Abstract
Summary: DNA-binding repressor proteins that govern transcription initiation in response to end products generally regulate bacterial biosynthetic genes, but this is rarely true for the pyrimidine biosynthetic (pyr) genes. Instead, bacterial pyr gene regulation generally involves mechanisms that rely only on regulatory sequences embedded in the leader region of the operon, which cause premature transcription termination or translation inhibition in response to nucleotide signals. Studies with Escherichia coli and Bacillus subtilis pyr genes reveal a variety of regulatory mechanisms. Transcription attenuation via UTP-sensitive coupled transcription and translation regulates expression of the pyrBI and pyrE operons in enteric bacteria, whereas nucleotide effects on binding of the PyrR protein to pyr mRNA attenuation sites control pyr operon expression in most gram-positive bacteria. Nucleotide-sensitive reiterative transcription underlies regulation of other pyr genes. With the E. coli pyrBI, carAB, codBA, and upp-uraA operons, UTP-sensitive reiterative transcription within the initially transcribed region (ITR) leads to nonproductive transcription initiation. CTP-sensitive reiterative transcription in the pyrG ITRs of gram-positive bacteria, which involves the addition of G residues, results in the formation of an antiterminator RNA hairpin and suppression of transcription attenuation. Some mechanisms involve regulation of translation rather than transcription. Expression of the pyrC and pyrD operons of enteric bacteria is controlled by nucleotide-sensitive transcription start switching that produces transcripts with different potentials for translation. In Mycobacterium smegmatis and other bacteria, PyrR modulates translation of pyr genes by binding to their ribosome binding site. Evidence supporting these conclusions, generalizations for other bacteria, and prospects for future research are presented.
INTRODUCTION
The pyrimidine nucleotides UTP and CTP and their derivatives are essential for all living organisms, but pyrimidine bases and nucleosides, the transportable precursors of the nucleotides, are often unavailable as exogenous nutrients. It is not surprising, therefore, that all sequenced bacterial genomes, except certain intracellular parasites, encode the enzymes required for de novo biosynthesis of pyrimidine nucleotides. The enzymatic steps of the pyrimidine nucleotide biosynthetic pathway are the same in all bacteria. However, the genomic organization of the genes encoding the pyrimidine biosynthetic enzymes and the mechanisms controlling the expression of these genes vary greatly from gene to gene and across the phylogenetic spectrum. The study of mechanisms that regulate the expression of pyrimidine biosynthetic (pyr) genes, which has been a major focus of research in our laboratories for many years, has proven to be a rich source for the discovery of novel biochemical strategies for coordination of gene expression with the intracellular levels of pyrimidine nucleotides. We review here our current understanding of the mechanisms that regulate expression of pyr genes in bacteria. Studies of the regulation of pyr genes in Escherichia coli and Bacillus subtilis will be presented in most detail, because these systems have been by far the most thoroughly characterized. Examination of genomic sequences from many other bacteria, which will also be briefly presented here, indicates that the mechanisms found in E. coli and B. subtilis are operative, sometimes with variations, in a large number of other bacterial species.
It is a remarkable fact that, with rare exceptions, the many mechanisms known to regulate the expression of bacterial pyr genes do not involve the participation of a DNA-binding repressor or activator protein. Rather, as will be seen in this review, the information that specifies pyrimidine-responsive regulation of pyr gene expression is generally encoded within the promoter-leader region of the regulated downstream genes. (The leader region is defined as the DNA extending from the start of transcription to the first gene of an operon.) During transcription of the leader regions, alternative sequences and/or secondary structures in the leader-specified RNA determine whether transcripts will be prematurely terminated or fully elongated or, alternatively, whether an elongated transcript will be efficiently translated. In all cases except those involving the pyr mRNA-binding regulatory protein PyrR, the concentration of pyrimidine nucleotides is sensed directly by RNA polymerase. While the predominance of such mechanisms may result from their ancient evolutionary origins, their wide distribution and retention must also reflect their efficiency and sensitivity. The importance of the novel regulatory mechanisms described in this review extends beyond pyr genes, however. Their implications for the mechanism of transcription in bacteria in general and for the ways that transcription can be harnessed for regulation of other genes will be discussed in the course of this review.
REGULATORY MECHANISMS IN ENTERIC BACTERIA
History and Overview
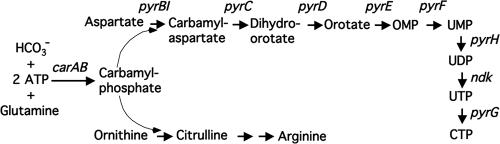
From classic experiments in the 1950s, the operon model emerged to explain regulation of lactose utilization in E. coli and, optimistically, all gene regulation in living cells (61). In this model, the rate of protein synthesis was controlled by a repressor, later shown to be a protein (47), which was either inactivated (induction) or activated (repression) by specific metabolites. The active repressor bound to a DNA operator to prevent the synthesis of mRNA, which served as a short-lived intermediate that in association with a ribosome directed the synthesis of the encoded protein(s). The operon model was so compelling that scientists studying the regulation of many different genes in various bacteria in the 1960s and 1970s eagerly searched for their repressors. One of these early studies focused on pyr gene expression in E. coli and the closely related bacterium Salmonella enterica serovar Typhimurium (12, 128). These studies concentrated on the six operons encoding the enzymes required for the biosynthesis of UMP, the precursor of all pyrimidine nucleotides (Fig. 1). These operons, designated carAB, pyrBI, pyrC, pyrD, pyrE, and pyrF, were shown to be genetically unlinked and scattered on the chromosome (124). These operons were also found to be subject to complex regulation. Expression of the pyrBI, pyrE, and pyrF operons was repressed by a uridine nucleotide, whereas expression of the pyrC and pyrD operons was repressed predominantly by a cytidine nucleotide (81, 132, 145). Expression of the carAB operon, which is essential for both pyrimidine and arginine biosynthesis (Fig. 1), was subject to cumulative repression by a pyrimidine nucleotide and arginine (1, 132). These results suggested that at least two repressors controlled transcription of the pyrimidine biosynthetic operons. However, attempts to isolate mutants lacking the putative repressors failed (75, 127). Additional experiments showed that under conditions of pyrimidine limitation, derepression of pyrimidine biosynthetic operon expression was noncoordinate (124). This observation suggested that the expression of each operon was regulated by an independent mechanism.
FIG. 1.
Pyrimidine nucleotide biosynthetic pathway of E. coli and Salmonella. Gene names are used to represent the encoded biosynthetic enzymes. The genes shown in the figure and the encoded proteins are as follows: carA, glutaminase subunit of carbamylphosphate synthetase; carB, catalytic subunit of carbamylphosphate synthetase; pyrB, catalytic subunit of aspartate transcarbamylase; pyrI, regulatory subunit of aspartate transcarbamylase; pyrC, dihydroorotase; pyrD, dihydroorotate dehydrogenase; pyrE, orotate phosphoribosyltransferase; pyrF, OMP decarboxylase; pyrH, UMP kinase; ndk, nucleoside diphosphokinase; and pyrG, CTP synthetase.
By the early 1980s, the iconoclastic discoveries of activator proteins (35, 179) and attenuation control mechanisms of amino acid biosynthetic operons (175) in E. coli and S. enterica serovar Typhimurium raised the possibility that expression of the pyrimidine biosynthetic operons in these bacteria was controlled by novel mechanisms. However, nothing could have prepared us for the number of new mechanisms that would emerge. These mechanisms were elucidated once investigators began to focus on the regulation of individual operons. The first unique mechanism was attenuation control of pyrBI expression in E. coli, which employed a previously unrecognized method of controlling transcription termination at an attenuator (159). An analogous mechanism was also found to control pyrE expression in E. coli (13). Next was the discovery that pyrC expression in E. coli and S. enterica serovar Typhimurium was mediated by CTP-sensitive transcription start site switching, which produced alternative transcripts with different potentials for translation (153, 169). The expression of pyrD appeared to be similarly regulated (40). Perhaps the most surprising discovery was a second pyrBI control mechanism that employed the unusual reiterative transcription reaction during transcription initiation. This reaction results in the repetitive addition of UMP to the growing end of the nascent transcript. This transcript, with poly(U) at its 3′ end, can no longer be productively elongated and is eventually released from the transcription initiation complex (97). Reiterative transcription was then found to participate in the regulation of carAB expression (53) and that of the pyrimidine salvage operons codBA (137) and upp-uraA (157). The latter two mechanisms provided additional surprises, integrating two of the newly discovered paradigms.
ATTENUATION CONTROL BY COUPLED TRANSCRIPTION AND TRANSLATION
First Examples of Attenuation Control: a Mold To Be Broken
The concurrent pioneering studies of Charley Yanofsky, Bruce Ames, and their collaborators in the 1970s led to the discovery of transcription attenuation control mechanisms for the trp operon of E. coli and the his operon of S. enterica serovar Typhimurium (reviewed in reference 86). The hallmark of these regulatory mechanisms is control over transcript elongation at a conditional intrinsic transcription terminator, called the attenuator, within the leader region of each operon. An intrinsic transcription terminator specifies a G+C-rich RNA hairpin (stem-loop) followed typically by an eight-residue poly(U) tract, and termination requires that the hairpin form while RNA polymerase is completing the synthesis of the poly(U) tract (52, 150). In addition to the attenuator, the common regulatory elements in each leader region are a peptide-encoding open reading frame (ORF) that contains multiple adjacent codons for the regulating amino acid (i.e., tryptophan with the trp operon, etc.) and a leader transcript with segments capable of forming alternative RNA hairpins. (The leader transcript is defined as the RNA specified by the leader region of an operon.) The upstream-most hairpin (1:2 hairpin for trp) forms part of a transcription pause site used to synchronize leader transcription and translation, and the downstream-most hairpin (3:4 hairpin for trp) is the terminator hairpin specified by the attenuator. Formation of an alternative hairpin, called the antiterminator hairpin (2:3 hairpin for trp), precludes formation of the terminator hairpin, allowing transcription through the structural genes of the operon and production of the encoded enzymes. The peptide-encoding ORF of the leader transcript overlaps the upstream segment of the first hairpin (segment 1 for trp).
According to the model for regulation and using the trp operon as an example, transcription is initiated at the promoter and proceeds through the leader region specifying transcript segments 1 and 2, which then form the 1:2 hairpin. The transcribing RNA polymerase pauses at this point, permitting a ribosome to bind to the nascent transcript and initiate translation of a 14-codon ORF that encodes the leader peptide. Early in translation the ribosome releases the stalled RNA polymerase by disrupting hairpin 1:2, and then the ribosome proceeds to codons 10 and 11 of the leader ORF, which encode tandem tryptophan (Trp) residues. When Trp is limiting and the level of Trp-tRNATrp in the cell is low, the ribosome pauses at this site and covers transcript segment 1. During this time, the reengaged RNA polymerase continues transcription and synthesizes transcript segment 3, permitting formation of the 2:3 antiterminator hairpin. Continuing transcription extends the leader transcript through segment 4 and the poly(U) tract (without formation of the 3:4 terminator hairpin necessary for transcription termination) and eventually through the entire operon. Translation of the full-length trp mRNA produces the enzymes that increase the cell's capacity to make more Trp. On the other hand, when there is ample Trp and Trp-tRNATrp in the cell, the translating ribosome does not pause at the tandem Trp codons but proceeds to the stop codon at the end of the leader ORF. At this point, the ribosome covers transcript segments 1 and 2. As reengaged transcription continues, transcript segments 3 and 4 and the poly(U) tract are synthesized, allowing the 3:4 terminator hairpin to form and transcription termination to occur. As a consequence, the synthesis of more Trp biosynthetic enzymes is prevented when there is sufficient Trp-tRNATrp to support optimal cell growth. Regulation of the his operon occurs by an analogous mechanism in which seven adjacent histidine codons are used as the control codons in the leader region (72).
Soon after the elucidation of the attenuation control mechanisms of the trp and his operons, similar mechanisms were discovered for several other amino acid biosynthetic operons in enteric bacteria (86). Each example employed ribosome stalling at control codons as a regulatory signal and an alternative transcript secondary structure as a means of preventing terminator hairpin formation. These similarities raised the possibility that attenuation control was limited to amino acid biosynthetic operons and to a single mechanism for regulating transcription termination. However, this idea was soon dispelled by studies of the regulation of pyrBI expression in E. coli, which revealed an attenuation control mechanism that was fundamentally different from that described for the amino acid biosynthetic operons (159).
UTP-Sensitive Attenuation Control of pyrBI Expression in E. coli
In the in vivo studies of pyr operon expression in enteric bacteria described below, conditions of pyrimidine excess and limitation were typically produced by growing a pyrimidine auxotroph (usually carrying a mutation in the carAB operon) in a phosphate-buffered minimal medium with uracil or UMP as the pyrimidine source (140). Under these conditions, uracil is a good pyrimidine source and allows the auxotrophic cells to maintain pyrimidine nucleotide levels similar to those found in wild-type cells. In contrast, UMP is a poor pyrimidine source because it must be dephosphorylated to produce uridine, which unlike UMP can be transported into the cell. However, dephosporylation of UMP is a slow process when cells are grown with ample phosphate in the medium, and thus uridine production is restricted (168). As a consequence, pyrimidine nucleotide levels are lower and cell growth is slower in comparison to the case for wild-type cells.
The pyrBI operon of E. coli encodes the catalytic (pyrB) and regulatory (pyrI) subunits of the allosteric enzyme aspartate transcarbamylase, which catalyzes the first committed step in the de novo synthesis of pyrimidine nucleotides (Fig. 1). Expression of the pyrBI operon is negatively regulated over a wide range by pyrimidine availability, specifically by the intracellular concentration of UTP (98, 158). The regulatory role of UTP was first established in studies employing an in vitro DNA-dependent coupled transcription-translation system in which the levels of nucleotides and other small molecules could be varied (158). The discovery that a substrate for transcription was a regulatory effector of pyrBI expression suggested that the pyrBI control mechanism did not sense UTP levels per se but instead detected the effects of these levels on the rate of transcription of a regulatory site within the operon. This regulatory site was most likely located within the leader region. For this and other reasons, several laboratories determined the DNA sequence of the pyrBI leader region (120, 141, 159).
These studies identified the sequence of a putative intrinsic transcription terminator, or more specifically an attenuator, located 23 base pairs (bp) before the pyrB structural gene. Additional in vitro studies indicated that pyrBI transcription was initiated at either of two promoters located upstream of the attenuator and that this transcription was efficiently (∼98%) terminated at the attenuator (159). These results strongly indicated that pyrBI expression was regulated by a transcription attenuation control mechanism. However, the sequence of the pyrBI leader region revealed that the leader transcript could not adopt alternative secondary structures to regulate terminator hairpin formation, implying that attenuation control of pyrBI expression was mechanistically different from that described for the amino acid biosynthetic operons. The construction of a model for this new mechanism for attenuation control required the identification of two additional elements in the pyrBI leader region. The first was a 44-codon ORF that extends through the leader region and ends 6 base pairs before the pyrB gene (Fig. 2). In the leader transcript, this ORF is preceded by an apparent ribosome binding site, indicating that it can be translated. The second element was UTP-sensitive transcription pausing, i.e., pausing caused by low UTP levels, in the pyrBI leader region upstream of the attenuator. This pausing was detected in vitro (at 20 μM UTP) and initially appeared to be limited to a small cluster of sites at which UTP (or UMP after pyrophosphate release) was incorporated into the leader transcript (159). The pause site region was located approximately 20 nucleotides before the terminator hairpin.
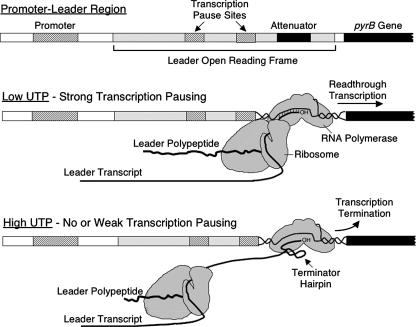
FIG. 2.
Model for attenuation control of pyrBI expression in E. coli. The diagram shows the relative positions of RNA polymerase and the translating ribosome within the leader region when UTP concentrations are either low or high. See the text for additional details. (Modified from reference 86 with permission.)
Based on these features and assuming that only the downstream in vitro promoter was physiologically significant, which was subsequently confirmed (31, 91, 96), the following model was proposed for UTP-mediated regulation of pyrBI expression (Fig. 2) (159). Transcription is initiated at the pyrBI promoter and proceeds into the 158-bp leader region. When the intracellular concentration of UTP is low, RNA polymerase stalls at the UTP-sensitive transcription pause sites, which provides time for a ribosome to initiate translation of the leader transcript and translate up to the stalled polymerase. When the RNA polymerase eventually escapes the pause region and transcribes the attenuator, formation of the terminator hairpin by the nascent transcript is blocked by the presence of the adjacent translating ribosome. In this case, transcription termination at the attenuator is precluded, and RNA polymerase continues transcription into the pyrBI genes. In contrast, when the intracellular concentration of UTP is high, RNA polymerase transcribes the leader region without stalling at the UTP-sensitive pause sites. In this instance, there is insufficient time for a ribosome to establish tight coupling with RNA polymerase (or perhaps even bind to the leader transcript) before the formation of the terminator hairpin. The result is transcription termination at the attenuator and no transcription of the pyrBI genes. The hallmark of this regulatory mechanism is that tight coupling between transcription and translation in the leader region allows a translating ribosome to disrupt or preclude the formation of the terminator hairpin by steric hindrance. In this mechanism, the extent of coupling reflects the intracellular concentration of UTP. Overall, this regulatory mechanism coordinates the synthesis of aspartate transcarbamylase with the level of UTP needed by the cell for optimal growth.
For a decade after this model was proposed, numerous studies that confirmed its key features were conducted. The central role of transcription termination at the pyrBI attenuator was established by biochemical analysis of cellular pyrBI transcripts and characterization of deletion mutations in the pyrBI leader region (31, 91, 92, 96, 98, 140). These studies clearly showed that pyrBI transcripts, initiated at a single physiologically relevant promoter, were subject to UTP-sensitive termination at the pyrBI attenuator in vivo. These studies also indicated that attenuation control accounted for most, although not all, pyrimidine-mediated regulation of pyrBI expression. To determine the contribution of attenuation control to this regulation, pyrBI expression was measured in a mutant E. coli strain containing a 9-bp chromosomal deletion that removes the run of eight T·A base pairs at the end of the pyrBI attenuator plus an adjacent base pair to maintain the reading frame of the leader polypeptide (98). All intrinsic transcription termination is abolished at this mutant attenuator. When the mutant strain was grown under conditions of pyrimidine excess, pyrBI expression was approximately 50-fold higher than that in an isogenic pyrBI+ strain. When growth of the mutant was limited for pyrimidines, operon expression increased an additional sevenfold. Growth of the pyrBI+ strain under the same pyrimidine-limiting conditions resulted in a 300- to 350-fold increase in operon expression. These results indicate that attenuation control is responsible for pyrimidine-mediated regulation over a 50-fold range, while additional regulation occurs over a sevenfold range through another control mechanism. The latter mechanism, which involves reiterative transcription, will be described in detail below.
In the pyrBI attenuation control model, translation of the 44-codon ORF of the leader transcript plays a critical regulatory role. To confirm that the leader ORF was indeed translated in vivo, a gene fusion was constructed in which the pyrBI promoter-leader region through codon 11 was fused in frame to codon 9 of the lacZ gene. An E. coli strain carrying this gene fusion synthesized a β-galactosidase fusion protein with the amino-terminal sequence of the leader polypeptide (140). To show that regulation of the pyrBI operon requires translation of the leader ORF, the in vivo effects of mutations that either strongly inhibit translation initiation of the ORF or introduce stop codons early in the ORF, well before the attenuator, were measured (26, 139, 140). Each mutation greatly reduced operon expression, especially under conditions of pyrimidine limitation, and significantly reduced the range of pyrimidine-mediated regulation. Furthermore, mutant (i.e., rpsL) strains containing slowly translating ribosomes exhibited reduced pyrBI expression, apparently due to reduced coupling of transcription and translation in the pyrBI leader region (64). Although translation of the leader ORF is clearly important for regulation, the sequence of the encoded polypeptide is not. A mutant strain carrying a frameshift mutation that changes the sequence of the leader polypeptide, while still allowing translation of the entire leader region, exhibited essentially normal attenuation control (26).
One of the major assumptions of the model is that under conditions of pyrimidine limitation, tight coupling of transcription and translation in the pyrBI leader region allows the ribosome to physically prevent the formation of the terminator RNA hairpin. To test this assumption, stop codons were individually introduced at numerous sites within the 44-codon leader ORF to determine the distance that a ribosome must translate to suppress transcription termination at the attenuator (139). Based on the size of the ribosome footprint on its RNA template, translation would have to proceed to a codon located within approximately 15 nucleotides of the terminator hairpin sequence to permit the ribosome to interact directly with this sequence (79, 154). Examination of the strains carrying separate stop codon mutations showed that translation termination at or before codon 24, which is 16 nucleotides upstream of the terminator hairpin, limited operon expression to approximately 5% of the wild-type level under pyrimidine-limiting conditions. In contrast, translation termination at codon 25, which should be the first stop codon at which ribosome binding overlaps the sequence of the terminator hairpin, allowed expression at 64% of the wild-type level. The level of operon expression generally increased to near-wild-type levels as the stop codon was moved further downstream, perhaps reflecting greater disruption of the terminator hairpin. These results provide strong support for the proposed role of the ribosome. In addition, the observation that pyrBI expression increased as the stop codon mutations were moved downstream of codon 25 suggests that pyrBI expression is enhanced by coupling of translation of the leader ORF and the pyrB cistron. In the wild-type pyrBI operon, it is likely that such coupling occurs due to the close proximity of these ORFs (159).
The model requires only a single round of translation of the leader transcript to allow readthrough transcription, and more translation would presumably be wasteful. Such wasteful translation appears to be limited by the use of a relatively weak ribosome binding site preceding the leader ORF (86). In addition, sequences in the downstream half of the leader transcript are complementary to the leader ribosome binding site (120, 140). Formation of a secondary structure by these sequences could block multiple rounds of translation of readthrough transcripts and perhaps all translation of attenuated transcripts.
