devlin R H, Donaldson E M. In: Transgenic Fish. Hew C L, Fletcher G L, editors. Singapore: World Scientific; 1992. pp. 229–265. [Google Scholar]
- 4.Knibb W. Transgenic Res. 1997;6:59–67. [Google Scholar]
- 5.Regal P J. Recomb DNA Tech Bull. 1987;10:67–85. [PubMed] [Google Scholar]
- 6.Levin M A, Israeli E. Engineered Organisms in Environmental Settings: Biotechnological and Agricultural Applications. Boca Raton, FL: CRC; 1996. pp. 13–17. [Google Scholar]
- 7.Houdebine L M, editor. Transgenic Animals: Generation and Use. Amsterdam: Harwood Academic; 1996. [Google Scholar]
- 8.Devlin R H. In: Transgenic Animals: Generation and Use. Houdebine L M, editor. Amsterdam: Harwood Academic; 1996. pp. 105–117. [Google Scholar]
- 9.Devlin R H, Yesaki T Y, Blagl C A, Donaldson E M. Nature (London) 1994;371:209–210. [Google Scholar]
- 10.Du S, Gong Z, Fletcher G, Shears M, King M, Idler D, Hew C L. Bio-Technology. 1992;10:176–181. doi: 10.1038/nbt0292-176. [DOI] [PubMed] [Google Scholar]
- 11.Devlin R H, Yesaki T Y, Donaldson E M, Du S J, Hew C L. Can J Fish Aquat Sci. 1995;52:1376–1384. [Google Scholar]
- 12.Andersson M. Sexual Selection. Princeton, NJ: Princeton Univ. Press; 1994. [Google Scholar]
- 13.Muir W M, Howard R D, Bidwell C A. In: Proceedings of the Biotechnology Risk Assessment Symposium. Levin M, Grim C, Angle J S, editors. College Park, MD: Univ. Maryland Biotechnology Institute; 1994. pp. 170–197. [Google Scholar]
- 14.Muir W M, Martens R S, Howard R D, Bidwell C A. In: Proceedings of the Biotechnology Risk Assessment Symposium. Levin M, Grim C, Angle J S, editors. College Park, MD: Univ. Maryland Biotechnology Institute; 1995. pp. 140–149. [Google Scholar]
- 15.Muir W M, Howard R D, Martens R S, Schulte S, Bidwell C A. In: Proceedings of the Biotechnology Risk Assessment Symposium. Levin M, Grim C, Angle J S, editors. College Park, MD: Univ. Maryland Biotechnology Institute; 1996. pp. 354–356. [Google Scholar]
- 16.Howard R D, Martens R S, Innes S A, Drnevich J M, Hale J. Anim Behav. 1998;55:1151–1163. doi: 10.1006/anbe.1997.0682. [DOI] [PubMed] [Google Scholar]
- 17.Quinn T P, Foote C J. Anim Behav. 1988;48:751–761. [Google Scholar]
- 18.Fleming I A. Rev Fish Biol Fish. 1996;6:379–416. [Google Scholar]
- 19.Mjolnerod I B, Fleming I A, Refseth U H, Hindar K. Can J Zool. 1998;76:70–76. [Google Scholar]
- 20.Lande R. Evolution. 1980;34:292–305. doi: 10.1111/j.1558-5646.1980.tb04817.x. [DOI] [PubMed] [Google Scholar]
- 21.Maynard Smith J, Brown R L W. Theor Popul Biol. 1986;30:166–179. doi: 10.1016/0040-5809(86)90031-6. [DOI] [PubMed] [Google Scholar]