The discovery of UTP-sensitive transcription pausing in the pyrBI leader region was key in developing the attenuation control model. This pausing provided the regulatory sensor, equivalent to control codons in the amino acid biosynthetic operons, that responds to different levels of UTP in a way that influences transcription termination at the attenuator. In E. coli, the UTP concentration varies from approximately 50 μM in cells grown under conditions of severe pyrimidine limitation to 1 mM or slightly above in cells grown under conditions that provide ample pyrimidines (3, 124, 158). The first in vitro experiments to detect UTP-sensitive transcription pausing in the pyrBI leader region revealed only a small cluster of pause sites that correspond to a uridine-rich region located approximately 20 nucleotides before the terminator hairpin in the leader transcript (159). Subsequent in vitro transcription studies employing a more sensitive assay provided a different view of pausing in the leader region preceding the attenuator (32). Instead of one cluster of pause sites, there is a large number of sites throughout the leader transcript at which RNA polymerase pauses when the UTP concentration is low. Nearly all of these sites correspond to positions where UMP is added to the leader transcript. Pausing at these sites decreases with increasing UTP concentrations (from 20 to 200 μM) and is no longer detectable at a concentration of 400 μM. Although some degree of pausing apparently can occur before the addition of every UMP in the leader transcript at 20 μM UTP, the strength of individual pause sites is variable. This variability presumably reflects the effects of DNA sequence and RNA secondary structure (18, 19). In this regard, an upstream RNA hairpin enhances pausing within the originally identified cluster of UTP-sensitive transcription pause sites (86, 159). Although some pause sites within the leader region may be stronger than others, the large number of these sites indicates that the cumulative effect of pausing at multiple positions is the key factor in controlling coupling between RNA polymerase and the ribosome translating the pyrBI leader transcript. Consistent with this view, replacing all seven uridines in the originally identified pause cluster with adenines causes only a twofold reduction in the range of pyrimidine-mediated regulation of pyrBI expression (K. Mixter-Mayne and C. L. Turnbough, Jr., unpublished data).
In contrast to transcription pausing observed at low UTP concentrations, extensive pausing in the pyrBI leader region was not induced when the concentration of ATP, GTP, or CTP was low (i.e., 20 μM) (32). This difference appears to be due, at least in part, to a difference in the Km values for these nucleotides during transcription elongation. The apparent Km for UTP during elongation appears to be significantly higher than the Km values for the other nucleoside triphosphates (NTPs) (66, 85). This higher Km apparently results in nonsaturating binding of UTP to an elongating RNA polymerase at all physiological concentrations of UTP (i.e., in cells with limiting or ample pyrimidines). This situation appears to be unique because the physiological concentrations of the other NTPs are typically well above their Km values for transcription elongation (124, 135). Thus, the rate of transcription elongation is uniquely sensitive to the intracellular concentration of UTP, which makes UTP an ideal regulatory effector for a control mechanism based on coupling of transcription and translation.
Additional noteworthy support for the proposed role of UTP-sensitive transcription pausing in attenuation control came from studies of pyrBI regulation in S. enterica serovar Typhimurium, which is similar to that in E. coli (see below). A mutant strain was isolated that carries an altered RNA polymerase that exhibits an approximately sixfold-higher Km for the binding of UTP (and ATP) during transcription elongation (66). This mutant displayed constitutive expression of the pyrBI operon at high intracellular levels of UTP, indicating that transcription pausing during the addition of UMP (or another nucleotide) to the pyrBI leader transcript, and not the UTP level, is the key determinant in regulation. In related studies with E. coli, it was shown that the transcription elongation factor NusA enhances UTP-sensitive pausing within the pyrBI leader region in vitro and appears to be important in determining the level of pyrBI expression in vivo (3, 32). Presumably, NusA plays a key role in establishing a rate of transcription elongation that permits tight coupling of transcription and translation in cells limited for pyrimidines. These results indicate that the activity of NusA or of any factor that influences the rate of transcription elongation can affect the expression of the pyrBI operon or of similarly regulated operons.
Attenuation Control of pyrBI Expression in Other Enteric Bacteria
The earliest studies of pyrimidine biosynthetic gene expression in bacteria indicated that pyrBI expression was regulated similarly in E. coli and S. enterica serovar Typhimurium, which are closely related enteric bacteria. This similarity was confirmed with the determination of the sequence of the pyrBI operon of S. enterica serovar Typhimurium (117). The leader region of this operon is identical in length and very similar in sequence to that of E. coli, and it contains all the regulatory elements described above for UTP-sensitive attenuation control. Deletion of two T·A base pairs at the end of the pyrBI attenuator, which greatly reduces transcription termination efficiency, resulted in a 30-fold increase in pyrBI operon expression in S. enterica serovar Typhimurium, confirming the central regulatory role of transcription attenuation (117). The most notable difference between the pyrBI leader regions of E. coli and S. enterica serovar Typhimurium is that the latter contains a 33-codon ORF. This shorter ORF is due to a sequence difference that introduces an earlier in-frame stop codon in the leader transcript of S. enterica serovar Typhimurium. However, this stop codon is located near the middle of the sequence for the terminator hairpin, and translation to this point would still preclude formation of this hairpin. In fact, a mutation that introduces a stop codon at an equivalent site in the pyrBI leader transcript of E. coli allows for nearly normal levels of expression and regulation (139). On the other hand, the shorter ORF in the S. enterica serovar Typhimurium pyrBI leader transcript may preclude translation coupling with the pyrB cistron. Such coupling, which likely occurs in E. coli, would presumably enhance pyrBI expression.
The attenuation control mechanisms of the pyrBI operons of E. coli and S. enterica serovar Typhimurium were elucidated the old-fashioned way, i.e., by doing many experiments. These experiments identified readily recognizable regulatory sequences. Today, it is possible to inspect a large number of bacterial genomes for these regulatory sequences and thereby identify other operons that are likely to be regulated by attenuation control mechanisms similar to those described above. Although many search formats can be used, even limited searches reveal interesting information about the prevalence of particular control mechanisms. For example, a BLAST search of currently available bacterial genome sequences using the amino acid sequence of the pyrBI leader polypeptide as the query (with CLUSTAL W alignment) produced 14 matches. All matches correspond to polypeptides encoded by the pyrBI leader regions of five strains of E. coli (i.e., K-12 MG1655, K-12 W3110, O157 EDL933, O157 Sakai, and CFT073), five strains of Shigella (i.e., S. flexneri 301 and 2457T, S. dysenteriae, S. boydii, and S. sonnei), and four strains of Salmonella (i.e., S. enterica serovar Typhimurium LT2, S. enterica serovar Typhi CT18 and Ty2, and S. enterica serovar Paratyphi A). All 10 of the E. coli and Shigella polypeptides contain 44 amino acids; eight of the polypeptide sequences are identical, and two (from the S. flexneri strains) contain a single amino acid difference. All four of the Salmonella polypeptides contain 33 amino acids, due to the shorter leader ORF described above, and their sequences are identical. These four sequences differ at only five residues compared to the other 10 polypeptides. These results and further inspection of leader sequences indicate that the 14 strains listed above employ an essentially identical attenuation control mechanism for pyrimidine-mediated regulation of pyrBI expression. The results are also consistent with the established evolutionary relationships among strains of Escherichia, Shigella, and Salmonella (41).
In the search for matches to the E. coli pyrBI leader polypeptide, the misses are as interesting as the hits. For example, no matches were found in the genome sequences of many other enteric bacteria. This result may indicate that the mechanisms for regulating pyrBI expression in these bacteria are different from that described for E. coli. However, inspection of selected “missed” enteric pyrBI operons indicates that they may still be regulated by an E. coli-like attenuation control mechanism—one that employs comparable regulatory elements with distinct sequences. This situation appears to be the case for Yersinia pestis CO92 and Erwinia cartovora, which have all the regulatory elements found in the E. coli leader region, including 41- and 40-codon ORFs, respectively. These ORFs encode leader polypeptides with no sequence similarity to the leader polypeptide of E. coli and modest sequence similarity with each other. However, the leader ORFs of Y. pestis and E. cartovora both stop at the same position near the middle of the sequence for the terminator hairpin, which is similar to the situation described for S. enterica serovar Typhimurium. Interestingly, the sequence of the leader polypeptide of Y. pestis is very similar (i.e., 57% identical) to that of a 37-amino-acid leader polypeptide encoded by the pyrBI leader ORF of Serratia marcesens. On the other hand, the leader region of S. marcesens does not appear to contain the sequence for an E. coli-like intrinsic transcription terminator, suggesting another regulatory twist. It should also be noted that the search for matches to the E. coli pyrBI leader polypeptide missed all nonenteric gram-negative bacteria. Nonetheless, inspection of selected genomic sequences, e.g., that of Vibrio cholerae, suggests again that E. coli-like regulation of pyrBI expression may occur but with divergent (and perhaps some new) regulatory elements.
Attenuation Control of pyrE Expression in E. coli
Early studies suggested that each E. coli pyrimidine biosynthetic operon would be regulated by an independent mechanism, which later research would show to be true. However, some of these independent control mechanisms are analogous. A case in point is the regulation of pyrE expression. The pyrE gene encodes the pyrimidine biosynthetic enzyme orotate phosphoribosyltransferase (Fig. 1). Expression of this gene is regulated over a 30-fold range almost entirely by an attenuation control mechanism that is analogous to that described for the pyrBI operon (13, 64, 134-136). However, there is a striking difference. The pyrE “leader ORF” contains 238 codons and is actually the rph gene, which encodes the tRNA-processing exoribonuclease RNase PH (129). Thus, the pyrE gene is the second gene of an rph-pyrE operon, and the cell uses UTP-sensitive transcription along with translation of the rph gene to control transcription termination at an attenuator preceding the pyrE gene. Another interesting contrast to the pyrBI story is that in the rph-pyrE transcript, the rph cistron ends 10 bases before the terminator hairpin sequence specified by the pyrE attenuator. Even so, based on the size of the ribosome footprint, translation to the end of the rph cistron would permit disruption of the terminator hairpin, thereby allowing readthrough transcription. Although it is now clear that the number of mechanistic variants used by bacteria to control gene expression by transcription attenuation is nearly endless (56, 86), especially with the recent discovery of riboswitches (50, 173), the studies of pyrBI and pyrE expression in enteric bacteria provided an exciting preview of coming attractions.
CONTROL OF TRANSLATION INITIATION VIA NUCLEOTIDE-SENSITIVE SELECTION OF TRANSCRIPTION START SITES
Promoters and Transcription Start Sites
Transcription of pyrimidine biosynthetic operons in enteric bacteria is initiated at promoters recognized by RNA polymerase containing the primary sigma factor σ70. This sigma factor recognizes −10 and −35 regions for which the consensus sequences are 5′-TATAAT and 5′-TTGACA, respectively (118). The spacing between the −10 and −35 regions is typically 17 ± 1 bp, and transcription is usually initiated at one or more sites located 7 ± 1 bp downstream from the −10 region (55, 147). At about 75% of promoters, transcription is initiated with ATP or GTP (95). In some molecular genetics textbooks, this preference is used to imply that initiation with CTP or UTP is of little importance. However, initiation with pyrimidine NTPs is often an essential element in gene expression. This fact was first demonstrated in studies of pyrC expression.
CTP-Sensitive Regulation of pyrC Expression
In E. coli and Salmonella enterica serovar Typhimurium, the pyrC gene encodes the pyrimidine biosynthetic enzyme dihydroorotase (Fig. 1). The primary pyrimidine regulatory effector of pyrC expression was identified as a cytidine nucleotide, probably CTP (145), and additional studies suggested that pyrC expression was regulated by the ratio of the intracellular concentrations of CTP and GTP (65). In early studies to define the mechanism controlling pyrC expression, it was found that the steady-state levels of pyrC transcripts and dihydroorotase activity changed coordinately in response to pyrimidine availability in E. coli, suggesting regulation at the transcriptional level (170). Furthermore, a highly conserved operator-like sequence was identified in the promoter regions of the pyrC and other pyrimidine biosynthetic (i.e., pyrD and carAB) operons whose expression appeared to be negatively regulated by CTP. This discovery suggested that pyrC expression was regulated by a pyrimidine repressor that employed CTP as a corepressor (170). However, subsequent studies provided different explanations for the circumstantial evidence for this model. The pyrimidine-mediated changes in the levels of pyrC transcripts were due not to changes in the rate of synthesis of these transcripts but to changes in their stability because of differential translation (99, 169). The operator-like sequence was in fact shown to be an operator but not one for a pyrimidine repressor. Instead, this operator was the binding site for the purine regulon repressor, PurR, which controls pyrC expression over a modest twofold range in response to purine availability in E. coli (25, 171) and S. enterica serovar Typhimurium (123). PurR is not involved in pyrimidine-mediated regulation of pyrC expression, which occurs over an approximately 15-fold range.
The experiments that eventually led to the correct mechanism for pyrimidine-mediated regulation of pyrC expression began with the determination of the sequence of the pyrC operon and primer extension mapping of its transcription start sites (9, 122, 170). Transcription initiation occurs at four adjacent sites in the initially transcribed region (ITR) of the promoter (170). The nontemplate strand sequence of these sites is 5′-TCCG, which is located 6 to 9 bp downstream from the −10 region (Fig. 3). These sites are designated T6, C7, C8, and G9, and the transcripts initiated at these sites are called the U6, C7, C8, and G9 transcripts, respectively (99). Inspection of the pyrC sequence also revealed a hyphenated dyad symmetry that includes the ITR of the promoter and a downstream region specifying part of the Shine-Dalgarno (SD) sequence of the pyrC ribosome binding site (Fig. 3) (100). This sequence indicates that U6 transcripts would form a hairpin with a 6-bp stem in which the upstream segment includes the first six nucleotides of the transcript and the downstream segment includes most of the pyrC SD sequence. Transcripts starting further downstream (i.e., at C7, C8, and G9) would form progressively shorter hairpins, with the shortest being a 3-bp hairpin formed by G9 transcripts. However, the calculated free energy of formation of the shortest possible hairpin suggests that it would not be stable in cells (39, 111), a supposition that was later confirmed experimentally (99).
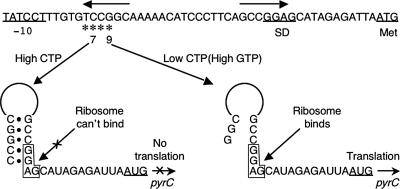
FIG. 3.
Model for transcription start site switching and translational control of pyrC expression in E. coli and Salmonella. The nucleotide sequence of the pyrC promoter-regulatory region of E. coli is shown, with the −10 region, SD sequence, and pyrC initiation (Met) codon underlined and labeled. Asterisks indicate the four transcription start sites at the pyrC promoter, and the two major start sites, C7 and G9, are indicated. Inverted horizontal arrows indicate the region of dyad symmetry. The sequence and structure of transcripts initiated at start sites C7 (high CTP) and G9 (low CTP) are shown, with the SD sequence boxed. Only C7 transcripts form the hairpin that includes the SD sequence and prevents translation initiation.
The final parts of the puzzle included the demonstration that point mutations in the hyphenated dyad symmetry, which were expected to destabilize the encoded hairpin, cause constitutive pyrC expression (82). In the same study, it was shown that expression of a transcriptional pyrC::galK fusion constructed with a short fragment of the pyrC operon is not regulated by pyrimidine availability, while expression of a translational fusion containing the same pyrC fragment is regulated. These observations led Kelln and Neuhard to propose that pyrC expression is regulated at the level of translation initiation through modulation of the secondary structure of the leader transcript (82). The regulatory input of intracellular CTP levels in this mechanism was suggested by the discovery that the selection of the pyrC transcription initiation site is affected by pyrimidine availability (153, 169, 170). Under conditions of pyrimidine excess, position C7 is the dominant start site; under conditions of pyrimidine limitation, the dominant start site is G9. This feature and those described above, which are identical in E. coli and S. enterica serovar Typhimurium, gave rise to the current model for regulation of pyrC expression (153, 169).
According to the model (Fig. 3), nucleotide-sensitive selection of transcription start sites is used to produce alternative transcripts with different potentials for translation. When the intracellular level of CTP is high (e.g., during growth with excess pyrimidines), C7 transcripts are synthesized predominantly. These transcripts are not translated, however, because they form a stable hairpin at their 5′ ends that blocks ribosome binding to the pyrC SD sequence. In contrast, when the CTP level is low and the GTP level is high, conditions found in cells limited for pyrimidines (142), G9 transcripts are synthesized primarily. The shorter G9 transcripts are unable to form the inhibitory hairpin and are readily translated. Thus, this mechanism allows the level of pyrC expression to change according to the cell's requirement for pyrimidine nucleotides. Furthermore, in this model changes in pyrC expression can be gradual in response to incremental changes in the intracellular CTP (and GTP) concentrations.
The key aspects of the model have been confirmed. The importance of the hairpin at the 5′ end of the pyrC transcript was shown by using pairs of mutations in the hyphenated dyad symmetry of the pyrC leader region. Individually, these mutations cause constitutive pyrC expression. However, when a pair of complementary mutations capable of restoring complete base pairing in the leader transcript hairpin was introduced into a strain, it exhibited nearly normal levels of pyrimidine-mediated regulation of pyrC expression (153, 169). In related experiments, direct evidence for the predicted secondary structure at the 5′ end of C7 transcripts and the absence of this structure in G9 transcripts was obtained by chemical and enzymatic probing of pyrC transcripts isolated from cells grown under conditions of pyrimidine excess or limitation (151). The importance of start site switching was demonstrated by showing that a strain carrying a mutant pyrC promoter unable to switch start sites (e.g., containing a C7-to-A or C7-to-G mutation [see below]) fails to exhibit pyrimidine-mediated regulation of pyrC expression (99). In addition, nucleotide (CTP/GTP)-sensitive selection of transcription starts sites was demonstrated in vitro using a transcription assay containing only highly purified RNA polymerase, DNA template, NTP substrates, and salts. These results closely mimicked those observed in vivo, indicating that additional regulatory factors are not required for transcription start site switching at the pyrC promoter (169). One seemingly wasteful feature of the model is the synthesis of untranslated C7 transcripts. It was suggested that these transcripts would be prematurely terminated, as observed in polarity (82). Such a fate for C7 transcripts is indeed likely, because multiple Rho-dependent termination sites exist early in the pyrC ORF (J. Liu and C. L. Turnbough, Jr., unpublished data). Perhaps the most intriguing feature of the model for regulation of pyrC expression was nucleotide-sensitive selection of transcription start sites. Characterizing transcription initiation at mutant pyrC promoters provided rules for this selection process.
Rules for Selecting Transcription Start Sites and a Revised Model for pyrC Regulation
In E. coli and S. enterica serovar Typhimurium growing exponentially in minimal-glucose or rich media, the intracellular concentrations of CTP and GTP are approximately 0.7 mM and 1.1 mM, respectively. These cells also contain approximately 1.4 mM UTP and 2.7 mM ATP (110, 124). When cells are grown under conditions that severely limit pyrimidine availability, the CTP and UTP levels decrease about 3-fold and 20-fold, respectively. In contrast, under these conditions the GTP and ATP levels each increase approximately threefold (142). These changes seem sufficient to explain the initial step in pyrimidine-mediated regulation of pyrC expression, namely, CTP/GTP-sensitive selection of transcription start sites. Assuming that CTP and GTP are competing initiating nucleotides, CTP would “win” when CTP and GTP concentrations were similar, and GTP would “win” when its concentration was much greater than the CTP concentration. However, this simple solution implies that CTP is a better initiating nucleotide than GTP. If this is true, then it seems peculiar that many more E. coli and S. enterica serovar Typhimurium transcripts are initiated with GTP than with CTP. These observations indicated that more experiments were needed to establish the basis for transcription start site selection. The pyrC promoter-leader region was well suited for use in quantitative primer extension mapping experiments to determine preferred initiating NTPs and transcription start sites (99).
The nontemplate strand sequence of the pyrC ITR is 5′-TCCGG, located 6 to 10 bases downstream of the −10 region (Fig. 3). Transcription at the wild-type promoter can occur at the first four positions, as described above. Therefore, if context effects are ignored and corrections are made for different transcript stabilities, the levels of C7 and C8 transcripts in cells can be used to calculate the frequency of in vivo transcription initiation at positions C7 and C8 (99). Such an experiment demonstrated that C7 was a fivefold better start site than C8. If a single-base deletion that removes the T residue immediately downstream of the −10 region is introduced into the pyrC promoter, the possible start sites are now CCGG at “new” positions 6 to 9. Repeating the experiment described above with the mutant promoter revealed that C7 was a much better start site than C6, with C6 transcript levels so low that they could not be measured. Likewise, it was possible to use the tandem G8/G9 sites to show that G8 was a 13-fold-better start site than G9. Additional mutant promoters were then constructed in which other deletions (i.e., ΔTT and ΔTTG) or a T insertion were introduced immediately downstream of the −10 region. These promoters created more possible positions for the CC and GG pairs, and the transcripts initiated at these sites were analyzed as described above. Combining all of the results permitted the assignment of the following preferences for start site positions: 7 > 8 > 6 > 9 > 10. Similar analyses were performed to determine preferences for the initiating nucleotide, using a different set of mutant pyrC promoters. These promoters contain single-base substitutions at the best initiation position, 7, and at a relatively poor initiation position, 9. Specifically, C7 was changed to a T, G, or A, and G9 was changed to a C or A. Measuring the frequency of initiation at these sites revealed the following preferences for the initiating nucleotide: ATP ≥ GTP > UTP ≫ CTP. The actual difference between the initiation efficiencies of UTP and CTP was sixfold, making CTP the poorest initiating nucleotide by far. Although the experiments described above were done with E. coli (99), the same preferences were observed at wild-type and mutant pyrC promoters in S. enterica serovar Typhimurium (152).
The preferences or “rules” for selecting transcription start sites suggest a somewhat revised version of the model for pyrC regulation. Specifically, these rules provide the basis for nucleotide-sensitive start site switching at the wild-type pyrC promoter. The worst initiating nucleotide (CTP) is used to start transcripts at the best start location (position 7), and a good initiating nucleotide (GTP) is used to start transcripts at a weak start location (position 9). These combinations establish competition between initiation at positions C7 and G9, which can be influenced dramatically by changes in intracellular levels of CTP and GTP that reflect pyrimidine availability. The same rules restrict transcription initiation at positions T6 and C8, which utilizes the combination of a suboptimal start position and a poor initiating nucleotide.
It appears that the rules for selecting transcription start sites identified with the pyrC promoter apply in general to other σ70 promoters. Examination of several hundred well-characterized E. coli promoters shows frequencies for selecting initiating nucleotides (A [47%], G [28%], T [15%], and C [10%]) (57, 95) and start site positions (7 [40%], 8 [24%], 6 [11%], 9 [10%], and other sites [≤5%]) (55) that reflect the preferences identified above. These results suggest that most transcripts start with efficient initiating nucleotides and favored positions to maximize transcript synthesis. It also suggests that the use of inefficient initiating nucleotides and less favored positions is evolutionarily selected to reduce or control transcript synthesis.
Finally, the rules described above for selecting transcription start sites ignore context effects. However, the local DNA sequence can be an important factor in start site selection (17, 69, 93, 166). Of particular importance is the sequence at position +2 of the transcript, which accounts for the so-called second-nucleotide effect. It was demonstrated many years ago (113, 126), and again in a clear fashion during the analysis of mutant pyrC promoters (152), that high concentrations of both the first and second NTP substrates are required for highly efficient initiation of transcription. Apparently, after formation of the first internucleotide bond, the dinucleotide product stabilizes the transcription initiation complex. Lower concentrations of NTP substrates are required for transcript extension beyond position +2, though the relaxation of the requirement for high NTP concentrations may occur gradually until promoter clearance (2). Based on these observations and the fact that the +2 nucleotide in pyrC C7 transcripts is a C, it appears necessary to make a final modification to the model for pyrC regulation. Namely, synthesis of C7 transcripts is restricted at low CTP concentrations because of insufficient levels of the first and second NTPs required to initiate transcription.
CTP-Sensitive Regulation of pyrD Expression
In E. coli and Salmonella enterica serovar Typhimurium, the pyrD gene encodes the membrane-associated flavoprotein dihydroorotate dehydrogenase (88), which catalyzes the fourth step in the de novo pyrimidine nucleotide biosynthetic pathway (Fig. 1). Pyrimidine-mediated regulation of pyrD expression occurs over an approximately 20-fold range (124) through a mechanism analogous to that described for the pyrC gene (40, 152). The only noteworthy difference is that the nontemplate strand sequence of the pyrD transcription start region is 5′-CCCG (instead of 5′-TCCG). Transcription initiation at the pyrD promoter appears to occur primarily at positions C6 and C7 under conditions of pyrimidine excess and at position G9 under conditions of pyrimidine limitation. The longer C6 and C7 transcripts are capable of forming a stable hairpin at their 5′ ends that blocks ribosome binding to the pyrD SD sequence, while shorter G9 transcripts cannot form this hairpin and are readily translated (151). Also, as described for the pyrC operon, the purine repressor PurR controls pyrD expression over an approximately twofold range in response to purine availability (163).
Inspection of published bacterial promoter sequences reveals many other transcription initiation regions at which nucleotide-sensitive start site switching is predicted. Such switching can produce transcripts with minor differences in sequence at their 5′ ends, which produce major differences in the ability of the transcripts to be translated. This effect may be due to formation of secondary structures that inhibit translation initiation as seen with the pyrC and pyrD regulatory mechanisms. However, nucleotide-sensitive start site switching can generate sequence differences at the 5′ ends of transcripts that alter gene expression in many other ways, some of which were also discovered by studying genes of pyrimidine metabolism (see below).
REGULATION BY UTP-SENSITIVE REITERATIVE TRANSCRIPTION
Reiterative Transcription
Reiterative transcription, which is also known as pseudotemplated transcription, transcriptional slippage, and RNA polymerase stuttering, is a reaction catalyzed by a number of RNA polymerases, including bacterial, phage, viral, and eukaryotic enzymes (62, 68, 94, 107, 137). In this reaction, nucleotides are repetitively added to the 3′ end of a nascent transcript because of slippage between the transcript and DNA (or viral RNA) template. Typically, slippage occurs between a homopolymeric sequence in the transcript and at least three complementary bases in the template (23, 174). The mechanism apparently involves one or more rounds of a one-base upstream shift of the transcript so that the same nucleotide in the template specifies multiple residues in the transcript (10, 51). Reiterative transcription can occur during initiation or elongation, resulting in transcripts that can be immediately released from the transcription complex (11, 97) or extended by normal elongation after a switch to nonreiterative nucleotide addition (87, 164). Although reiterative transcription can involve the addition of any nucleotide, at least under certain conditions, addition of U or A residues appears to occur most frequently. This preference presumably reflects a requirement in the reaction for disruption of the RNA-DNA hybrid, which would be facilitated by relatively weak U·A or A·T base pairing (34).
Second Mechanism to Regulate pyrBI Expression in E. coli
As described above, characterization of the transcription attenuation control mechanism of the pyrBI operon of E. coli revealed that pyrimidine (UTP)-mediated regulation of pyrBI expression also occurs through a second mechanism, which independently controls operon expression over a sevenfold range. Several studies indicated that this second mechanism requires only the pyrBI promoter region and functions at the level of transcription initiation (31, 96, 98). Other observations suggested that this second mechanism involves a run of three T·A base pairs (nontemplate strand T residues) in the ITR of the pyrBI promoter. The pyrBI promoter region contains the sequence 5′-TATAATGCCGGACAATTTGCCG, with the −10 region and the in vivo transcription start site (A8) underlined (31). It was discovered that RNA polymerase forms heparin-resistant, transcription-competent initiation complexes at the pyrBI promoter in the presence of ATP but not with ATP and UTP. This result suggested that the synthesis of a nascent transcript with the sequence AAUUU (but not AA) destabilizes the initiation complex or perhaps interferes with promoter clearance. It was proposed that this effect could be modulated by the intracellular concentration of UTP and thus contribute to pyrimidine-mediated regulation (31). These observations lingered, however, until a fortuitous encounter with a report of pseudotemplated transcription at a mutant sar promoter of phage P22 (63). This mutant promoter contained a G-to-T change at the transcription start site (+1), which created a run of four nontemplate strand T residues from −1 to +3 (i.e., TGTT to TTTT). Transcription from the mutant promoter in vitro produced poly(U) transcripts of various lengths, with abundance decreasing with length. The only requirement to detect the more abundant short poly(U) transcripts was separation of transcription products in a high-percentage polyacrylamide gel.
The sequence requirement for reiterative transcription at the mutant sar promoter, as well as at several other promoters (51, 54, 59, 106), appeared to be a short (i.e., ≥3-bp) tract in the ITR that specified a homopolymeric run in the nascent transcript. Thus, the run of three T residues at positions +3 to +5 in the ITR of the pyrBI promoter appeared to be a possible site for reiterative transcription. To investigate this possibility, the pyrBI promoter-leader region was transcribed in vitro in reaction mixes containing high (≥200 μM) or low (20 μM) concentrations of UTP, with high concentrations of [γ-32P]ATP, GTP, and CTP. The transcripts produced were separated in a 25% polyacrylamide gel (a procedure never employed in the many previous analyses of pyrBI transcripts synthesized in vitro) and visualized by autoradiography. The results revealed a ladder of transcripts generated at high UTP concentrations, with the longest transcript containing over 30 nucleotides. Synthesis of this ladder was greatly reduced at the low UTP concentration. The sequences of the transcripts in the ladder were shown to be AAUUUn (with n = 1 to >30), which established that reiterative transcription indeed occurs at the T3 tract within the pyrBI ITR. Furthermore, transcripts containing extra (i.e., >3) U residues were always released from the transcription initiation complex without switching to normal transcript elongation (which was also demonstrated in vivo), and synthesis of the AAUUUn transcripts inhibited the production of full-length pyrBI transcripts (97). These results suggested that reiterative transcription could be involved in UTP-sensitive regulation of transcription initiation at the pyrBI promoter.
To examine the role of reiterative transcription in regulation of pyrBI expression, base substitutions were introduced into the T3 tract within the pyrBI ITR. Transcription in vitro of DNA templates carrying these substitutions showed that any change in the T3 tract abolished reiterative transcription (L. Heath and C. L. Turnbough, Jr., unpublished data). Using a mutant strain carrying one of these base substitutions, it was shown that pyrBI expression was sevenfold greater that that observed in a pyrBI+ strain when cells were grown under conditions of pyrimidine excess. When this base substitution was introduced into a strain carrying a defective pyrBI attenuator, pyrimidine-mediated regulation of pyrBI expression was effectively eliminated (97). These results demonstrate the regulatory role of reiterative transcription at the pyrBI promoter and show that UTP-dependent reiterative transcription and UTP-sensitive transcription attenuation are sufficient to account for all pyrimidine-mediated regulation of pyrBI expression in E. coli.
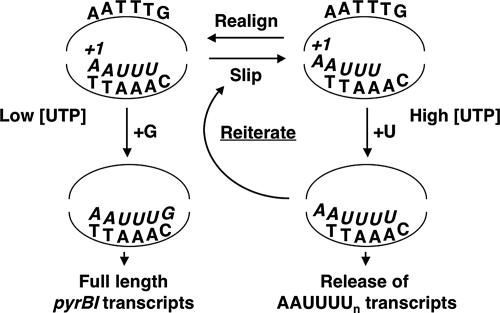
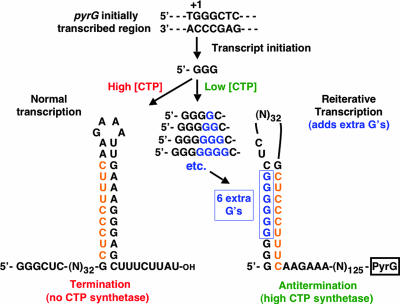
According to these observations, the following model was proposed for regulation of pyrBI expression by reiterative transcription (Fig. 4) (97). After the synthesis of the nascent transcript AAUUU, weak base pairing between the transcript and its DNA template allows a rapid and reversible one-base upstream shift (or slip) of the nascent transcript. When the intracellular level of UTP is high and the transcript is in the “slipped” position, the last (i.e., 5′) A in the AAA tract in the DNA template efficiently directs the addition of another U residue to the 3′ end of the transcript. This transcript can be released from the transcription initiation complex or it can shift again. The cycle of slippage and U addition can occur repeatedly, resulting in transcripts with progressively longer runs of U residues. However, all AAUUUUn transcripts are eventually released from the initiation complex, thereby preventing productive transcription of the pyrBI operon. On the other hand, when the intracellular level of UTP is low, slippage (if it occurs) and correct repositioning of the AAUUU transcript—without addition of extra U residues—occurs predominantly. Correct positioning of the RNA-DNA hybrid permits the addition of a G residue to the 3′ end (i.e., position +6) of the transcript. Once this addition occurs, more stable base pairing between the transcript and template precludes further slippage. The AAUUUG transcript either is released from the initiation complex, as a simple aborted transcript, or is extended by the addition of a C residue, which apparently commits the transcription complex to the elongation mode (97). Therefore, high levels of full-length pyrBI transcripts are produced only when their encoded enzyme, aspartate transcarbamylase, is needed to synthesize more UTP. In this model, regulation of pyrBI expression can occur gradually, over a range of intracellular UTP concentrations, by corresponding adjustments in the efficiency of reiterative transcription.
FIG. 4.
Model for the regulation of pyrBI expression by UTP-sensitive reiterative transcription. DNA sequences in the transcription bubble are shown, and the sequence of the nascent transcript, starting at position +1, is italicized. For details, see the text.
Distribution of the “TTT Motif”
Comparison of the sequences of the pyrBI promoter and other promoters at which UTP-dependent reiterative transcription occurs (63, 71, 174) suggested that the only requirement for this reaction during transcription initiation is a run of at least three nontemplate strand T residues located at or very near the beginning of the ITR. If in fact these conditions were sufficient to permit reiterative transcription, it would seem likely that other operons with promoters containing the TTT motif would be subject to regulation similar to that described for the pyrBI operon. Inspection of approximately 500 well-characterized E. coli promoters (57, 95) revealed that approximately 10% of these contain a run of three to eight nontemplate strand T residues starting at positions +1 to +3 relative to the transcription start site. Interestingly, several of the promoters containing this motif are in operons involved in nucleic acid metabolism, some of which are negatively regulated by a pyrimidine effector. Included in this group is the carAB operon.
Regulation of carAB Expression in E. coli
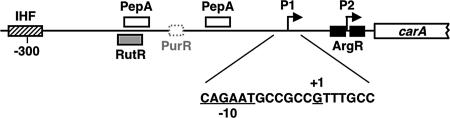
The carAB operon encodes the two subunits of carbamylphosphate synthetase. This enzyme (and only this enzyme in enteric bacteria) catalyzes the formation of carbamylphosphate, an intermediate in both the pyrimidine nucleotide and arginine biosynthetic pathways (Fig. 1). Expression of the carAB operon is subject to cumulative repression by the end products of each pathway (27). Transcription of the operon is initiated at two tandem promoters designated P1 and P2 (Fig. 5). Initiation at promoter P2, the more downstream promoter, is negatively regulated by arginine-dependent binding of the hexameric arginine repressor, ArgR, to two operator sequences that flank the transcription start site (Fig. 5) (15, 133). The molecular details of the ArgR-operator interactions have been described (22, 167). Initiation at promoter P1, the more upstream promoter, is negatively regulated by pyrimidines and to a lesser extent by purines, with the latter occurring by PurR-mediated repression (15, 101, 133). The purine-mediated regulation and part of the pyrimidine-mediated regulation require a nucleoprotein complex that forms upstream of promoter P1 (30). This complex includes integration host factor (IHF), PepA (aminopeptidase A), and PyrH (UMP kinase). The binding site for IHF and two binding sites for PepA have been mapped upstream of promoter P1 (Fig. 5); UMP kinase appears to be recruited to the complex by protein-protein contacts (29). UMP kinase was initially assumed to be the pyrimidine sensor of the complex; however, recent results indicate that this role is played by a protein called RutR, which appears to be a uracil/thymine-binding master regulator for genes involved in pyrimidine synthesis and degradation (146). Apparently, low intracellular levels of pyrimidines allow RutR to bind upstream of promoter P1, at a site that overlaps one of the PepA binding sites (Fig. 5). Without PepA bound to this site, repression of transcription initiation at promoter P1 is prevented (146). In this mechanism, uracil and thymine act as regulatory surrogates for pyrimidine nucleotides.
FIG. 5.
Promoter-regulatory region of the carAB operon of E. coli. Promoters P1 and P2 and the binding sites for IHF, PepA, RutR, PurR, and ArgR are shown. The partial sequence of promoter P1 includes the −10 region and transcription start site (+1), which are underlined.
Although IHF/PepA/PyrH/RutR-mediated regulation is unusually complex, pyrimidine-mediated regulation of carAB expression involves yet another independent control mechanism. As suggested by the presence of a TTT motif in the ITR of promoter P1, this other mechanism requires reiterative transcription. Promoter P1 contains the sequence 5′-CAGAATGCCGCCGTTTGCC, with the −10 region and the transcription start site (G7) underlined (53). Analysis of transcription initiation at promoter P1 in vitro demonstrated reiterative transcription within the T3 tract of the ITR, which increased with higher concentrations of UTP, essentially as observed at the pyrBI promoter (53). The analysis of transcripts initiated at promoter P1 in vivo showed that transcripts containing one or more extra U residues (i.e., GUUUUn, where n ≥ 1) were not extended to include sequences specified by the carAB genes (53). Finally, +3T-to-G or +3T-to-C mutations were shown to prevent reiterative transcription at promoter P1 while increasing the production of normally elongated, full-length carAB transcripts. Each mutation also caused an approximately threefold reduction in pyrimidine-mediated regulation of carAB expression, which was independent of regulation involving IHF and PepA. Pyrimidine-mediated regulation involving IHF and PepA occurs over a six- to ninefold range (53).
Taken together, these results indicate that regulation of carAB expression by UTP-sensitive reiterative transcription occurs by a mechanism analogous to that described for the pyrBI operon (Fig. 4). In this mechanism, transcription is initiated at the G7 start site in a manner independent of the UTP concentration. After the nascent transcript is extended normally to include four bases and has the sequence GUUU, weak base pairing between the transcript and DNA template permits reversible one-base slippage. With a high UTP concentration and the nascent transcript in the slipped position, an extra U residue is added to the 3′ end of the transcript. Either this transcript can be released from the initiation complex or another round of slippage and U addition can occur. Repeating this cycle generates transcripts with long runs of U residues; however, these transcripts are excluded from the normal mode of transcription elongation. With a low UTP concentration, slippage (if it occurs) and correct repositioning of the GUUU transcript—without extra U addition—permit normal template-directed insertion of a G residue at position +5. This addition results in a more stable RNA-DNA hybrid and the loss of alternative alignments for the 3′ end of the transcript, which precludes further slippage. The GUUUG transcript is either released as a simple aborted transcript or extended downstream with a high probability that it will become a full-length carAB transcript. In this model, the level of carAB expression is inversely proportional to UTP-sensitive reiterative transcription, and the production of carbamylphosphate synthetase corresponds to the cell's need for pyrimidine nucleotides. Although not included in this (or the pyrBI) model, it is possible that intracellular GTP levels affect operon expression by influencing the addition of a U or G residue at position +5 (or +6 in the case of pyrBI) of the nascent transcript (70). Physiological conditions that allow GTP levels to modulate reiterative and productive transcription at the carAB P1 and pyrBI promoters remain to be established. However, this possibility seems likely because pyrimidine limitation typically results in both a decrease in the UTP level and an increase in the GTP level in the cell (142).
The full range of pyrimidine-mediated regulation of carAB expression requires two independent control mechanisms that respond to the same or comparable (i.e., UTP and uracil) small-molecule effectors. Similar situations exist for UTP-sensitive regulation of pyrBI expression (i.e., transcription attenuation and reiterative transcription), Trp-sensitive regulation of the trpEDCBA expression (TrpR-mediated repression and transcription attenuation) (176), and numerous other operons in E. coli and other bacteria (48). The major advantage of such multiple control mechanisms is that regulation can respond to a wide range of concentrations of a particular effector molecule, with each control mechanism sensitive to a different range of effector concentrations. In the case of the carAB operon, it appears that IHF/PepA/RutR-mediated regulation occurs when UTP levels are relatively high (i.e., between 0.9 and 1.4 mM), while regulation by reiterative transcription occurs when UTP levels are lower (i.e., between 0.9 mM and 50 μM) (53). The lowest intracellular levels of UTP may be experienced by pyrimidine auxotrophs grown under pyrimidine-limiting conditions or by prototrophs following a shift from a pyrimidine-rich to a pyrimidine-poor environment.
Finally, an interesting difference between the reiterative transcription control mechanisms of the carAB and pyrBI operons is that the range of regulation provided by UTP-sensitive reiterative transcription at the carAB P1 promoter is smaller (by a factor of two to three) than that observed with the pyrBI promoter. This difference is presumably due to differences in the carAB and pyrBI promoter sequences. Mutational variants of the carAB P1 promoter were constructed to examine this assumption (X. Han and C. L. Turnbough, Jr., unpublished data). One variant showed that changing the G at the 5′ end of the transcript to an A increases the range of regulation nearly twofold. This result suggests that stronger G/C base pairing between the 5′ end of the transcript and the DNA template suppresses reiterative transcription and restricts the range of regulation. Another variant showed that changing the location of the transcription start site from position 7 to position 8 increases the range of regulation 2.5-fold. The reason for this enhancement is not obvious. However, the observations with the mutant P1 promoters indicate that promoter sequences and the architecture of the transcription initiation complex can significantly affect reiterative transcription. As a corollary, a TTT motif is necessary but not sufficient for UTP-dependent reiterative transcription. The dependence of reiterative transcription on additional promoter elements—sometimes an absolute dependence—was clearly established by the following examples of gene regulation in E. coli.
COMPOUND MECHANISMS FOR NUCLEOTIDE-SENSITIVE REGULATION
Salvage of Pyrimidine Bases
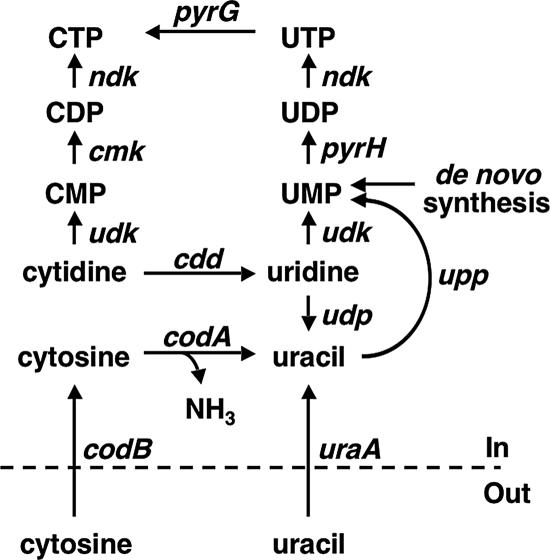
In addition to de novo synthesis, pyrimidine nucleotides can be synthesized from pyrimidine bases and nucleosides via salvage pathways in enteric bacteria (121). The pyrimidine salvage pathways can assimilate exogenous bases and nucleosides or can use bases and nucleosides produced inside the cell by normal nucleotide degradation. The pathways for uracil and cytosine salvage are shown in Fig. 6. Exogenous uracil and cytosine are transported into the cell by the cytoplasmic membrane proteins uracil permease and cytosine permease, respectively (5, 28). Intracellular uracil is converted directly to UMP by the enzyme uracil phosphoribosytransferase. In contrast, intracellular cytosine is rapidly deaminated to uracil and ammonia by the enzyme cytosine deaminase. The uracil produced in this reaction is also converted to UMP by uracil phosphoribosytransferase. The UMP formed by uracil and cytosine salvage is converted to UDP, UTP, and CTP as described for de novo nucleotide biosynthesis.
FIG. 6.
Salvage pathways for uracil and cytosine. Gene names are used to represent the encoded proteins. Excluding those shown in Fig. 1, the genes and their encoded proteins are as follows: cdd, cytidine deaminase; cmk, CMP kinase; codA, cytosine deaminase; codB, cytosine permease; udk, uridine kinase; udp, uridine phosphorylase; upp, UPRTase; and uraA, uracil permease.
Regulation of codBA Expression in E. coli
In E. coli, the pyrimidine salvage proteins cytosine permease and cytosine deaminase are encoded by the codB and codA genes (Fig. 6), which are included in the codBA operon (28). Regulation of codBA expression is complex, including control by pyrimidine, purine, and nitrogen availability. Control by purines occurs through PurR-mediated repression (4, 84), and nitrogen control involves the Ntr system acting through the nitrogen assimilation control protein NAC (4, 119). Pyrimidine-mediated regulation occurs through a mechanism involving UTP-sensitive reiterative transcription, but this mechanism is fundamentally different from the original reiterative transcription mechanism described for the pyrBI operon (137).
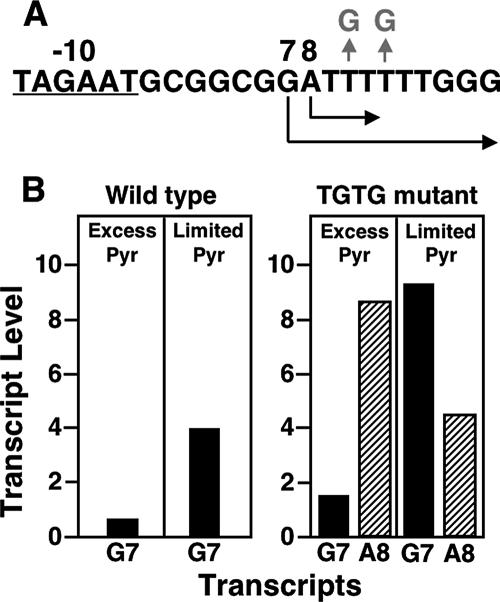
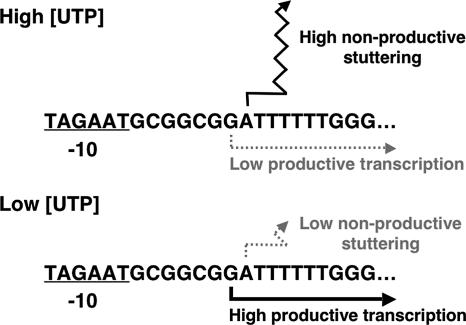
The first indication that codBA expression might be regulated by UTP-sensitive reiterative transcription was the discovery that the codBA promoter contains a T6 tract in its ITR, at a position that resembles the location of the T tract in the pyrBI promoter. The sequence of the codBA promoter region containing the ITR is 5′-TAGAATGCGGCGGATTTTTTGGG, with the −10 region and predicted transcription start sites underlined. However, the T tract in the codBA promoter is twice as long as the T tract of the pyrBI operon, which suggested that regulation of reiterative transcription at the codBA promoter would be different from that at the pyrBI promoter. Specifically, it was not clear how low UTP levels would inhibit reiterative transcription. With six U residues specified by the codBA ITR and only three U residues required for reiterative transcription, it appeared that extra U residues would be added to essentially every nascent codBA transcript, regardless of the UTP level. Furthermore, if extra U addition prevented productive transcript elongation, as observed with nascent pyrBI transcripts, then transcription of the codBA operon would be precluded. Obviously, something was missing in this scenario.
To examine reiterative transcription at the codBA promoter, a DNA template containing the codBA promoter region was transcribed in vitro in reaction mixtures containing various physiological concentrations (from 20 μM to 1 mM) of UTP (137). Analysis of the transcription products showed that codBA transcripts are initiated at two sites: a G residue and an A residue located seven and eight bases downstream from the −10 region (and designated G7 and A8), respectively (Fig. 7A). Most transcripts initiated at position G7 appeared to be elongated normally (i.e., they did not engage in reiterative transcription). In contrast, all transcripts initiated at position A8 appeared to engage in reiterative transcription that produced AUUUUn (where n = 1 to >15) transcripts. These transcripts were released from the transcription initiation complex without further downstream extension. Varying the UTP concentration did not affect the extent of the reiterative transcript; i.e., AUUUUn transcript ladders were always comparable in length. Although the UTP concentration did not affect reiterative transcription, it had a major effect on start site selection: higher concentrations of UTP favored initiation at position A8. At 1 mM UTP, initiation at position A8 was strongly favored; below 100 μM UTP, nearly all initiation occurred at position G7. Thus, the UTP concentration controlled start site selection, even though UTP was not used as the initiating NTP (which will be explained below).
FIG. 7.
Transcription from wild-type and mutant codBA promoters. (A) Sequence of the wild-type codBA promoter region with the −10 region underlined and the G7 and A8 start sites labeled with the number 7 or 8, respectively. Horizontal arrows indicate transcription initiation at the two start sites. The two T-to-G substitutions that created the TGTG mutant promoter are shown in gray. (B) Levels of codB::lacZ transcripts initiated at the wild-type and TGTG mutant promoters. Cells carrying either the wild-type or TGTG mutant codB::lacZ fusion were grown under conditions of pyrimidine excess or limitation, and fusion transcript levels were measured by quantitative primer extension mapping (137). Transcript levels are in arbitrary units. (Modified from reference 137 with permission from Elsevier.)
To investigate the role of reiterative transcription in pyrimidine-mediated regulation of codBA expression, a mutant codBA promoter was constructed in which the T6 tract of the ITR was changed to TGTGTT (Fig. 7A) (137). This mutation was shown to eliminate UTP-dependent reiterative transcription initiated at the codBA promoter in vitro, which resulted in a sevenfold increase in the synthesis of full-length codBA transcripts at 1 mM UTP. To measure the effects of the mutant promoter on codBA expression and pyrimidine-mediated regulation in vivo, this promoter was incorporated into a codB::lacZ gene fusion, which was inserted into the chromosome of a pyrimidine auxotrophic (i.e., car-94 ΔcodBA-lacZYA) strain of E. coli. This strain and an isogenic control strain with a wild-type codB::lacZ fusion were grown under conditions of pyrimidine limitation and excess, and β-galactosidase levels in these cells were compared. Wild-type codB::lacZ expression was regulated over an approximately 30-fold range, while mutant codB::lacZ expression was unregulated (excluding ∼1.5-fold pyrimidine-independent basal regulation). When cells were grown under conditions of pyrimidine excess, expression of the mutant codB::lacZ fusion was approximately 30-fold higher than that of the wild-type fusion. These results indicated that UTP-dependent reiterative transcription at the codBA promoter was required for all pyrimidine-mediated regulation of codBA expression.
Using the same four cultures (i.e., mutant and wild-type codB::lacZ fusion strains grown with limiting or excess pyrimidines), the steady-state levels and transcription start sites of the codB::lacZ transcripts were determined by quantitative primer extension mapping (137). The primer used in these experiments was complementary to codB sequences included in the codB::lacZ fusion. In the case of the wild-type strain, essentially all detectable transcripts were initiated at position G7, and the level of G7 transcripts in cells grown under conditions of pyrimidine limitation was at least 10-fold higher than that in cells grown with excess pyrimidines (Fig. 7B). Transcripts initiated at position A8 were not detected in cells grown under either condition. In the case of the mutant strain, in which reiterative transcription at the codBA promoter is precluded, both G7 and A8 transcripts were detected in cells grown under either condition (Fig. 7B). In both cultures, the levels of total (i.e., G7 plus A8) transcripts were similar, i.e., only 1.4-fold higher in cells grown under conditions of pyrimidine limitation. Also, the levels of total transcripts in the mutant cells (grown with either pyrimidine excess or limitation) were roughly threefold higher than that in wild-type cells grown with limiting pyrimidines, with a large part of this increase due to A8 transcripts. These results indicated that position A8 was a major transcription start site at the codBA promoter and that in the wild-type fusion strain, A8 transcripts were not detected because they were not extended downstream to include codB sequences. Presumably, these A8 transcripts were produced by nonproductive reiterative transcription and contain the sequence AUUUUn. In addition, the results with the mutant strain showed a high level of pyrimidine-mediated switching between the G7 and A8 start sites. In mutant cells grown with excess pyrimidines, approximately 75% of the transcripts were initiated at position A8. In contrast, in mutant cells grown with limiting pyrimidines, the level of G7 transcripts was twice that of A8 transcripts (Fig. 7B). These responses suggest that in the case of the wild-type codBA promoter, pyrimidine (presumably UTP)-mediated switching between productive transcription initiation at position G7 and nonproductive transcription initiation at position A8 plays a major role in regulation.
Based on the in vitro and in vivo results, the following model involving both UTP-dependent reiterative transcription and UTP-sensitive transcription start site switching was proposed for pyrimidine-mediated regulation of codBA expression (Fig. 8) (137). When UTP levels are high, RNA polymerase initiates transcription primarily at position A8—the preferred start site—and synthesizes a nascent transcript with the sequence AUUU (or perhaps AUUUU). At this point, weak base pairing between the transcript and its DNA template allows slippage between the two strands, resulting in a one-base, upstream shift of the transcript. RNA polymerase then switches to the reiterative mode of transcription and adds an extra U residue, which thereafter excludes this transcript from normal elongation. This transcript can be released from the initiation complex or the slippage/extra U addition cycle can be repeated many times, producing longer AUUUUn transcripts that have a fixed probability of release after each cycle. Furthermore, synthesis of the AUUUn transcripts precludes initiation at position G7, resulting in a low level of productive transcription and operon expression. Alternatively, when UTP levels are low, initiation at position A8 is inefficient, which restricts the synthesis of AUUUn transcripts. This restriction allows efficient transcription initiation at the secondary start site G7, which results in the synthesis of high levels of full-length codBA transcripts. Translation of these transcripts provides the proteins required for cytosine uptake and conversion to pyrimidine nucleotides when the nucleotides are needed by the cell.
FIG. 8.
Model for UTP-sensitive regulation of codBA expression. The model shows the effects of UTP concentration on productive transcription and nonproductive reiterative transcription (or stuttering), which occurs following transcription initiation at start sites G7 and A8, respectively. (Modified from reference 137 with permission from Elsevier.)
This model introduces two key regulatory elements that require additional explanation. The first element is UTP-sensitive selection of the transcription start site. This process apparently depends on two effects, namely, the inherent preference of RNA polymerase for position A8 as the start site (see above for rules) and the second-nucleotide effect (i.e., high concentrations of both the first and second NTP substrates are required for highly efficient initiation of transcription). The second-nucleotide effect is relevant in this case because UTP is the second nucleotide added to A8 transcripts. Furthermore, ample ATP is present in cells grown with excess pyrimidines (122, 135). Accordingly, high UTP levels support efficient initiation of A8 transcripts. On the other hand, when UTP levels are low, initiation at position A8 is restricted by the second-nucleotide effect and RNA polymerase selects the next best start site, position G7. Initiation of G7 transcripts can proceed relatively efficiently under these conditions because ATP, not UTP, is used as the second nucleotide. Additionally, initiation at position G7 might be facilitated by the two- to threefold increase in the GTP level that occurs in cells limited for pyrimidines (142). The second regulatory element in need of further explanation is the avoidance of reiterative transcription by G7 transcripts. The apparent explanation is that nascent G7 transcripts, from GAUUU through GAUUUUUU, form an RNA-DNA hybrid that is stable enough to preclude slippage and thus avoid reiterative transcription. Remarkably, this stability is imparted by the single G·C base pair formed by the first nucleotide of the nascent transcript.
Regulation of upp-uraA Expression in E. coli
In E. coli, the pyrimidine salvage proteins uracil permease and uracil phosphoribosytransferase are encoded by the uraA and upp genes, respectively (Fig. 6). These genes are included in the upp-uraA operon (5), which hereafter will be referred to as the upp operon for simplicity. Expression of the upp operon is negatively regulated over an approximately sixfold range by pyrimidine availability (7, 157). The sequence of the upp promoter region containing the ITR is 5′-TATAATCCGTCGATTTTTTTTGTG, with the −10 region and the initially reported transcription start site (7) underlined. This region is remarkably similar to the comparable region of the codBA operon, although there are two curious differences (Fig. 9). First, the T tract of the upp operon contains two more residues. The second difference is that in the upp promoter the GA residues preceding the T tract are one base closer to the −10 region. Thus, positions G6 and A7 in the upp promoter correspond to positions G7 and A8 in the codBA promoter. In spite of these differences, the strong similarities between the ITRs of the codBA and upp operons suggested that pyrimidine-mediated regulation of the operons would occur through analogous mechanisms.
FIG. 9.
Promoter region sequences of the upp and codBA operons. The −10 regions are underlined, and asterisks indicate the two transcription start sites at each promoter. The start sites are numbered according to their position downstream from the −10 region.
The characterization of pyrimidine-mediated regulation of upp expression was essentially as described for the codBA operon (157). Initially, reiterative transcription at the upp promoter was examined in vitro, using reaction mixtures containing various physiological UTP concentrations. The results showed that upp transcripts are initiated at positions G6 and A7; most G6 transcripts were elongated normally, while all A7 transcripts appeared to engage in reiterative transcription without further downstream extension. The UTP concentration did not affect the extent of reiterative transcription, which produced a ladder of AUUUn transcripts with n = 1 to >50. However, the UTP concentration had a major effect on start site selection, with lower concentrations favoring initiation at position G6. At 1 mM UTP, initiation at positions G6 and A7 was comparable, but at UTP concentrations of below 100 μM, all detectable initiation occurred at position G6.
The regulatory role of reiterative transcription at the upp promoter was investigated by constructing isogenic E. coli strains carrying a chromosomal upp::lacZ gene fusion with either a wild-type upp promoter or a mutant promoter in which the T tract was altered (e.g., T8 to TTGTTTTT) to eliminate reiterative transcription. These strains were grown under conditions of pyrimidine limitation or excess, and upp::lacZ expression levels were measured. The results showed that wild-type upp::lacZ expression was regulated over a sixfold range and that this regulation was effectively abolished by the elimination of reiterative transcription. In addition, elimination of reiterative transcription at the upp promoter caused constitutive upp::lacZ expression. Quantitative primer extension mapping of upp::lacZ transcripts isolated from these cultures showed that in the case of the wild-type fusion strain, only G6 transcripts were detected and the level of G6 transcripts in cells grown with pyrimidine limitation was sevenfold higher than that in cells grown with excess pyrimidines. (Note that with the primer used here, AUUUUn transcripts could not be detected.) In the case of the mutant fusion strain, both G6 and A7 transcripts were detected in cells grown under either condition; however, pyrimidine availability dramatically affected the relative amounts of the two transcripts. For cells grown with excess pyrimidines, 40% of the transcripts were initiated at position G6 and 60% were initiated at position A7. For cells grown with limiting pyrimidines, 90% of the transcripts were initiated at position G6 and 10% were initiated at position A7. Under both conditions, the levels of total (G7 plus A8) transcripts were similar, i.e., only 1.4-fold higher in cells grown under conditions of pyrimidine limitation. Taken together, these results revealed UTP-sensitive selection of alternative transcription start sites and different fates of the transcripts initiated at these sites that mirror the key regulatory elements of the codBA operon. Thus, a model for pyrimidine-mediated regulation of upp expression was proposed that is completely analogous to that for codBA expression (Fig. 8).
Briefly, according to the model, when intracellular levels of UTP are high, RNA polymerase preferentially initiates transcription at position A7. The resulting nascent transcript is extended until it contains three or perhaps four U residues, at which point weak base pairing in the RNA-DNA hybrid permits the transcript to slip one base upstream. RNA polymerase then adds a U residue to the 3′ end of transcript, which irreversibly directs the transcript into a nonproductive transcription pathway. The transcript can be released from the initiation complex, or another round of slippage and U addition can occur. This process can be repeated many times, with a similar probability of transcript release after every U addition. Synthesis of the resulting AUUUUn transcripts occludes the promoter, thereby reducing the opportunity for initiation at position G6 and the production of full-length upp transcripts. In contrast, when intracellular levels of UTP are low, RNA polymerase initiates transcription almost exclusively at position G6. The resulting transcripts, in general, avoid reiterative transcription due to the formation of a more stable hybrid between the transcript and DNA template. These nonstuttering transcripts are elongated normally and can be extended downstream to generate translatable upp transcripts. Consequently, high levels of the proteins required for uracil salvage are produced only under conditions of pyrimidine limitation.
An Interesting Difference between the upp and codBA Operons
Although there are many similarities between the mechanisms of pyrimidine-mediated regulation of upp and codBA expression, there is one striking difference. The range of regulation observed with the upp operon (6-fold) is much smaller than that with the codBA operon (30-fold). One factor that could contribute to this difference is the length of the T tract in the ITR. For example, the longer T tract of the upp operon (eight versus six residues) could promote a higher level of reiterative transcription with G-initiated transcripts, thereby restricting the maximum level of productive transcription and the range of regulation. However, this explanation was excluded by examining the effects of systematic, single-base deletions in the T tract of the upp operon (23). Reducing the number of residues in the T tract from eight to six did not increase the range of pyrimidine-mediated regulation but decreased it from 5.7-fold to 4.0-fold. In fact, the range of regulation gradually decreased as the T tract was shortened from eight to three residues, at which point only basal (1.5-fold) regulation remained. The decreases in the range of regulation were due to increases in the fractions of both G6 and A7 transcripts that avoid reiterative transcription and are elongated normally. These results indicate that the long T tract of the upp operon and presumably the codBA operon is required to ensure a high level of nonproductive reiterative transcription with the A-initiated transcripts, which is necessary for the widest range of regulation. Furthermore, the results indicate that although the tract of three T residues in an ITR with the sequence ATTT is necessary for reiterative transcription, transcription of these residues does not ensure reiterative transcription.
Other factors that could contribute to the different ranges of upp and codBA regulation are the different positions of the transcription start sites and the preferences for selecting these sites (99). The G and A start sites are located at positions 6 and 7 in the upp promoter and at positions 7 and 8 in the codBA promoter. At the upp promoter, the G6 start site is predicted to be a much weaker start site than the A7 site. In contrast, at the codBA promoter, both start sites are predicted to be highly efficient. Therefore, much stronger competition between the start sites should occur at the codBA promoter. Consistent with this prediction, the observed pyrimidine-mediated start site switching at the codBA promoter is much more extensive than that at the upp promoter, which should permit a wider range of regulation. To examine this explanation, a mutant upp::lacZ operon was constructed by inserting a C residue into the sequence between the −10 region and transcription start sites of the upp promoter (Fig. 9), which changes the wild-type sequence CCGTC to CCGTCC. This +C mutation changes the positions of the start sites from G6 and A7 to G7 and A8, which are the same as the start sites in the codBA promoter. According to the previous arguments, the +C mutation should change the range of pyrimidine-mediated regulation of upp expression from 6-fold to 30-fold. However, the +C mutation did something quite unexpected: it completely eliminated pyrimidine-mediated regulation of upp::lacZ expression. The loss of regulation occurred because transcription was initiated almost exclusively at position G7 (i.e., without competition from position A8) in cells grown with either limiting or excess pyrimidines (E. Várady and C. L. Turnbough, Jr., unpublished data). Interestingly, partial regulation—and competition from the A8 start site—could be restored by a second mutation in the −10 region of the upp promoter. These surprising results suggest that interactions between distinct promoter elements can alter the conformation of the transcription initiation complex in ways that strongly influence the selection of transcription start sites. Additional studies are needed to provide the rules for these interactions.
REGULATORY MECHANISMS IN GRAM-POSITIVE BACTERIA
History and Overview
Only the gram-negative enteric bacteria E. coli and Salmonella were intensively studied in early investigations of the mechanisms of gene regulation, and for a time it was thought that the regulatory mechanisms found in these species were applicable to all bacteria. Subsequent research has shown that this is rarely the case. While the biochemical pathways of central metabolism are generally the same in gram-negative and gram-positive bacteria, the mechanisms adopted to regulate the expression of the genes for these pathways are usually quite different. Investigations with gram-positive bacteria have uncovered a fascinating variety of novel regulatory mechanisms (37, 172). The mechanisms for regulation of pyr gene expression described in this section of the review have been intensively studied in gram-positive bacteria only; however, as described under the heading Phylogenetic Distribution of pyrR Genes and Mechanisms of PyrR-Mediated Gene Regulation, variations on these mechanisms are found in gram-negative phyla.
The study of pyrimidine biosynthetic gene regulation in B. subtilis in the Switzer laboratory originated from an interest in the regulation of aspartate transcarbamylase activity by intracellular proteolysis (112). B. subtilis aspartate transcarbamylase differs from its enteric homolog in that it contains no regulatory subunit capable of mediating allosteric control of enzyme activity (16); allosteric regulation of de novo UMP biosynthesis is exerted at the level of a pyrimidine-repressible carbamylphosphate synthetase (131). The B. subtilis pyrB locus was sequenced to determine the amino acid sequence of aspartate transcarbamylase (90). The DNA sequences flanking the pyrB gene showed that, in contrast to the case in enteric bacteria, the pyrB gene was part of a 10-gene pyr operon that encoded all of the enzymes required for the de novo synthesis of UMP (Fig. 10) (89, 138, 161). The sequence of the pyr operon revealed three putative intrinsic transcription terminators in the promoter-proximal region, and the positions of these terminators suggested their involvement in a transcription attenuation control mechanism (Fig. 10). The characterization of this mechanism is described in detail in this review. The B. subtilis pyrG gene, which as in enteric bacteria encodes CTP synthetase, is not located in the multigene pyr operon. Regulation of B. subtilis pyrG expression is also regulated by an attenuation control mechanism, but this mechanism, which is described below, is quite different from that of the pyr operon.
FIG. 10.
Map of the B. subtilis pyr operon. Shaded bars indicate ORFs, and the bent arrow denotes the pyr promoter. The proteins encoded by the genes shown were identified in Fig. 1, except as follows: pyrR, pyr mRNA-binding attenuation regulatory protein; pyrP, uracil permease; pyrAA, glutamine-utilizing subunit of carbamylphosphate synthetase equivalent to carA; pyrAB, catalytic subunit of carbamylphosphate synthetase equivalent to carB; pyrK, electron-transferring accessory protein to dihdroorotate dehydrogenase. Numbers indicate the positions of nucleotides in the B. subtilis genome. (Modified from reference 155 with permission from Elsevier.)
TRANSCRIPTION ATTENUATION BY PyrR, AN mRNA-BINDING PROTEIN
Regulation of the Bacillus subtilis pyr Operon
In B. subtilis, the genes encoding all enzymes required for de novo synthesis of UMP lie in a single coordinately regulated pyr operon (130). The structure of this operon (Fig. 10) was established by determination of its nucleotide sequence (138, 161). The functions of its individual genes were assigned by comparison of their sequences to those of pyr genes of known function (138) and were confirmed by complementation of E. coli pyr mutations (89). The functions of three B. subtilis pyr genes, pyrR, pyrP, and pyrK (previously called pyrDII), were not immediately recognized from their sequences. Subsequent analysis demonstrated that these genes encode the regulatory protein for the pyr operon (161), a uracil permease that is homologous to the permease encoded by uraA in E. coli (44, 161), and an electron-transferring accessory subunit of dihydroorotate dehydrogenase (77, 78), respectively. The last eight genes of the operon overlap by 1 to 32 base pairs, and the operon contains three short noncoding segments that are involved in regulation of operon expression.
The B. subtilis pyr operon is transcribed from a single promoter, which appears to be constitutive (105, 161). Operon expression is controlled by an attenuation mechanism in which transcription termination at three sites in the 5′ region of the operon is regulated by uridine and guanosine nucleotides via the PyrR regulatory protein. It was evident from the earliest studies that each of the three untranslated segments of the operon contains a typical intrinsic transcription terminator, which specifies a terminator hairpin (the 3:4 stem-loops in Fig. 11, left) followed immediately by a run of U residues in the mRNA. These untranslated segments are the 5′ pyr leader (attenuation region 1, 151 nucleotides), the pyrR-pyrP intercistronic region (attenuation region 2, 173 nucleotides), and the pyrP-pyrB intercistronic region (attenuation region 3, 145 nucleotides) (Fig. 10 and 11). Clearly, if the action of these terminators is not suppressed, little expression of the downstream genes, which include all of the enzymes of de novo pyrimidine biosynthesis, can occur. It was also recognized that the RNA from all three untranslated segments is capable of folding into an alternative secondary structure, which is a large hyphenated stem-loop (Fig. 11, right) (161). These structures were named antiterminators because they prevent formation of the downstream terminator hairpin by sequestering residues of the 5′ segment of the terminator stem-loop via base pairing with upstream sequences. The key to regulation of the pyr operon lies in the ability of PyrR to favor formation of the terminator hairpins, which results in premature termination of transcription and reduced expression of the downstream genes. PyrR does this by binding to pyr mRNA when the protein is activated by binding of uridine nucleotides. Furthermore, binding of PyrR to pyr mRNA is antagonized by guanosine nucleotides, which effectively activates pyr operon expression. The segment of pyr mRNA to which PyrR binds was first identified from conserved regions within the nucleotide sequences of the three B. subtilis attenuator regions. These conserved regions were always found in the upstream segment of the antiterminator structure, so that binding by PyrR would be predicted to prevent base pairing in the antiterminator RNA and allow the downstream segment of the antiterminator to fold into the alternate terminator hairpin (Fig. 11).
FIG. 11.
Predicted secondary structures of the regions of pyr transcripts specified by attenuation regions 1 (5′ leader), 2 (pyrR-pyrP), and 3 (pyrP-pyrB). Nucleotides are numbered from the start of transcription (i.e., +1). (Left side) RNA is shown folded into the binding loop, formed by base pairing of segments 1 and 2, and the terminator hairpin, formed by base pairing of segments 3 and 4, conformation. Bases involved in the formation of the alternative antiterminator stem-loop conformation are circled. (Right side) RNA is shown folded into the antiterminator stem-loop conformation. (Modified from reference 178.)
Further analysis of the potential secondary structures formed by the attenuator region RNAs led to the prediction that the conserved PyrR binding sequences were embedded in a third stem-loop, which was named the antiantiterminator or the binding loop (104) (Fig. 11, left, stem-loops labeled 1:2). This stem-loop is formed by base pairing of nucleotides from the upstream strand of the antiterminator stem with sequences that lie still further upstream. Formation of the binding loop disrupts the lower stem of the antiterminator and allows its 3′ strand to fold into the terminator stem-loop. Thus, the regulation of the B. subtilis pyr operon can be conceived as involving two alternative conformations of each of the three attenuator RNA segments. One is the antiterminator conformation, which predominates when PyrR is not bound (Fig. 11, right) and leads to transcription readthrough of the downstream pyr genes. The other is the PyrR-stabilized antiantiterminator-plus-terminator conformation (Fig. 11, left), which results in termination and reduced expression of the downstream genes. The binding of PyrR to the binding loop within this conformation is crucial to regulation of the pyr operon. Since the binding of PyrR is stimulated by UMP and UTP (14, 103) and is antagonized by GMP, GDP, and GTP (14, 21, 73), an effective means of metabolic regulation of transcription of the pyr operon by the ratio of uridine to guanosine nucleotides is provided. This mechanism is illustrated in schematic form in Fig. 12.
FIG. 12.
Mechanism of PyrR-mediated transcription attenuation control of pyr operon expression in B. subtilis. For simplicity, only transcription of the pyr 5′-leader attenuation region is shown. For details, see the text. (Modified from reference 178.)
The secondary structure of the binding loop is conserved (Fig. 11). In its 5′ strand, it contains a purine-rich tract within an internal loop or bulge. The beginning of the internal loop or bulge contains the conserved sequence 5′-UUUAA. In addition, the consensus sequence 5′-ARUCCNGNGAGGYU is located in the terminal stem and loop of the binding loop. Experimental evidence that this secondary structure forms and that the regions of conserved sequence are important for PyrR binding to the RNA is presented below.
Regulatory Function of PyrR: Genetic Evidence
The first clues that the product of the B. subtilis pyrR gene acts as a regulatory protein for pyr gene expression were obtained from studies with high-copy-number plasmids carrying a fusion between either the pyr promoter-leader region or the pyr promoter-leader region plus pyrR and a downstream reporter gene (161). When transformed into B. subtilis, the plasmid carrying just the pyr promoter-leader region caused elevated expression of both the reporter gene and the chromosomal pyrB gene. The elevated expression was not repressible by exogenous uracil. However, when the plasmid carrying the pyr promoter-leader region plus pyrR was transformed into cells, regulation of both the reporter gene and the chromosomal pyrB gene was normal and repressible. This result suggested that multiple copies of the pyr leader region, or more likely its specified RNA, sequestered the PyrR formed from the chromosomal pyrR gene, thereby eliminating PyrR-mediated repression. Furthermore, normal regulation was restored by producing elevated levels of PyrR from the high-copy-number plasmid. Subsequent studies confirmed that the plasmid-specified pyr leader transcripts were responsible for the titration of PyrR (102). In fact, within a limited range there was a correlation between the amount of pyr transcript specified by any of the three noncoding/attenuation regions of the pyr operon and the extent of depression of the chromosomal pyr operon (102).
A second line of genetic evidence for a role of PyrR in regulation of the pyr operon came from characterization of 12 point mutations that caused constitutive expression of the pyr operon (46). All of the mutations were located in or near the pyrR gene. Two were premature chain termination mutations, one altered the pyrR ribosome binding site, and the others were missense mutations in the pyrR gene. The most direct demonstration of the crucial role of PyrR in the regulation of pyr operon expression came from the analysis of a B. subtilis strain carrying an in-frame deletion in the chromosomal pyrR gene (161). In the mutant strain, expression of the pyrB gene was 250-fold greater than the repressed levels found in pyrR+ cells, and the elevated expression was completely refractory to repression by exogenous pyrimidines. The regulation of pyrB expression in the mutant strain was restored to normal by the introduction of a plasmid-borne copy of pyrR.
Regulatory Function of PyrR: Biochemical Evidence
To demonstrate directly the regulatory function of PyrR, highly purified protein was included in an in vitro transcription system consisting of B. subtilis DNA templates containing the pyr promoter fused to each attenuation region, purified B. subtilis RNA polymerase, and the four ribonucleoside triphosphates. In this system RNA polymerase initiated transcription at the pyr promoter and produced transcripts that either were terminated at an attenuator or were extended to the end of the template. As would be predicted if PyrR mediates repression of pyr operon expression in the presence of pyrimidines, addition of PyrR plus UMP or UTP substantially increased the fraction of terminated transcripts. (GMP did not affect termination in these experiments, probably because the high concentrations of GTP used as a transcription substrate obscured its effects.) Regulation of transcription termination by PyrR plus uridine nucleotides was demonstrated with templates from all three pyr attenuation regions. However, the quantitative effects of PyrR on attenuation region 1 (the pyr leader) most closely recapitulated the effects observed in vivo with a chromosomal pyr::lacZ fusion containing a selected attenuator region (105). This result may indicate that trailing ribosomes translating upstream ORFs (i.e., pyrR and pyrP) alter the behavior of attenuation regions 2 and 3 in vivo.
Purified B. subtilis PyrR has been shown to possess two other properties required by the regulatory model: it binds with high specificity to pyr binding loop RNA sequences, and its affinity for these RNAs is increased by UMP and UTP (14, 160). More recent studies with purified PyrR from Bacillus caldolyticus led to the previously unrecognized finding that guanosine nucleotides cause greatly reduced affinity of PyrR for pyr RNA and antagonize the effects of uridine nucleotides (21, 73). Similar opposing effects of uridine nucleotides and guanosine nucleotides were observed in the allosteric regulation of the pyrAA/pyrAB-encoded carbamylphosphate synthetase from B. subtilis (131). Such effects have been proposed to provide a means of coordinating rates of pyrimidine biosynthesis with the size of intracellular purine nucleotide pools (21, 73).
Importance of RNA Secondary Structures in Regulation
The secondary structures of the RNAs from the three pyr attenuation regions shown in Fig. 11 were derived from computer-based folding programs (180). The parameters of these programs are updated from time to time, so the structures in Fig. 11 should be regarded as approximate; numerous variant, but functionally equivalent, structures can exist. Biochemical structural mapping of these RNA segments has been carried out only in the case of the binding loop from attenuation region 2, as described below (14). However, there can be little doubt that all binding loop, terminator, and antiterminator stem-loop structures form and play the regulatory roles assigned to them. The most convincing evidence for these roles came from studies of the effects of antisense oligodeoxynucleotides on the frequency of transcription termination within the three pyr attenuation regions in vitro (104). Oligodeoxynucleotides that were designed to disrupt each of the stem-loops by base pairing with their upstream segments consistently had the predicted effects on transcription. For example, disruption of the antiterminator stem-loop caused greater termination at the downstream attenuator, whereas disruption of the binding loop or terminator stem-loop caused increased readthrough transcription. Interestingly, only oligodeoxynucleotides that base pair with the upstream strands of the target stem-loops were effective; oligodeoxynucleotides of equal length that base pair with their downstream strands had little effect. This observation indicates that stem-loop formation occurs very rapidly in solution and that intramolecular base pairing to form the stem-loop competes very effectively with intermolecular base pairing. This led to the suggestion that kinetic aspects of transcription and RNA folding are important to the proper functioning of this attenuation control mechanism. Specifically, the binding loop hairpin must fold and bind PyrR before the synthesis of downstream sequences that direct folding of the more stable antiterminator stem-loop.
The properties of a set of deletion mutations introduced into a gene fusion in which the pyr promoter-leader region was joined to lacZ also illustrated the importance of RNA secondary structures in regulation (161). Deletion of the terminator in the leader region resulted in high and constitutive lacZ expression, whereas deletion of the antiterminator region eliminated expression under all growth conditions. A detailed, systematic deletion analysis of the pyr leader region has not been conducted, however.
As noted above, the secondary structure of binding loop 2 of the pyrR-pyrP intercistronic attenuation region has been studied by nuclease digestion (14). The patterns of cleavage by single-strand-specific and double-strand-specific nucleases were consistent with the secondary structure shown in Fig. 13B, although these results do not establish this structure unequivocally. Weak cleavage of the RNA in the terminal loop regions provided an indication that this loop may fold into a relatively compact structure, as is known for some other terminal RNA loops (38, 58). Unexpectedly strong single-strand cleavages in the region of A·U base pairs upstream of the bulge also suggested that the single-stranded segment of the lower 5′ strand of the stem-loop is longer than predicted from computer analysis of RNA folding. The other predicted RNA secondary structures shown in Fig. 11 have not been subjected to nuclease digestion analysis.
FIG. 13.
Elements in the pyr binding loop RNA that are important for PyrR binding and pyr operon regulation. Nucleotides are numbered as in Fig. 11. (A) Mutations in the pyr 5′ leader RNA that cause a loss of repression by pyrimidines. (B) Protection against hydroxyl radical cleavage of binding loop 2 RNA by PyrR and mapping of secondary structure by nuclease digestion. Sites adjacent to nucleotides shown in blue were strongly protected against hydroxyl radical cleavage, the site adjacent to the nucleotide in red was moderately protected, and sites adjacent to nucleotides in green were weakly protected. Arrows with circles indicate sites of cleavage by a single-strand-specific nuclease (RNase I), arrows with squares indicate sites of cleavage by a double-strand-specific nuclease (RNase V1), and S and W denote strong and weak cleavage, respectively. Suggested alternative structures for the terminal loop of the RNA hairpin are shown in circles. (C) Specificity of PyrR binding to binding loop 2 RNA determined by gel mobility shift analysis. Residues shown in red cannot be replaced without loss of binding, residues in green can be replaced as long as the secondary structure of the RNA is preserved, and residues in black can be replaced or deleted. (D) Consensus sequence and structure of the PyrR binding site. The consensus structure was derived from 20 binding loops identified in pyr operons of gram-positive bacteria. Secondary structures were predicted by MFOLD (R = A or G, Y = U or C, and N = any nucleotide). Parentheses indicate nucleotides present in only some species. In 8 of 20 examples, the U nucleotide shown in a dashed circle is part of the internal loop, which directs the predicted alternative base pairing shown with dashed lines. The sequences and base pairs shown in boxes are highly but not universally conserved. (Panel A is reprinted from reference 155 with permission from Elsevier; panels B and D are reprinted from reference 14 with permission from Oxford University Press.)
Regulation of pyr Operon Expression as Deduced from pyr::lacZ Fusions
The model of PyrR-mediated regulation of the B. subtilis pyr operon predicts that four pyr transcripts will be formed from the operon under derepressing conditions (161). These transcripts are 0.12, 0.65, 2.3, and 12 kb in length and correspond to termination at attenuation region 1, attenuation region 2, attenuation region 3, and the terminator at the end of the operon, respectively. All except the first transcript are predicted to become less abundant when cells are grown with exogenous pyrimidines. The cumulative diminution of readthrough transcription at the multiple attenuators should produce larger reductions in the levels of the longer transcripts. Presumably, the 12-kb transcript is formed and translated largely as a single unit, because the genes that it encodes have been shown to be coordinately regulated in vivo (130). The qualitative predictions of this model were confirmed by Northern hybridization analysis (161). All four pyr transcripts were detected, and the abundance of all but the 0.12-kb transcript decreased sharply in cells grown with pyrimidines. However, this approach was not suitable for quantitative analysis of pyr transcription.
For the quantitative analysis of pyr transcription, Lu et al. analyzed a series of chromosomal pyr::lacZ fusions expressed in cells under conditions that repress or derepress pyr operon expression (105). The first set of transcriptional fusions included the pyr promoter followed by DNA containing either attenuator region 1, attenuator region 2, or attenuator region 3. For the latter two fusions, the native intervening upstream attenuators and ORFs were excised. Expression of all fusions was repressed to about the same extent by exogenous pyrimidines. Repression was approximately 4-fold relative to that in cells grown on minimal medium and 20-fold relative to that in cells starved for pyrimidines by slow growth on orotate, a poor pyrimidine source. Repression was completely dependent on a wild-type pyrR gene. In addition, the expression levels of the three pyr::lacZ fusions were similar. In another set of transcriptional fusions, two or three attenuators were linked in tandem. In these cases, repression of lacZ expression was cumulative. That is, the extents of repression by excess uracil relative to that in cells starved for pyrimidines were 18-fold, 136-fold, and 200-fold when the fusions contained one, two, and three attenuators, respectively. These observations were entirely consistent with the proposed regulatory model. Studies with other pyr::lacZ fusions provided clear evidence that the DNA sequences upstream of the pyr promoter are not involved in regulation of operon expression and that possible translation of a small ORF in the pyrP-pyrB intercistronic region is not required for attenuation control.
Occurrence and Significance of Transcription Pausing in the pyr Operon
Our studies with antisense oligodeoxynucleotides suggested a key role for rapid folding of the RNA secondary structures involved in attenuation control of the pyr operon. To examine this possibility, Zhang and Switzer analyzed the pausing during the transcription of the pyr attenuation regions and attempted to determine whether pausing was important for regulation (178). According to the model for attenuation control, transcription pausing at a site or sites in the downstream strand of the antiterminator stem-loop would allow the time needed for the antiantiterminator stem-loop to form and bind to uridine nucleotide-activated PyrR. Such binding would then preclude formation of the antiterminator stem-loop. Pausing within the three pyr attenuation regions was measured in vitro by using a two-step, single-round transcription assay developed to examine the kinetics of transcript elongation. With each DNA template carrying a particular attenuation region, one or more discrete, NusA-enhanced pause sites were detected. In each case, the site of pausing was located within the upstream region of the antiterminator sequence, at a position that would allow pausing to play its proposed role in the timing of RNA folding and PyrR binding. These findings demonstrated that transcription pausing could play an important role in the regulation of the pyr operon, but they did not demonstrate that it actually does so in vivo.
In a subsequent study, Zhang et al. attempted to evaluate the function of transcription pausing in the pyr operon in vivo by constructing and analyzing mutations that greatly reduce pausing (177). The mutations changed selected pyrimidine nucleotides to purine nucleotides near pause sites, which was shown to substantially reduce transcription pausing at the mutant site in vitro. These mutations were then incorporated into pyr::lacZ fusions, and their effect on pyrimidine-mediated regulation of pyr::lacZ expression was determined. No consistent correlation between elimination of transcription pausing in vitro and defects in cellular regulation was observed. A major complication in these experiments was the inability to separate the effects of the mutations on transcription pausing from their effects on transcription termination at an attenuator.
BIOCHEMICAL CHARACTERIZATION OF PyrR
PyrR Is a UPRTase
An unexpected property of PyrR, first discovered by Ghim and Neuhard (44) in studies of B. caldolyticus pyrR, is that it catalyzes the uracil phosphoribosyltransferase (UPRTase) reaction. This activity was surprising because Bacillus species also produce a highly active UPRTase of the ubiquitous upp family (108, 155). Outside of a short segment in the phosphoribosyl pyrophosphate(PRPP)/UMP binding site, PyrR homologs have no significant sequence similarity to other phosphoribosyltransferases, including the upp-encoded UPRTases (155). Both pyrR-encoded and upp-encoded UPRTase activities are functional in vivo (108), but the affinity of PyrR for uracil is much lower than the affinity of upp-encoded UPRTases for this substrate (160). For this reason it is unlikely that PyrR plays an important role in uracil salvage in vivo.
The three-dimensional structure of PyrR demonstrates that the protein is a member of the type I phosphoribosyltransferase structural family in spite of its low sequence relatedness to them (149). The kinetic mechanism of the UPRTase reaction catalyzed by B. subtilis PyrR has been characterized (49). In most respects, this enzyme is a typical phosphoribosyltransferase. Other than the requirement for the binding of nucleotides to the UPRTase active site, there is no obvious relationship between the protein's enzymatic and regulatory activities. Mutant forms of PyrR that fail to bind RNA and regulate pyr transcription, but which have normal UPRTase activity, have been described (143). A mutant PyrR that has lost catalytic activity while retaining the ability to regulate pyr expression has not been isolated, but it has been reported that Lactococcus lactis PyrR lacks UPRTase activity (109). Thus, it is unlikely that PyrR must be able to catalyze this reaction to exert its regulatory function. We believe that the UPRTase activity of PyrR reflects its evolutionary origin as a phosphoribosyltransferase that later acquired the ability to bind to RNA. Of all the members of the type I phosphoribosyltransferase family, PyrR most strongly resembles, in both sequence and tertiary structure, hypoxanthine guanine phosphoribosyltransferase (76). This similarity suggests that PyrR did not evolve from the upp family (156). The recent observation of a dimer of B. caldolyticus PyrR with GMP in one active site and UMP in the other reinforces the idea that the protein is evolved from the hypoxanthine guanine phosphoribosyltransferases (21). The purine repressor PurR of B. subtilis provides another example of a phosphoribosyltransferase structural domain that has acquired a regulatory function (148). The binding of PurR to pur operator DNA is regulated by PRPP, which binds to the conserved phosphoribosyltransferase active site of the protein. PurR apparently does not catalyze an enzymatic reaction, however, and the nucleic acid binding site resides in an additional helix-turn-helix domain that is not found in PyrR or other phosphoribosyltransferases (148).
RNA Sequence and Structure Required for PyrR Binding
Several experimental methods have been used to identify the RNA sequence and secondary structure required for tight binding of B. subtilis PyrR. In an early genetic study, Ghim and Switzer isolated cis-acting mutations that resulted in constitutive expression of pyr::lacZ fusions (45). Most of the mutations were mapped to a short region of conserved sequence in the terminal loop and upper stem of the binding loop RNA (Fig. 13A), which implicated this region in the regulation of pyr operon expression. A reasonable conclusion was that this region is involved in PyrR binding, but this was not directly demonstrated by these studies.
Hydroxyl radical footprinting experiments with an RNA specifying B. subtilis binding loop 2, which is known to bind tightly to PyrR, were used to map the portions of the RNA that were protected by PyrR (14). Three segments were protected from hydroxyl radical cleavage (Fig. 13B). The terminal loop and upper stem were strongly protected, as were three nucleotides at the top of the lower stem opposite the conserved element that initiates the purine-rich bulge. The purine-rich bulge itself was weakly protected.
Electrophoretic gel mobility shift experiments were used to examine the binding of purified PyrR to 37 structural variants of B. subtilis binding loop 2 (14). The results are summarized in Fig. 13C. The requirements for tight PyrR binding to pyr RNA appeared to be quite exacting. Numerous single-nucleotide substitutions or deletions led to a reduction in the apparent affinity by as much as three orders of magnitude. The importance of maintaining the RNA secondary structure shown in Fig. 13C was documented. The requirement for specific nucleotide residues in positions in the upper stem and loop and in the lower stem just below the purine-rich bulge was confirmed, as was the requirement for the bulge itself. The smallest RNA that bound well to PyrR contained the 28 nucleotides from position 708 to 735 in Fig. 13C (14). Recently, examination of the specificity of PyrR binding to all three B. subtilis binding loop RNAs in vivo using a yeast three-hybrid method confirmed the generalizations shown in Fig. 13C (60).
The results of genetic, footprinting, and RNA binding studies yielded a consistent picture of the PyrR binding site. However, several aspects of PyrR binding suggested by gel shift assays were puzzling. Specifically, PyrR binding to binding loops 1 and 3 appeared to be much weaker and less affected by nucleotides than binding to binding loop 2. These differences were inconsistent with the essentially equivalent regulation in vivo of pyr::lacZ fusions containing individual attenuation regions. For this reason, a detailed study of the binding of B. caldolyticus PyrR to the three B. caldolyticus pyr attenuation regions was undertaken using a filter binding assay (73). (For technical reasons, B. subtilis PyrR binding to RNA could not be measured with this assay.) The apparent dissociation constants (0.1 to 1 nM) for PyrR binding and the effects of nucleotides on this binding were similar for all three binding loops. These results were consistent with regulation of all three attenuators by physiological concentrations of nucleotides. The ratio of uridine to guanine nucleotides appeared to be the primary determinant of PyrR binding, a conclusion supported by the effects of exogenous uridine and guanosine on pyr operon expression in growing cells of B. subtilis. Recent studies by Jørgensen et al. revealed a requirement for Mg2+ in the gels used for gel shift assays, which may have led to substantial artifacts in the previous determinations of binding constants for PyrR binding to B. subtilis attenuation regions 1 and 3 (73). Furthermore, some of the conclusions from the study of RNA structural variants summarized in Fig. 13C should be reexamined, especially those results with variants that appeared to bind very poorly to PyrR. Many of the conclusions of Bonner et al. (14) concerning the RNA sequence and secondary structure required for binding to PyrR were confirmed, however.
How well do the studies of B. subtilis PyrR binding to attenuation regions predict the binding requirements of PyrR proteins from other species? A mutational analysis of PyrR binding to attenuation regions in Lactobacillus plantarum indicates that the required RNA features are similar to those found in B. subtilis (125). A phylogenetic comparison of 20 known or probable PyrR binding loops from nine different species suggests that there is little variation in PyrR selectivity among these species (14). As seen in Fig. 13D, the loops vary in the lengths of the upper and lower stems and the size of the bulge/internal loop but not in the overall secondary structure or identity of critical nucleotides. The structure of the binding loop can vary as much from attenuator to attenuator within a given species (e.g., in B. subtilis or L. lactis) as it does from species to species. We have relied on this conservation of structure and sequence to identify likely modes of action of PyrR in other bacteria (see below).
High-Resolution Structures of PyrR and PyrR Complexes with Nucleotides
A detailed understanding of how PyrR binds to RNA and how the binding of nucleotides alters the affinity of PyrR for RNA requires the determination of the structure of PyrR with and without nucleotides and RNA bound to it. Considerable progress has been made toward that goal. High-resolution structures of unliganded PyrR from B. subtilis (156) and B. caldolyticus (21) have been obtained by X-ray diffraction analysis of crystals. The structure of B. caldolyticus PyrR with bound nucleotides has been obtained (20, 21), as have the structures of unliganded Mycobacterium tuberculosis PyrR (80) and Thermus thermophilus PyrR (PDB code 1UFR).
PyrR folds into a core domain consisting of a curved central sheet formed by five parallel β-strands and flanked by three α-helices (Fig. 14B). A small subdomain called the “hood,” which is made up of three antiparallel β-strands, caps the major core domain. The core domain strongly resembles the architecture found in many other type I phosphoribosyltransferases (149). The conserved amino acid residues of the PRPP/nucleotide binding site in all type I phosphoribosyltransferases are located in the central β-strand of the core domain and the following α-helix; PyrR obeys this generalization. Binding of a sulfate ion in this site in unliganded B. subtilis PyrR and subsequent determination of the location of nucleotides bound to B. caldolyticus PyrR confirmed this identification of the nucleotide binding site (21). The “hood” domain varies greatly from one phosphoribosyltransferase to another; residues in this domain are involved in determining the specificity of nucleotide binding.
FIG. 14.
Ribbon diagram of the crystal structure of PyrR from B. caldolyticus. Each polypeptide chain is shown in a distinct color. (A) The native tetrameric structure. (B) One of the two identical dimeric structures that combine to form the tetramer. Very similar dimeric structures are found in PyrR from other species. The black circles indicate the location of bound Mg2+ ions. The stick structures indicate the locations of 5′-UMP bound to the active site of the green subunit, 5′-GMP bound to the active site of the purple subunit, and 3′-GMP bound in a crystal contact lattice. (Reprinted from reference 21.)
B. subtilis PyrR was crystallized in dimeric and hexameric forms. In the hexameric form, dimeric units that are virtually identical to the dimeric crystal are arranged around a threefold central axis with a small solvent-filled central cavity. Both unliganded and liganded B. caldolyticus PyrR crystallized as a tetramer (Fig. 14A) made up of dimeric units with a structure that is very similar to that of the B. subtilis dimer. The structure of unliganded M. tuberculosis PyrR is very similar to that of PyrR of B. caldolyticus. PyrR from T. thermophilus is also a tetramer made up of dimeric units that are very similar to those of the Bacillus PyrR proteins. However, the mode of association of the dimeric units is very different from that for B. caldolyticus PyrR. The finding of three different quaternary structures indicates that PyrR probably functions as the dimeric form. The strong interactions between subunits in the dimer make its dissociation to native monomers unlikely. Recently, the stoichiometry of RNA binding to B. caldolyticus PyrR was shown by analytical ultracentrifugation to correspond to one RNA to two PyrR monomers (i.e., one RNA binds per PyrR dimer), as predicted (73).
Crystals of B. subtilis PyrR with UMP bound could not be obtained under conditions in which unliganded PyrR crystallizes, and the addition of UMP to PyrR crystals led to dissolution of the crystals (J. L. Smith, personal communication). These observations suggest that substantial conformational changes accompany binding of uridine nucleotides to PyrR. Such conformational changes are of particular interest because they might help to explain how the binding of nucleotides increases the affinity of PyrR for RNA. Crystals of B. caldolyticus PyrR with nucleotides bound were first obtained inadvertently in attempts to obtain a PyrR-RNA cocrystal (21). An RNase contaminant led to degradation of the RNA, and PyrR was obtained with UMP in the active site of one monomer of the dimeric unit and with GMP in the active site of the other monomer (Fig. 14B). These nucleotides interact with Mg2+ and with conserved active-site amino acid residues in a manner that differs in interesting ways from that observed with other phosphoribosyltransferases (21). Specifically, the magnesium ions in the active sites of PyrR are not ligated to the phosphate moiety or to the vicinal 2′, 3′ hydroxyls of the nucleotide, as is usual in the substrate complexes of other phosphoribosyltransferases. UMP and GMP are bound in a very similar manner to the active sites; the same amino acid residues are hydrogen bonded to the uracil and guanine bases. A third nucleotide, probably 3′-GMP, was found located between tetramers in the crystal lattice. More recently, the structure of B. caldolyticus PyrR with only 5′-UMP bound to the active sites was solved (20). The structure of this form of the protein was identical to the structure of the unliganded (but sulfate-bound) state and the UMP- plus GMP-bound state of PyrR. Curiously, no protein conformational changes among unliganded and two nucleotide-bound PyrR crystal structures were detectable, so the structures do not reveal how nucleotide binding alters the affinity of PyrR for RNA. Perhaps the association of PyrR subunits in the tetrameric state in the crystal obscures the changes that are induced by nucleotides in the dimeric RNA binding form of PyrR.
Characterization of the RNA Binding Site of PyrR
The electrostatic surface potential map of B. subtilis PyrR revealed a large concave surface on the dimer that is lined with positively charged and hydrophilic residues (156). It was proposed that this surface was the most likely site for binding of the negatively charged pyr binding loop RNA. A similar surface is also found on the B. caldolyticus, T. thermophilus, and M. tuberculosis PyrR dimers, but otherwise the electrostatic surface potential maps of these PyrR homologs are quite different from one another (21). This observation reinforces the suggestion that the concave basic surface of PyrR is the RNA binding site. In the hexameric state of B. subtilis PyrR and the tetrameric states of B. caldolyticus and M. tuberculosis PyrR, the basic surface is located in a central cavity that is too small to accommodate the pyr binding loop. Only the dimeric forms of these proteins would be capable of binding this RNA, which is consistent with the idea that the PyrR dimer is the physiologically functional form.
Site-directed mutagenesis of B. subtilis PyrR was used to test the hypothesis that conserved amino acid residues whose side chains lie on the concave basic surface are required for RNA binding and regulation of the pyr operon (143). Glutamine substitution mutations in four residues that lie in this surface (Thr18, His22, Arg141, and Arg146) clearly identified them as involved in normal RNA binding and pyr regulation; the mutants had no detectable loss of UPRTase activity or structural integrity. Two other residues (Arg27 and Lys152) were similarly implicated, with the reservation that small changes in their average apparent native molecular weight were observed, which might indicate that these mutant proteins did not fold into fully native tertiary structure (143). These six residues are generally conserved in bacterial PyrR sequences. It seems likely that they form part of the RNA binding surface, although one cannot conclude that they interact directly with RNA. The elucidation of a detailed map of PyrR-pyr RNA interactions must await a high-resolution X-ray diffraction analysis of PyrR-RNA cocrystals.
PHYLOGENETIC DISTRIBUTION OF pyrR GENES AND MECHANISMS OF PyrR-MEDIATED GENE REGULATION
Distribution of pyrR Genes
The sequences of pyrR genes are well conserved, so the identification of species carrying pyrR has rapidly followed the flood of new genome sequences. As of October 2007, genes believed to encode PyrR proteins had been identified in 245 discrete bacterial species. No doubt more will be found as additional genome sequences are reported. In addition, probable PyrR binding sequences in RNA can be readily identified, as can likely attenuator and antiterminator sequences upstream of pyr genes. Probable modes of PyrR action deduced from this information fall into several classes. However, it should be noted that genetic or biochemical experiments implicating PyrR in the regulation of pyr genes have been reported only for B. subtilis (155, 161), B. caldolyticus (21, 44), Enterococcus faecalis (43), L. lactis (109), L. plantarum (125), Mycobacterium smegmatis (36), and M. tuberculosis (C. J. Fields and R. L. Switzer, unpublished data).
PyrR-Mediated Transcription Attenuation of pyr Operons
In all 15 species in the genus Bacillus for which genomic sequences are available, pyr genes are organized in the same order within a single pyr operon. All but one of these operons contain three attenuation regions that are located as described for the pyr operon of B. subtilis (Fig. 10). The one exception, the pyr operon of Bacillus clausii, lacks the pyrP-pyrB intercistronic attenuation region. Presumably, in each case the pyr operon is regulated in the same way as described for B. subtilis. The pyr operon in E. faecalis is organized in a similar fashion, but the pyrR-pyrP and pyrP-pyrB intercistronic regions are very short and do not contain PyrR binding sequences, attenuators, or antiterminators (43). In this case, only a single attenuation region in the 5′ leader is used to control pyr operon expression, but the attenuation mechanism is still the same as in B. subtilis. L. plantarum pyr genes are also found in a single operon in the same order as in Bacillus, but the pyrP (uracil permease) gene is absent and only two attenuation regions are found (5′ leader and pyrR-pyrB intercistronic regions) (33); the mechanism of regulation by PyrR is essentially the same in L. plantarum as in B. subtilis (125). Thus, pyr operons can apparently be adequately regulated in arrangements involving one, two, or three attenuation regions. Without further study it is impossible to conclude whether a significant physiological advantage is conferred by pyr operons containing multiple attenuation regions. Organization of pyr genes into a single PyrR-regulated operon has been deduced from the genome sequences of a number of other low-G+C gram-positive species (e.g., Listeria monocytogenes and Listeria innocua). In many of these species, one or two genes, usually pyrP, pyrK, pyrD, or pyrE and pyrF, are located elsewhere on the chromosome and often appear not to be regulated by PyrR.
PyrR-Mediated Transcription Attenuation of Unlinked pyr Genes
The clustering of all of the pyrimidine biosynthetic genes into a single operon is limited to a small number of low-G+C gram-positive genera. More commonly in the gram-positive organisms, pyr genes are scattered around the chromosome in multiple operons. It appears that most, but not all, of the unlinked operons are regulated by PyrR-mediated transcriptional attenuation. L. lactis presents the best-characterized example. The pyr genes of L. lactis are scattered in at least five transcription units. Four of these, pyrRPB-carA, pyrKDbF, pyrEC, and carB, have obvious attenuation regions in their 5′ leader regions; there is evidence that PyrR regulates them in much the same way as shown for B. subtilis (83, 109). The fifth gene, pyrDa, encodes a second dihydroorotate dehydrogenase, which may be involved in catabolism of orotate, a pyrimidine that is abundant in bovine milk (6). Kilstrup et al. reviewed the organization and regulation of unlinked pyr operons in lactic acid bacteria and their close relatives (83); numerous variations on the patterns found in Bacillus and Lactococcus are seen, but the fundamental mechanism of regulation by PyrR appears to be the same in all of the species. On the other hand, the nature of regulatory mechanisms governing expression of pyr genes that are not subject to PyrR-dependent attenuation in these species is unknown. Future investigations of these mechanisms are likely to yield novel findings.
The chromosomes of L. plantarum and a number of other members of the Lactobacillus genus contain two pyrR genes (8). The product of one of these, PyrR1, mediates regulation of the L. plantarum pyr operon in response to pyrimidines, as shown for B. subtilis and other species described above (125). Recently, the product of the second gene, PyrR2, was shown to regulate expression of the L. plantarum pyr operon and the unlinked pyrP operon in response to the CO2/HCO3− level in the medium (8). The two PyrR proteins operate independently and respond to different physiological signals. The biochemical mechanism of PyrR2 action is not yet clear. Two possible mechanisms have been discussed (8). PyrR2 could act by forming heterodimers with PyrR1 that are unable to repress pyr expression, or PyrR2 could interact with pyr RNA regulatory sequences to promote antitermination instead of favoring termination, the known action of PyrR1. This discovery of dual regulation of pyr expression in lactobacilli adds a fascinating new chapter to the study of PyrR function in metabolic regulation.
PyrR as an Inhibitor of pyr Gene Translation
Analysis of the genomes of a number of bacteria has led to the suggestion that PyrR acts in some species as a translational repressor (155). In mycobacteria, for example, pyr operons contain a pyrR gene at their 5′ end, and this gene is preceded by a consensus PyrR binding sequence that overlaps the putative pyrR ribosome binding site. No attenuator or antiterminator sequences are found in the 5′ leader region of the operon (i.e., near the predicted PyrR binding site). This arrangement suggests that PyrR could act to inhibit translation of the pyr operon by occluding the translation initiation site when intracellular uridine nucleotides are elevated. This binding would inhibit translation of the pyrR gene, and probably translation of the downstream pyr genes if their translation is coupled to translation of upstream genes. Experimental evidence for this conclusion has now been published (36). Plasmids containing translation fusions that join the M. smegmatis pyr promoter-leader region to lacZ (i.e., those that link the mycobacterial pyrR ribosome binding site to the lacZ ORF) were repressed by exogenous uracil in M. smegmatis, but transcription fusions, in which the lacZ ribosome binding site is retained, were not repressed. Repression by uracil was shown to require both the M. smegmatis pyrR gene and an intact PyrR RNA binding loop sequence. Furthermore, PyrR proteins from M. tuberculosis and M. smegmatis have been purified and shown to bind specifically to the predicted pyr RNA sequences; binding is enhanced by uridine nucleotides and antagonized by guanosine nucleotides. These results demonstrate that PyrR from mycobacteria is biochemically capable of regulating pyr operon expression in response to nucleotide levels. Further characterization of this system is in progress (C. J. Fields and R. L. Switzer, unpublished experiments).
The combination of pyrR genes and PyrR binding sequences that overlap the ribosome binding sites for pyr genes, which we take to be suggestive of translational repression of those genes by PyrR, is quite widespread in other bacterial species. Numerous species have been identified in which ORFs for putative pyrP (uraA) genes and/or another putative transport protein of the major facilitator protein superfamily fit this pattern (C. J. Fields, unpublished data). In some species (e.g., Bacillus, Clostridium, Lactobacillus, and Streptococcus spp.), this arrangement coexists with regulation of pyr operon expression regulated by PyrR-mediated transcription attenuation. In other cases, translational repression of the genes for transport proteins appears to be the only mode of PyrR regulation (e.g., in Haemophilus influenzae and Pasteurella maltocida).
A very interesting hybrid regulatory mechanism appears to exist in Thermus strain ZO5 and related Thermus species (162). In Thermus strain ZO5 an ORF for short leader polypeptide precedes the pyrR gene and other downstream genes of a pyr operon. The ribosome binding site in the mRNA for the leader polypeptide overlaps a consensus PyrR binding sequence and could be subject to translational repression by PyrR. The RNA encoding the leader polypeptide is also capable of forming a transcription terminator. Van de Casteele et al. (162) have suggested that attenuation at this terminator is regulated by the rate of translation of the leader polypeptide, which is in turn responsive to pyrimidines via PyrR-mediated translational repression. Furthermore, 6 of 28 codons in the leader polypeptide encode arginine, which might account for stimulation of pyr gene expression in Thermus strain ZO5 by this amino acid. This interesting model has not yet been tested by genetic or biochemical means, however.
Species in Which the Function of PyrR Is Unclear
There are a few species in which genes encoding PyrR homologs are clearly recognizable but in which the function of PyrR is obscure. In the genomes of the cyanobacteria Synechocystis and Synechococcus, pyrR genes are found isolated from pyr genes, but no predicted RNA structures have been found that implicate PyrR in regulation of the pyr or other genes. Cho et al. have reported that a plasmid-borne copy of pyrR from H. influenzae could complement a B. subtilis ΔpyrR mutant strain but that pyrR from Synechocystis sp. strain PCC 6803 could not (24). This result indicates that H. influenzae PyrR is capable of binding to B. subtilis pyr RNA in a pyrimidine-dependent manner; we have suggested above that it may do so to regulate genes for uracil transport proteins. In the case of Synechocystis, the role of PyrR in regulation, if any, remains obscure. It is conceivable that PyrR serves only as a phosphoribosyltransferase in some species, even though they also have upp genes.
The function of the PyrR homolog in Pseudomonas presents a particularly interesting unsolved problem. In the Pseudomonas aeruginosa and Pseudomonas putida genomes, a pyrR gene lies at the 5′ end of the operon pyrRBC′. Genetic evidence indicates that the product of the pyrR gene possesses UPRTase activity and is involved in repression of pyrB by uracil (A. P. Kumar, C. J. Fields, and G. A. O'Donovan, personal communication). However, none of the RNA structures involved in PyrR binding or transcription attenuation as in B. subtilis can be identified in the pyrRBC′ operon. Furthermore, the deduced sequences of Pseudomonas PyrR homologs differ from that of Bacillus PyrR in numerous residues that are thought to be important for RNA recognition and binding. If PyrR is involved in the regulation of pyr gene expression in Pseudomonas, its mechanism of action must be quite different from the previously characterized mechanisms. In preliminary studies by O'Donovan and colleagues, it was proposed that the P. putida PyrR acts as a DNA-binding protein to activate pyr gene expression, but detailed experiments have not been published. Future research on this system will be of great interest.
Species in Which pyrR Genes Are Not Identifiable
pyrR genes are readily identifiable in most gram-positive bacteria and are somewhat unpredictably scattered among many gram-negative phyla. A pyrR gene has been found so far in only one mycoplasma species, Mycobacterium penetrans. pyrR genes have not yet been identified in the sequenced genomes of enteric bacteria, bacteroides, alphaproteobacteria, epsilonproteobacteria, spirochetes, chlamydiae, or any archaea or eukaryotes.
REGULATION OF pyrG EXPRESSION BY A NOVEL MECHANISM BASED ON CTP-SENSITIVE REITERATIVE TRANSCRIPTION
pyrG Is Regulated by CTP Levels
CTP synthetase, which catalyzes the glutamine- and ATP-dependent amination of UTP to form CTP, is encoded by the pyrG gene (Fig. 1), which in B. subtilis is not part of the pyr operon and is not regulated by PyrR (115). It is not surprising that this gene would be regulated by the end product CTP, but such regulation is not readily demonstrated with wild-type strains for three reasons. First, the only exogenous cytosine-containing metabolite that can be used to increase internal CTP pools is cytidine, and cytidine is readily converted to uridine by cytidine deaminase. Specific repression of pyrG by cytidine can be demonstrated in mutants in which cytidine deaminase is inactive (115). Second, repressive effects of cytidine are small because the formation of CTP by the endogenous biosynthetic pathway maintains significant CTP pools. Only when de novo biosynthesis of pyrimidine nucleotides is impaired in mutants that are defective in the pyr operon or in pyrG can full derepression of pyrG be demonstrated (115). The third problem in characterizing pyrG regulation is that it is extremely difficult to assay CTP synthetase activity in crude extracts, a circumstance that has required the use of pyrG::lacZ transcription fusions (115). Such fusions are derepressed by 15- to 20-fold in pyrimidine auxotrophs that were grown on orotate, a poor pyrimidine source, compared to the same strain grown with excess cytidine. Similar studies with mutants in which interconversions between uridine and cytidine nucleotides were blocked, or in which cytidine uptake was inhibited, demonstrated that cytidine or a metabolite derived from cytidine, most likely CTP, specifically regulates pyrG expression (115).
These conclusions, which were based on studies with B. subtilis (115), were confirmed and extended in experiments performed with L. lactis by Jørgensen et al. (74). These investigators used similar genetic methods to manipulate nucleotide pools, and they employed pyrG::lacLM fusion strains to measure pyrG expression and to determine nucleotide pool levels. Their studies demonstrated that pyrG expression was directly correlated with the intracellular concentration of CTP.
pyrG Is Regulated by Transcription Attenuation
The pyrG promoter of B. subtilis has been mapped to the region between the similarly oriented rpoE and pyrG genes (115). The 5′ leader region of the pyrG operon contains 189 base pairs. This leader region contains an intrinsic transcription terminator, which also serves as the transcription terminator for the upstream rpoE operon. The analysis of deletions in the comparable pyrG leader regions of B. subtilis and L. lactis revealed that the intrinsic terminator is required for regulation; i.e., it is an attenuator (74, 115). However, leader sequences that could specify an antiterminator stem-loop could not be identified, and most of the B. subtilis leader region could be deleted without loss of normal regulation of pyrG expression (114, 115).
Comparison of the pyrG leader regions of several low-G+C gram-positive bacteria yielded valuable clues concerning the mechanism of attenuation control of pyrG expression (115). Only three short nucleotide sequences are conserved. These leader sequences specify GGGCUC at the 5′ end of the pyrG transcript and two complementary sequences, GCUCCC and GGGACG, at the bottom of the stem of the terminator hairpin of the attenuator. The nucleotides between the conserved sequences apparently do not play a role in regulation because they can be deleted (114). Systematic mutagenesis of the pyrG leader region of B. subtilis indicated that the conserved sequences 5′-GGGC at the start of the transcript and 5′-GCUCCC in the upstream segment of the terminator hairpin are the only cis-acting elements required for normal pyrG regulation (114). The lack of a requirement for an antiterminator stem-loop led Meng and Switzer to suggest that a regulatory protein might bind to critical transcript sequences at low CTP levels and prevent formation of the terminator hairpin (114). However, a search for the gene encoding such a protein by transposon mutagenesis was unsuccessful. Furthermore, Jørgensen et al. demonstrated that the regulation of the expression of pyrG::lacLM fusions in L. lactis was essentially the same whether the fusions were present as a single chromosomal copy or carried on a multicopy plasmid (74). This observation indicated either that a regulator protein was not titrated by multiple copies of pyrG mRNA or that no such protein was involved in regulation.
Regulation of pyrG by CTP-Sensitive Reiterative Transcription
A peculiar property of pyrG transcription was revealed by primer extension mapping of the 5′ ends of pyrG transcripts of B. subtilis (116). When cells were grown with excess cytidine, nearly all of the transcripts started with the sequence 5′-GGGCUC. However, when cells were grown under pyrimidine-limiting conditions, a ladder of transcripts from 1 to ∼10 nucleotides longer than the 5′-GGGCUC… transcripts was also detected. Copying of these extended transcripts by reverse transcription, followed by cloning and sequencing, demonstrated that these longer transcripts contained 5′-end extensions produced by the addition of a variable number of G residues (116).
To identify the source and function of the longer transcripts, mutant pyrG promoters were constructed in which one of the first four residues in the ITR (i.e., GGGC) was replaced by another base. Analysis of transcripts initiated at the mutant promoters in cells grown under conditions of pyrimidine excess and limitation revealed that substitution of any residue in the GGG sequence eliminated the formation of poly(G) extensions. Furthermore, the elimination of poly(G) extensions caused very low and essentially unregulated pyrG (actually pyrG::lacZ) expression (116). Analysis of a mutant promoter in which the C at position +4 in the ITR (i.e., +4C) was changed to a T (specifying a U in the transcript) showed that pyrG expression and regulation were similar to those observed with the wild-type pyrG promoter. However, a +4C-to-A substitution in the pyrG promoter abolished regulation. Finally, a mutant pyrG promoter was constructed in which four G residues were inserted after position +G3 in the ITR, creating a promoter with an ITR with the sequence 5′-GGGGGGGCUC. A pyrG::lacZ fusion containing this mutant promoter exhibited constitutive expression (116).
These observations are accounted for by conditional reiterative transcription at the pyrG promoter, which provides the basis for the current model for CTP-sensitive regulation of pyrG expression in B. subtilis (Fig. 15). When the intracellular level of CTP is high, pyrG transcripts are faithful copies of the DNA template and transcription elongation continues until termination at the attenuator. Therefore, when CTP is plentiful, transcription of the pyrG gene is suppressed (Fig. 15). On the other hand, when the intracellular level of CTP is low, pyrG transcription pauses after the synthesis of the nascent transcript 5′-GGG (and before position +4C) because of insufficient substrate. This pause provides time for the nascent transcript to slip upstream (relative to the DNA template) and allow an extra G residue to be added to the nascent transcript. This process can be repeated multiple times (e.g., up to at least 10 times) until eventually a C residue is inserted. The transcript is then elongated normally until RNA polymerase transcribes the attenuator sequence that specifies the upstream segment of the terminator hairpin. The sequence of this segment in B. subtilis is 5′-GCUCCCUUUCAA, which includes a tract of nine pyrimidines. Because both C and U residues base pair with G residues, the run of pyrimidines will immediately base pair with the poly(G) tract at the 5′ end of the transcript, forming an antiterminator stem-loop. As RNA polymerase continues to elongate the pyrG transcript, formation of the terminator hairpin is precluded by the antiterminator secondary structure and full-length pyrG transcripts are formed (Fig. 15). These transcripts are translated to make CTP synthetase, which is needed to overcome the CTP deficiency. Although the model describes pyrG expression at high and low intracellular concentrations of CTP, regulation can occur continuously over a wide range of CTP concentrations that control the extent of pausing at position +4.
FIG. 15.
Model for CTP-mediated regulation of pyrG expression in B. subtilis. The figure shows the effects of CTP concentration on the fate of the pyrG transcript after the first three G residues have been incorporated into the nascent transcript. A high CTP concentration allows normal transcript elongation until intrinsic termination occurs in the pyrG leader region. A low CTP concentration induces a transcription pause that allows reiterative transcription and the addition of extra G residues, which participate in the formation of an antiterminator hairpin. The extra G residues are boxed. The figure shows the insertion of six extra G residues, but as many as 10 extra residues can be added. (Modified from reference 116 with permission of the publisher. Copyright 2004 National Academy of Sciences, U.S.A.)
This model also accounts for the effects of the +4C-to-T or +4C-to-A and the G4 insertion mutations. In the case of the +4C-to-T mutation, regulation of pyrG expression is nearly normal. Under the conditions used to measure regulation, the intracellular levels of both CTP and UTP vary in parallel. Therefore, the transcription pausing required to allow reiterative transcription can be induced at the mutant promoter at low UTP levels just as it is by low CTP levels at the wild-type promoter. Conversely, the +4C-to-A mutation abolishes regulation because pyrimidine starvation does not cause a decrease in the intracellular level of ATP, which would be needed to induce pausing before the addition of the nucleotide at position +4. Additionally, the constitutive expression caused by the G4 insertion mutation is consistent with the model, which predicts that extra G residues at the 5′ end of the pyrG transcript, and not reiterative transcription per se, is required to prevent transcription termination at the attenuator. Therefore, permanently adding the extra G residues leads to suppression of transcription termination regardless of the state of pyrimidine availability. Furthermore, the model predicts that it is base pairing between the poly(G) and polypyrimidine tracts that precludes the formation of the terminator hairpin and transcription termination. In support of this prediction, a mutation that introduces two G residues into the polypyrimidine tract (i.e., the mutant sequence is 5′-CUCGGUUUC) greatly reduces pyrG expression to similar low levels in cells grown with excess and limiting pyrimidines (114). (Note that in this experiment, the two residues in the downstream segment of the terminator hairpin that normally base pair with the two mutated positions in the polypyrimidine tract were changed to maintain complete base pairing in the stem of the terminator hairpin.) Finally, the model provides a clear explanation for why the pyrG attenuator functions as a nonconditional transcription terminator for rpoE transcripts: these transcripts do not contain poly(G) tracts.
Reiterative transcription is a central element in the regulation of several pyrimidine biosynthetic and salvage operons in enteric bacteria, as described above, but the reiterative transcription reaction in these examples is fundamentally different from the reiterative transcription reaction that occurs at the pyrG promoter of B. subtilis. Specifically, in the case of the enteric promoters, all transcripts produced by reiterative transcription are aborted during the initiation phase of transcription, while transcripts that undergo reiterative transcription at the pyrG promoter switch to the normal mode of elongation, which allows transcription through the pyrG gene. A major part of this difference clearly involves the substrate for reiterative transcription, as discussed above. Reiterative transcription with UTP produces aborted transcripts, while reiterative transcription with non-UTP substrates produces transcripts that can be productively extended. The ability to make this distinction appears to be an intrinsic property of all RNA polymerases, although not much else is known about it.
The conserved sequences required for regulation of pyrG expression in B. subtilis—namely, the GGGC sequence that starts the ITR and the leader sequence specifying the polypyrimidine tract in the leader transcript—are found in the pyrG leader regions of at least 17 low-G+C gram-positive bacteria, including the genera Bacillus, Listeria, Lactococcus, Enterococcus, Staphylococcus, and Streptococcus (116). While reiterative transcription that produces runs of G residues has been demonstrated only for the pyrG operon of B. subtilis and CTP-sensitive regulation of pyrG expression has been shown only for the pyrG operons of B. subtilis and L. lactis, it seems highly likely that many species of the genera listed above will share these capabilities. On the other hand, the pyrG operons of enteric bacteria, which also encode CTP synthetase, possess leader regions that do not contain these conserved elements and are regulated by other mechanisms.
Further Characterization of pyrG Regulation
CTP-sensitive reiterative transcription at the pyrG promoter of B. subtilis and poly(G)-mediated suppression of transcription termination at the pyrG attenuator do not require any protein other than RNA polymerase. These processes have been recapitulated in vitro using a minimal assay for transcription, requiring only B. subtilis RNA polymerase, pyrG template DNA, ribonucleoside triphosphates, salts, and a buffered reaction mixture (67). Reiterative transcription producing poly(G) tracts and suppression of transcription termination at the pyrG attenuator were specifically induced at low CTP concentrations but not at low concentrations of any other NTPs. Mutations in the pyrG template that altered reiterative transcription and attenuation in vivo caused comparable changes in the in vitro system (67). These findings provide strong support for the major regulatory elements in the model for regulation of pyrG expression in B. subtilis.
To determine the minimum number of G residues at the 5′ end of the pyrG transcript that is required to prevent formation of the terminator hairpin, a set of mutant pyrG promoters was constructed with the number of G residues in the ITR varied systematically (34). The effects of these mutations on pyrG expression and CTP-sensitive regulation were measured, with decreases in the range of regulation used to indicate the inability of the terminator hairpin to form. The range of regulation with a promoter containing four G residues was slightly less than half of that with the wild-type promoter (i.e., 5.6-fold instead of 14-fold). Regulation with a promoter containing five or more G residues was essentially absent, and pyrG expression was constitutive. These phenotypes indicate that five G residues are sufficient to form the antitermination stem—with the exclusion of the terminator hairpin—in essentially every pyrG transcript. The mutant pyrG promoters were also used to determine the number of G residues in the ITR that are needed to support maximum reiterative transcription in vivo. The results showed that a minimum of three G residues (as in the wild-type promoter) is required for reiterative transcription and that promoters with three or four G residues exhibit comparable levels of reiterative transcription. Compared to these levels, reiterative transcription is slightly reduced at a promoter with five G residues. In contrast, reiterative transcription is severely reduced or eliminated at promoters with six or more G residues, even in cells starved for pyrimidines. Apparently, an rG6·dC6 RNA-DNA hybrid is too stable to permit transcript slippage, even with extensive transcription pausing. Taken together, the results of this analysis reveal that the wild-type pyrG promoter, with its G3 tract in the ITR, permits the widest range of regulation by the reiterative transcription mechanism.
It is interesting to note that recent studies with yeast mitochondrial RNA polymerase suggest that progressively lower concentrations of NTP substrates are required for active-site binding as the nascent transcript is extended from position +3 through +11 during transcription initiation (2). The end of this gradient at position +11 apparently reflects the transition to the elongation phase of transcription, when even lower concentrations of NTP substrates are needed. If these findings can be generalized to the B. subtilis RNA polymerase—which seems likely—they suggest that the sequence of the pyrG ITR, which directs CTP to position +4 of the transcript, was selected to maximize the sensitivity of reiterative transcription to the intracellular concentration of CTP. If the site for reiterative transcription in the ITR were followed by a C residue located downstream of position +4, the affinity of RNA polymerase for CTP might be too great to permit pausing even at low levels of CTP in the cell, and reiterative transcription producing poly(G) tracts would be prevented.
CONCLUSIONS AND SPECULATION
This review describes seminal research on the regulation of pyr gene expression in bacteria that extends over more than 30 years. A hallmark of these studies is that expectations and attractive ideas were often proven wrong. However, negative results provided opportunities for the discovery of new regulatory mechanisms and concepts. These discoveries frequently followed unpredictable paths along which intuition and happenstance guided the formation of hypotheses. To emphasize this process, our descriptions of the mechanisms that regulate pyr gene expression were presented in an historical context. These descriptions also include the rigorous and critical experimental testing that confirm proposed regulatory models. These models describe simple and economical mechanisms designed to provide gradual and sensitive control of pyr gene expression that reflects the metabolic needs of the cell.
Although these models are unlikely to be modified significantly in the future, many questions remain regarding fundamental processes that underlie the regulatory mechanisms. For example, some of the mechanisms involve transcription start site switching, which to a first approximation can be predicted from promoter sequences. However, the rules for start site selection do not take into account context effects that can significantly influence the process, as illustrated with mutant upp promoters of E. coli. Clearly, more work is required to understand these context effects. In addition, some regulatory mechanisms involve reiterative transcription, but this reaction can have quite different outcomes. Reiterative transcription at the pyrBI promoter of E. coli produces AAUUUUn transcripts that are always released from the initiation complex, whereas reiterative transcription at the pyrG promoter of B. subtilis produces GGGGn transcripts that can return to the normal mode of transcription elongation. The basis of these alternative outcomes is presently a mystery. Furthermore, the extent of reiterative transcription at a promoter can be influenced by the location of the transcription start site, as observed with a mutant carAB promoter. Apparently, there are undefined architectural features of the transcription initiation complex that modulate the reiterative transcription reaction. Finally, the structure-function relationships of the PyrR-RNA complex remain undefined. In future studies, it will be important to describe amino acid-RNA interactions, conformational changes that occur in both PyrR and its RNA substrate upon binding, and the structural basis for the uridine nucleotide-mediated increase and guanosine nucleotide-mediated decrease in the affinity of PyrR for RNA. Analysis of high-resolution structures of PyrR with RNA and nucleotides bound would likely accomplish most of these goals.
In this review, we have emphasized that the mechanisms and concepts elucidated by the study of pyr gene regulation can serve as useful guides in the analysis of unknown regulatory mechanisms. One interesting example is the study of pyrimidine (CTP)-mediated regulation of pyrG expression in E. coli. It appears that the regulatory mechanism, which is clearly different from the mechanism controlling pyrG expression in B. subtilis, requires CTP-sensitive start site switching in much the same way as described for the pyrC and pyrD regulatory mechanisms (T. Bedekovics and C. L. Turnbough, Jr., unpublished data). However, the mechanism by which pyrG transcripts initiated at neighboring start sites are differentially expressed remains to be determined. Other intriguing examples are the unresolved and apparently novel mechanisms of regulation by PyrR. Included in this list are the mechanism of PyrR2-mediated regulation of pyr gene expression in response to inorganic carbon in L. plantarum and PyrR-mediated regulation of pyr gene expression in the absence of recognizable PyrR binding sequences in Pseudomonas species.
Importantly, the information garnered from the study of pyr gene regulation can also facilitate the analysis of regulatory mechanisms that are unrelated to pyrimidine biosynthesis. In fact, this information has already contributed to the discovery of such mechanisms. Two noteworthy examples are the regulation of expression in E. coli of the rRNA operons (42, 144) and the fis operon (165) by the concentration of the initiating NTP. In these cases, promoters contain sequences that restrict initiation to a single “unfavorable” position (e.g., A9 in the case of most rRNA operons), as defined by the rules established with mutant pyrC promoters. As a consequence, the efficiency of transcription initiation at the rRNA and fis promoters can be determined by the intracellular concentration of the initiating NTP (i.e., ATP or GTP for the rRNA promoters and CTP for the fis promoter). Furthermore, the concentrations of the initiating NTPs vary under different growth conditions in a manner that causes appropriate levels of rRNA and fis operon expression, providing simple yet elegant regulation of gene expression.
Many other bacterial operons that are unrelated to pyrimidine biosynthesis contain elements that are critical components in the mechanisms of pyr gene regulation described in this review, so it seems likely that at least some of these operons will employ these features in comparable ways. However, the number of combinations and permutations of these elements is large, and thus there may be a high degree of flexibility in the assembly of regulatory mechanisms. In addition, it seems highly likely that hybrid mechanisms have evolved that combine features described above with a different set of regulatory elements. For example, many of the pyr gene regulatory mechanisms rely on nucleotide-sensitive reiterative transcription, start site switching, or transcription pausing as the key regulatory event in conditional gene expression. It is certainly conceivable, especially with the nuances described in this review, that factors other than the intracellular levels of nucleotides could also modulate these phases of transcription. Activating or inactivating these other factors by a cellular metabolite or condition would then make gene expression responsive to a nonnucleotide signal. On the other hand, the analysis of bacterial genome sequences reveals many pyr genes that do not appear to be regulated by any of the mechanisms discussed in this review. It is not known whether expression of these genes is altered in response to intracellular nucleotide pools, but this seems to be a likely possibility. The identification and examination of such regulated pyr genes offers rich possibilities for the discovery of novel modes of metabolic control.
Bioinformatic analysis of genomic DNA sequences was a useful tool in the development of models for many of the regulatory mechanisms discussed in this review, particularly in the identification of transcription terminators, antiterminators, and transcription pause sites that play roles in these mechanisms. However, a major limitation to this approach was illustrated by the mechanism of pyrG regulation in B. subtilis. In this mechanism, the antiterminator RNA hairpin contains an essential tract of nucleotides that was added by reiterative transcription and could not have been predicted from the sequence of the pyrG operon. In general, regulatory mechanisms involving reiterative transcription rely on subtle features in the DNA sequence of the promoter-leader region. These features are not readily identified by current bioinformatic analyses.
It should also be noted that many of the regulatory features described in this review involve only the basic transcriptional machinery of the cell and simple DNA sequences. Therefore, it is reasonable to suspect that some of the mechanisms and regulatory elements described here are also operative in eukaryotic cells. In fact, it would be surprising if they were not. The obstacle in their discovery is likely to be the education of more investigators about the surprises provided by the study of pyrimidine nucleotide biosynthesis in bacteria.
Finally, this review is yet another example of the importance of collaboration with other scientists. The combination of independent, intelligent minds that bring divergent experiences and ways of analyzing problems, when brought to bear on an unsolved scientific puzzle, is almost always more likely find a solution that survives the most thorough experimental testing. We also emphasize our shared pleasure in discovery. The more unexpected the answer, the greater the joy in finding it.
Acknowledgments
We gratefully acknowledge the many important contributions of our collaborators, students, and postdoctoral research associates, past and present, to the research described in this review.
Research in the authors' laboratories was supported by NIH grants GM47112 to R.L.S. and GM29466 to C.L.T.
REFERENCES
- 1.Abd-el-Al, A., and J. L. Ingraham. 1969. Control of carbamyl phosphate synthesis in Salmonella typhimurium. J. Biol. Chem. 2444033-4038. [PubMed] [Google Scholar]
- 2.Amiott, E. A., and J. A. Jaehning. 2006. Sensitivity of the yeast mitochondrial RNA polymerase to +1 and +2 initiating nucleotides. J. Biol. Chem. 28134982-34988. [DOI] [PubMed] [Google Scholar]
- 3.Andersen, J. T., K. F. Jensen, and P. Poulsen. 1991. Role of transcription pausing in the control of the pyrE attenuator in Escherichia coli. Mol. Microbiol. 5327-333. [DOI] [PubMed] [Google Scholar]
- 4.Andersen, L., M. Kilstrup, and J. Neuhard. 1989. Pyrimidine, purine and nitrogen control of cytosine deaminase synthesis in Escherichia coli K 12. Involvement of the glnLG and purR genes in the regulation of codA expression. Arch. Microbiol. 152115-118. [DOI] [PubMed] [Google Scholar]
- 5.Andersen, P. S., D. Frees, R. Fast, and B. Mygind. 1995. Uracil uptake in Escherichia coli K-12: isolation of uraA mutants and cloning of the gene. J. Bacteriol. 1772008-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersen, P. S., P. J. G. Jansen, and K. Hammer. 1994. Two different dihydroorotate dehydrogenases in Lactococcus lactis. J. Bacteriol. 1763975-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andersen, P. S., J. M. Smith, and B. Mygind. 1992. Characterization of the upp gene encoding uracil phosphoribosyltransferase of Escherichia coli K12. Eur. J. Biochem. 20451-56. [DOI] [PubMed] [Google Scholar]
- 8.Arsène-Ploetze, F., V. Kugler, J. Martinussen, and F. Bringel. 2006. Expression of the pyr operon of Lactobacillus plantarum is regulated by inorganic carbon availability through a second regulator, PyrR2, homologous to the pyrimidine-dependent regulator PyrR1. J. Bacteriol. 1888607-8616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bäckström, D., R. M. Sjöberg, and L. G. Lundberg. 1986. Nucleotide sequence of the structural gene for dihydroorotase of Escherichia coli K12. Eur. J. Biochem. 16077-82. [DOI] [PubMed] [Google Scholar]
- 10.Baranov, P. V., A. W. Hammer, J. Zhou, R. F. Gesteland, and J. F. Atkins. 2005. Transcriptional slippage in bacteria: distribution in sequenced genomes and utilization in IS element gene expression. Genome Biol. 6R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barr, J. N., and G. W. Wertz. 2001. Polymerase slippage at vesicular stomatitis virus gene junctions to generate poly(A) is regulated by the upstream 3′-AUAC-5′ tetranucleotide: implications for the mechanism of transcription termination. J. Virol. 756901-6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beckwith, J. R., A. B. Pardee, R. Austrian, and F. Jacob. 1962. Coordination of the synthesis of the enzymes in the pyrimidine pathway of E. coli. J. Mol. Biol. 5618-634. [DOI] [PubMed] [Google Scholar]
- 13.Bonekamp, F., K. Clemmesen, O. Karlström, and K. F. Jensen. 1984. Mechanism of UTP-modulated attenuation at the pyrE gene of Escherichia coli: an example of operon polarity control through the coupling of translation to transcription. EMBO J. 32857-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonner, E. R., J. N. D'Elia, B. K. Billips, and R. L. Switzer. 2001. Molecular recognition of pyr mRNA by the Bacillus subtilis attenuation regulatory protein PyrR. Nucleic Acids Res. 294851-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bouvier, J., J.-C. Patte, and P. Stragier. 1984. Multiple regulatory signals in the control region of the Escherichia coli carAB operon. Proc. Natl. Acad. Sci. USA 814139-4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brabson, J. S., and R. L. Switzer. 1975. Purification and properties of Bacillus subtilis aspartate transcarbamylase. J. Biol. Chem. 2508664-8669. [PubMed] [Google Scholar]
- 17.Carpousis, A. J., J. E. Stefano, and J. D. Gralla. 1982. 5′ nucleotide heterogeneity and altered initiation of transcription at mutant lac promoters. J. Mol. Biol. 157619-633. [DOI] [PubMed] [Google Scholar]
- 18.Chan, C. L., and R. Landick. 1993. Dissection of the his leader pause site by base substitution reveals a multipartite signal that includes a pause hairpin. J. Mol. Biol. 23325-42. [DOI] [PubMed] [Google Scholar]
- 19.Chan, C. L., and R. Landick. 1994. New perspectives on RNA chain elongation and termination by E. coli RNA polymerase, p. 297-320. In R. Conaway and J. Conaway (ed.), Transcription: mechanisms and regulation. Raven Press, New York, NY.
- 20.Chander, P. 2006. Structural analysis of the regulatory protein of pyrimidine biosynthesis, PyrR. Ph.D. thesis. Purdue University, West Lafayette, IN.
- 21.Chander, P., K. M. Halbig, J. K. Miller, C. J. Fields, H. K. S. Bonner, G. K. Grabner, R. L. Switzer, and J. L. Smith. 2005. Structure of the nucleotide complex of PyrR, the pyr attenuation protein from Bacillus caldolyticus, suggests dual regulation by pyrimidine and purine nucleotides. J. Bacteriol. 1871773-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Charlier, D., M. Roovers, F. Van Vliet, A. Boyen, R. Cunin, Y. Nakamura, N. Glansdorff, and A. Piérard. 1992. Arginine regulon of Escherichia coli K-12. A study of repressor-operator interactions and of in vitro binding affinities versus in vivo repression. J. Mol. Biol. 226367-386. [DOI] [PubMed] [Google Scholar]
- 23.Cheng, Y., S. M. Dylla, and C. L. Turnbough, Jr. 2001. A long T·A tract in the upp initially transcribed region is required for regulation of upp expression by UTP-dependent reiterative transcription in Escherichia coli. J. Bacteriol. 183221-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cho, H.-S., K.-J. Seul, S.-H. Park, and S.-Y. Ghim. 2003. Characterization and functional study of PyrR orthologues from genome sequences of bacteria. Kor. J. Microbiol. Biotechnol. 31103-110. [Google Scholar]
- 25.Choi, K. Y., and H. Zalkin. 1990. Regulation of Escherichia coli pyrC by the purine regulon repressor protein. J. Bacteriol. 1723201-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clemmesen, K., F. Bonekamp, O. Karlström, and K. F. Jensen. 1985. Role of translation in the UTP-modulated attenuation at the pyrBI operon of Escherichia coli. Mol. Gen. Genet. 201247-251. [DOI] [PubMed] [Google Scholar]
- 27.Cunin, R., N. Glansdorff, A. Piérard, and V. Stalon. 1986. Biosynthesis and metabolism of arginine in bacteria. Microbiol. Rev. 50314-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Danielsen, S., M. Kilstrup, K. Barilla, B. Jochimsen, and J. Neuhard. 1992. Characterization of the Escherichia coli codBA operon encoding cytosine permease and cytosine deaminase. Mol. Microbiol. 61335-1344. [DOI] [PubMed] [Google Scholar]
- 29.Devroede, N., N. Huysveld, and D. Charlier. 2006. Mutational analysis of intervening sequences connecting the binding sites for integration host factor, PepA, PurR, and RNA polymerase in the control region of the Escherichia coli carAB operon, encoding carbamoylphosphate synthase. J. Bacteriol. 1883236-3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Devroede, N., T.-L. Thia-Toong, D. Gigot, D. Maes, and D. Charlier. 2004. Purine and pyrimidine-specific repression of the Escherichia coli carAB operon are functionally and structurally coupled. J. Mol. Biol. 33625-42. [DOI] [PubMed] [Google Scholar]
- 31.Donahue, J. P., and C. L. Turnbough, Jr. 1990. Characterization of transcriptional initiation from promoters P1 and P2 of the pyrBI operon of Escherichia coli K12. J. Biol. Chem. 26519091-19099. [PubMed] [Google Scholar]
- 32.Donahue, J. P., and C. L. Turnbough, Jr. 1994. Nucleotide-specific transcriptional pausing in the pyrBI leader region of Escherichia coli K-12. J. Biol. Chem. 26918185-18191. [PubMed] [Google Scholar]
- 33.Elagöz, A., A. Abdi, J.-C. Hubert, and B. Kammerer. 1996. Structure and organisation of the pyrimidine biosynthesis pathway genes in Lactobacillus plantarum: a PCR strategy for sequencing without cloning. Gene 18237-43. [DOI] [PubMed] [Google Scholar]
- 34.Elsholz, A. K. W., C. M. Jørgensen, and R. L. Switzer. 2007. The number of G residues in the Bacillus subtilis pyrG initially transcribed region governs reiterative transcription-mediated regulation. J. Bacteriol. 1892176-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Englesberg, E., J. Irr, J. Power, and N. Lee. 1965. Positive control of enzyme synthesis by gene C in the l-arabinose system. J. Bacteriol. 90946-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fields, C. J., and R. L. Switzer. 2007. Regulation of pyr gene expression in Mycobacterium smegmatis by PyrR-dependent translational repression. J. Bacteriol. 1896236-6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fisher, S. H. 1999. Regulation of nitrogen metabolism in Bacillus subtilis: vive la différence! Mol. Microbiol. 32223-232. [DOI] [PubMed] [Google Scholar]
- 38.Flodell, S., J. Schleucher, J. Cromsigt, H. Ippel, K. Kidd-Ljundggren, and S. Wijmenga. 2002. The apical stem-loop of the hepatitis B virus encapsidation signal folds into a stable tri-loop with two underlying pyrimidine bulges. Nucleic Acids Res. 304803-4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Freier, S. M., R. Kierzek, J. A. Jaeger, N. Sugimoto, M. H. Caruthers, T. Neilson, and D. H. Turner. 1986. Improved free-energy parameters for predictions of RNA duplex stability. Proc. Natl. Acad. Sci. USA 839373-9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frick, M. M., J. Neuhard, and R. A. Kelln. 1990. Cloning, nucleotide sequence and regulation of the Salmonella typhimurium pyrD gene encoding dihydroorotate dehydrogenase. Eur. J. Biochem. 194573-578. [DOI] [PubMed] [Google Scholar]
- 41.Fukushima, M., K. Kakinuma, and R. Kawaguchi. 2002. Phylogenetic analysis of Salmonella, Shigella, and Escherichia coli strains on the basis of the gyrB gene sequence. J. Clin. Microbiol. 402779-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gaal, T., M. S. Bartlett, W. Ross, C. L. Turnbough, Jr., and R. L. Gourse. 1997. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science 2782092-2097. [DOI] [PubMed] [Google Scholar]
- 43.Ghim, S.-Y., C. C. Kim, E. R. Bonner, J. N. D'Elia, G. K. Grabner, and R. L. Switzer. 1999. The Enterococcus faecalis pyr operon is regulated by autogenous transcriptional attenuation at a single site in the 5′ leader. J. Bacteriol. 1811324-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghim, S.-Y., and J. Neuhard. 1994. The pyrimidine biosynthesis operon of the thermophile Bacillus caldolyticus includes genes for uracil phosphoribosyltransferase and uracil permease. J. Bacteriol. 1763698-3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghim, S.-Y., and R. L. Switzer. 1996. Characterization of cis-acting mutations in the first attenuator region of the Bacillus subtilis pyr operon that are defective in regulation of expression by pyrimidines. J. Bacteriol. 1782351-2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ghim, S.-Y., and R. L. Switzer. 1996. Mutations in B. subtilis PyrR, the pyr regulatory protein with defects in regulation by pyrimidines. FEMS Microbiol. Lett. 13713-18. [DOI] [PubMed] [Google Scholar]
- 47.Gilbert, W., and B. Muller-Hill. 1966. Isolation of the lac repressor. Proc. Natl. Acad. Sci. USA 561891-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gollnick, P., P. Babitzke, A. Antson, and C. Yanofsky. 2005. Complexity in regulation of tryptophan biosynthesis in Bacillus subtilis. Annu. Rev. Genet. 3947-68. [DOI] [PubMed] [Google Scholar]
- 49.Grabner, G. K., and R. L. Switzer. 2003. Kinetic studies of the uracil phosphoribosyltransferase reaction catalyzed by the Bacillus subtilis pyrimidine attenuation regulatory protein PyrR. J. Biol. Chem. 2786921-6927. [DOI] [PubMed] [Google Scholar]
- 50.Grundy, F. J., and T. M. Henkin. 2006. From ribosome to riboswitch: control of gene expression in bacteria by RNA structural rearrangements. Crit. Rev. Biochem. Mol. Biol. 41329-338. [DOI] [PubMed] [Google Scholar]
- 51.Guo, H. C., and J. W. Roberts. 1990. Heterogeneous initiation due to slippage at the bacteriophage 82 late gene promoter in vitro. Biochemistry 2910702-10709. [DOI] [PubMed] [Google Scholar]
- 52.Gusarov, I., and E. Nudler. 1999. The mechanism of intrinsic transcription termination. Mol. Cell 3495-504. [DOI] [PubMed] [Google Scholar]
- 53.Han, X., and C. L. Turnbough, Jr. 1998. Regulation of carAB expression in Escherichia coli occurs in part through UTP-sensitive reiterative transcription. J. Bacteriol. 180705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harley, C. B., J. Lawrie, H. W. Boyer, and J. Hedgpeth. 1990. Reiterative copying by E. coli RNA polymerase during transcription initiation of mutant pBR322 tet promoters. Nucleic Acids Res. 18547-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harley, C. B., and R. P. Reynolds. 1987. Analysis of E. coli promoter sequences. Nucleic Acids Res. 152343-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Henkin, T. M., and C. Yanofsky. 2002. Regulation by transcription attenuation in bacteria: how RNA provides instructions for transcription termination/antitermination decisions. Bioessays 24700-707. [DOI] [PubMed] [Google Scholar]
- 57.Hershberg, R., G. Bejerano, A. Santos-Zavaleta, and H. Margalit. 2001. PromEC: an updated database of Escherichia coli mRNA promoters with experimentally identified transcriptional start sites. Nucleic Acids Res. 29277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heus, H. A., and A. Pardi. 1991. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science 253191-194. [DOI] [PubMed] [Google Scholar]
- 59.Heyden, B., C. Nüsslein, and H. Schaller. 1975. Initiation of transcription within an RNA-polymerase binding site. Eur. J. Biochem. 55147-155. [DOI] [PubMed] [Google Scholar]
- 60.Hobl, B., and M. Mack. 2007. The regulator protein PyrR of Bacillus subtilis specifically interacts in vivo with three untranslated regions within pyr mRNA of pyrimidine biosynthesis. Microbiology 153693-700. [DOI] [PubMed] [Google Scholar]
- 61.Jacob, F., and J. Monod. 1961. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 3318-356. [DOI] [PubMed] [Google Scholar]
- 62.Jacques, J. P., and D. Kolakofsky. 1991. Pseudo-templated transcription in prokaryotic and eukaryotic organisms. Genes Dev. 5707-713. [DOI] [PubMed] [Google Scholar]
- 63.Jacques, J. P., and M. M. Susskind. 1990. Pseudo-templated transcription by Escherichia coli RNA polymerase at a mutant promoter. Genes Dev. 41801-1810. [DOI] [PubMed] [Google Scholar]
- 64.Jensen, K. F. 1988. Hyper-regulation of pyr gene expression in Escherichia coli cells with slow ribosomes. Evidence for RNA polymerase pausing in vivo? Eur. J. Biochem. 175587-593. [DOI] [PubMed] [Google Scholar]
- 65.Jensen, K. F. 1989. Regulation of Salmonella typhimurium pyr gene expression: effect of changing both purine and pyrimidine nucleotide pools. J. Gen. Microbiol. 135805-815. [DOI] [PubMed] [Google Scholar]
- 66.Jensen, K. F., R. Fast, O. Karlström, and J. N. Larsen. 1986. Association of RNA polymerase having increased for Km for ATP and UTP with hyperexpression of the pyrB and pyrE genes of Salmonella typhimurium. J. Bacteriol. 166857-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jensen-MacAllister, I. E., Q. Meng, and R. L. Switzer. 2007. Regulation of pyrG expression in Bacillus subtilis: CTP-regulated antitermination and reiterative transcription with pyrG templates in vitro. Mol. Microbiol. 631440-1452. [DOI] [PubMed] [Google Scholar]
- 68.Jeong, S. W., W. H. Lang, and R. H. Reeder. 1996. The yeast transcription terminator for RNA polymerase I is designed to prevent polymerase slippage. J. Biol. Chem. 27116104-16110. [DOI] [PubMed] [Google Scholar]
- 69.Jeong, W., and C. Kang. 1994. Start site selection at lacUV5 promoter affected by the sequence context around the initiation sites. Nucleic Acids Res. 224667-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jin, D. J. 1996. A mutant RNA polymerase reveals a kinetic mechanisms for the switch between nonproductive stuttering synthesis and productive initiation during promoter clearance. J. Biol. Chem. 27111659-11667. [PubMed] [Google Scholar]
- 71.Jin, D. J. 1994. Slippage synthesis at the galP2 promoter of Escherichia coli and its regulation by UTP concentration and cAMP·cAMP receptor protein. J. Biol. Chem. 26917221-17227. [PubMed] [Google Scholar]
- 72.Johnston, H. M., W. M. Barnes, F. G. Chumley, L. Bossi, and J. R. Roth. 1980. Model for regulation of the histidine operon of Salmonella. Proc. Natl. Acad. Sci. USA 77508-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jørgensen, C. M., C. J. Fields, P. Chander, D. Watt, J. W. Burgner II, J. L. Smith, and R. L. Switzer. 2008. pyr RNA binding to the Bacillus caldolyticus PyrR attenuation protein. Characterization and regulation by uridine and guanosine nucleotides. FEBS J. 275655-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jørgensen, C. M., K. Hammer, and J. Martinussen. 2003. CTP limitation increases expression of CTP synthase in Lactococcus lactis. J. Bacteriol. 1856562-6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Justesen, J., and J. Neuhard. 1975. pyrR identical to pyrH in Salmonella typhimurium: control of expression of the pyr genes. J. Bacteriol. 123851-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kadziola, A., J. Neuhard, and S. Larsen. 2002. Structure of product-bound Bacillus caldolyticus uracil phosphoribosyltransferase confirms ordered sequential substrate binding. Acta Crystallogr. D 58936-943. [DOI] [PubMed] [Google Scholar]
- 77.Kahler, A. E., F. S. Nielsen, and R. L. Switzer. 1999. Biochemical characterization of the heteromeric Bacillus subtilis dihydroorotate dehydrogenase and its isolated subunits. Arch. Biochem. Biophys. 371191-201. [DOI] [PubMed] [Google Scholar]
- 78.Kahler, A. E., and R. L. Switzer. 1996. Identification of a novel gene of pyrimidine nucleotide biosynthesis, pyrDII, that is required for dihydrooroate dehydrogenase activity in Bacillus subtilis. J. Bacteriol. 1785013-5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kang, C., and C. R. Cantor. 1985. Structure of ribosome-bound messenger RNA as revealed by enzymatic accessibility studies. J. Mol. Biol. 181241-251. [DOI] [PubMed] [Google Scholar]
- 80.Kantardjieff, K. A., C. Vasquez, P. Castro, N. M. Warfel, B.-S. Rho, T. Lekin, C.-Y. Kim, B. W. Segelke, T. C. Terwillinger, and B. Rupp. 2005. Structure of PyrR (Rv1379) from Mycobacterium tuberculosis: a persistence gene and protein drug target. Acta Crystallogr. D 61355-364. [DOI] [PubMed] [Google Scholar]
- 81.Kelln, R. A., J. J. Kinahan, K. F. Foltermann, and G. A. O'Donovan. 1975. Pyrimidine biosynthetic enzymes of Salmonella typhimurium, repressed specifically by growth in the presence of cytidine. J. Bacteriol. 124764-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kelln, R. A., and J. Neuhard. 1988. Regulation of pyrC expression in Salmonella typhimurium: identification of a regulatory region. Mol. Gen. Genet. 212287-294. [DOI] [PubMed] [Google Scholar]
- 83.Kilstrup, M., K. Hammer, P. R. Jensen, and J. Martinussen. 2005. Nucleotide metabolism and its control in lactic acid bacteria. FEMS Microbiol. Rev. 29555-590. [DOI] [PubMed] [Google Scholar]
- 84.Kilstrup, M., L. M. Meng, J. Neuhard, and P. Nygaard. 1989. Genetic evidence for a repressor of synthesis of cytosine deaminase and purine biosynthesis enzymes in Escherichia coli. J. Bacteriol. 1712124-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kingston, R. E., W. C. Nierman, and M. J. Chamberlin. 1981. A direct effect of guanosine tetraphosphate on pausing of Escherichia coli RNA polymerase during RNA chain elongation. J. Biol. Chem. 2562787-2797. [PubMed] [Google Scholar]
- 86.Landick, R., C. L. Turnbough, Jr., and C. Yanofsky. 1996. Transcription attenuation, p. 1263-1286. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. American Society for Microbiology, Washington, DC. [Google Scholar]
- 87.Larsen, B., N. M. Wills, C. Nelson, J. F. Atkins, and R. F. Gesteland. 2000. Nonlinearity in genetic decoding: homologous DNA replicase genes use alternatives of transcriptional slippage or translational frameshifting. Proc. Natl. Acad. Sci. USA 971683-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Larsen, J. N., and K. F. Jensen. 1985. Nucleotide sequence of the pyrD gene of Escherichia coli and characterization of the flavoprotein dihydroorotate dehydrogenase. Eur. J. Biochem. 15159-65. [DOI] [PubMed] [Google Scholar]
- 89.Lerner, C. G., B. T. Stephenson, and R. L. Switzer. 1987. Structure of the Bacillus subtilis pyrimidine biosynthetic (pyr) gene cluster. J. Bacteriol. 1692202-2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lerner, C. G., and R. L. Switzer. 1986. Cloning and structure of the Bacillus subtilis aspartate transcarbamylase gene (pyrB). J. Biol. Chem. 26111156-11165. [PubMed] [Google Scholar]
- 91.Levin, H. L., K. Park, and H. K. Schachman. 1989. Attenuation in the regulation of the pyrBI operon in Escherichia coli. In vivo studies of transcriptional termination. J. Biol. Chem. 26414638-14645. [PubMed] [Google Scholar]
- 92.Levin, H. L., and H. K. Schachman. 1985. Regulation of aspartate transcarbamoylase synthesis in Escherichia coli: analysis of deletion mutations in the promoter region of the pyrBI operon. Proc. Natl. Acad. Sci. USA 824643-4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lewis, D. E. A., and S. Adhya. 2004. Axiom of determining transcription start points by RNA polymerase in Escherichia coli. Mol. Microbiol. 54692-701. [DOI] [PubMed] [Google Scholar]
- 94.Linton, M. F., M. Raabe, V. Pierotti, and S. G. Young. 1997. Reading-frame restoration by transcriptional slippage at long stretches of adenine residues in mammalian cells. J. Biol. Chem. 27214127. [DOI] [PubMed] [Google Scholar]
- 95.Lisser, S., and H. Margalit. 1993. Compilation of E. coli mRNA promoter sequences. Nucleic Acids Res. 211507-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu, C., J. P. Donahue, L. S. Heath, and C. L. Turnbough, Jr. 1993. Genetic evidence that promoter P2 is the physiologically significant promoter for the pyrBI operon of Escherichia coli K-12. J. Bacteriol. 1752363-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu, C., L. S. Heath, and C. L. Turnbough, Jr. 1994. Regulation of pyrBI operon expression in Escherichia coli by UTP-sensitive reiterative RNA synthesis during transcriptional initiation. Genes Dev. 82904-2912. [DOI] [PubMed] [Google Scholar]
- 98.Liu, C., and C. L. Turnbough, Jr. 1989. Multiple control mechanisms for pyrimidine-mediated regulation of pyrBI operon expression in Escherichia coli K-12. J. Bacteriol. 1713337-3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu, J., and C. L. Turnbough, Jr. 1994. Effects of transcriptional start site sequence and position on nucleotide-sensitive selection of alternative start sites at the pyrC promoter in Escherichia coli. J. Bacteriol. 1762938-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu, J., and C. L. Turnbough, Jr. 1994. Identification of the Shine-Dalgarno sequence required for expression and translational control of the pyrC gene in Escherichia coli K-12. J. Bacteriol. 1762513-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lu, C. D., S. Katzif, and A. T. Abdelal. 1995. Participation of the purine repressor in control of the carbamoylphosphate synthetase operon in Salmonella typhimurium. Mol. Microbiol. 17981-988. [DOI] [PubMed] [Google Scholar]
- 102.Lu, Y., and R. L. Switzer. 1996. Evidence that the Bacillus subtilis pyrimidine biosynthetic regulatory protein PyrR acts by binding to pyr mRNA at three sites in vivo. J. Bacteriol. 1785806-5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lu, Y., and R. L. Switzer. 1996. Transcriptional attenuation of the Bacillus subtilis pyr operon by the PyrR regulatory protein and uridine nucleotides in vitro. J. Bacteriol. 1787206-7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lu, Y., R. J. Turner, and R. L. Switzer. 1996. Function of the RNA secondary structures in the regulation of the Bacillus subtilis pyr operon expression. Proc. Natl. Acad. Sci. USA 9314462-14467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lu, Y., R. J. Turner, and R. L. Switzer. 1995. Roles of the three transcriptional attenuators of the Bacillus subtilis pyrimidine biosynthetic operon in the regulation of its expression. J. Bacteriol. 1771315-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Machida, C., Y. Machida, and E. Ohtsubo. 1984. Both inverted repeat sequences located at the ends of IS1 provide promoter functions. J. Mol. Biol. 177247-267. [DOI] [PubMed] [Google Scholar]
- 107.Martin, C. T., D. K. Muller, and J. E. Coleman. 1988. Processivity in early stages of transcription by T7 RNA polymerase. Biochemistry 273966-3974. [DOI] [PubMed] [Google Scholar]
- 108.Martinussen, J., P. Glaser, P. S. Andersen, and H. H. Saxild. 1995. Two genes encoding uracil phosphoribosyltransferase are present in Bacillus subtilis. J. Bacteriol. 177271-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Martinussen, J., J. Schallert, B. Andersen, and K. Hammer. 2001. The pyrimidine operon pyrRPB-carA from Lactococcus lactis. J. Bacteriol. 1832785-2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mathews, C. K. 1972. Biochemistry of deoxyribonucleic acid-defective amber mutants of bacteriophage T4. III. Nucleotide pools. J. Biol. Chem. 2477430-7438. [PubMed] [Google Scholar]
- 111.Mathews, D. H., M. D. Disney, J. L. Childs, S. J. Schroeder, M. Zuker, and D. H. Turner. 2004. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. USA 1017287-7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Maurizi, M. R., J. S. Brabson, and R. L. Switzer. 1978. Immunochemical studies of the inactivation of aspartate transcarbamylase by stationary phase Bacillus subtilis cells: evidence for selective, energy-dependent degradation. J. Biol. Chem. 2535585-5593. [PubMed] [Google Scholar]
- 113.McClure, W. R., C. L. Cech, and D. E. Johnston. 1978. A steady state assay for the RNA polymerase initiation reaction. J. Biol. Chem. 2538941-8948. [PubMed] [Google Scholar]
- 114.Meng, Q., and R. L. Switzer. 2002. cis-acting sequences of Bacillus subtilis pyrG mRNA essential for regulation by antitermination. J. Bacteriol. 1846734-6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Meng, Q., and R. L. Switzer. 2001. Regulation of transcription of the Bacillus subtilis pyrG gene encoding cytidine triphosphate synthetase. J. Bacteriol. 1835513-5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Meng, Q., C. L. Turnbough, Jr., and R. L. Switzer. 2004. Attenuation control of pyrG expression in Bacillus subtilis is mediated by CTP-sensitive reiterative transcription. Proc. Natl. Acad. Sci. USA 10110943-10948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Michaels, G., R. A. Kelln, and F. E. Nargang. 1987. Cloning, nucleotide sequence and expression of the pyrBI operon of Salmonella typhimurium LT2. Eur. J. Biochem. 16655-61. [DOI] [PubMed] [Google Scholar]
- 118.Mitchell, J. E., D. Zheng, S. J. Busby, and S. D. Minchin. 2003. Identification and analysis of ′extended −10′ promoters in Escherichia coli. Nucleic Acids Res. 314689-4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Muse, W. B., C. J. Rosario, and R. A. Bender. 2003. Nitrogen regulation of the codBA (cytosine deaminase) operon from Escherichia coli by the nitrogen assimilation control protein, NAC. J. Bacteriol. 1852920-2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Navre, M., and H. K. Schachman. 1983. Synthesis of aspartate transcarbamoylase in Escherichia coli: transcriptional regulation of the pyrB-pyrI operon. Proc. Natl. Acad. Sci. USA 801207-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Neuhard, J., and R. A. Kelln. 1996. Biosynthesis and conversions of pyrimidines, p. 580-599. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. American Society for Microbiology, Washington, DC. [Google Scholar]
- 122.Neuhard, J., R. A. Kelln, and E. Stauning. 1986. Cloning and structural characterization of the Salmonella typhimurium pyrC gene encoding dihydroorotase. Eur. J. Biochem. 157335-342. [DOI] [PubMed] [Google Scholar]
- 123.Neuhard, J., L. M. Meng, K. I. Sørensen, and R. A. Kelln. 1990. Purine control of pyrC expression in Salmonella typhimurium. Int. J. Purine Pyrimidine Res. 161-66. [Google Scholar]
- 124.Neuhard, J., and P. Nygaard. 1987. Purines and pyrimidines, p. 445-473. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, vol. 1. American Society for Microbiology, Washington, DC. [Google Scholar]
- 125.Nicoloff, H., A. Elagoz, F. Arsene-Ploetze, B. Kammerer, J. Martinussen, and F. Bringel. 2005. Repression of the pyr operon in Lactobacillus plantarum prevents its ability to grow at low carbon dioxide levels. J. Bacteriol. 1872093-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nierman, W. C., and M. J. Chamberlin. 1979. Studies of RNA chain initiation by Escherichia coli RNA polymerase bound to T7 DNA. Direct analysis of the kinetics and extent of RNA chain initiation at T7 promoter A 1. J. Biol. Chem. 2547921-7926. [PubMed] [Google Scholar]
- 127.O'Donovan, G. A., and J. C. Gerhart. 1972. Isolation and partial characterization of regulatory mutants of the pyrimidine pathway in Salmonella typhimurium. J. Bacteriol. 1091085-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.O'Donovan, G. A., and J. Neuhard. 1970. Pyrimidine metabolism in microorganisms. Bacteriol. Rev. 34278-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ost, K. A., and M. P. Deutscher. 1991. Escherichia coli orfE (upstream of pyrE) encodes RNase PH. J. Bacteriol. 1735589-5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Paulus, T. J., T. J. McGarry, P. G. Shekelle, S. Rosenzweig, and R. L. Switzer. 1982. Coordinate synthesis of the enzymes of pyrimidine biosynthesis in Bacillus subtilis. J. Bacteriol. 149775-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Paulus, T. J., and R. L. Switzer. 1979. Characterization of pyrimidine-repressible and arginine-repressible carbamyl phosphate synthetases from Bacillus subtilis. J. Bacteriol. 13782-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Piérard, A., N. Glansdorff, D. Gigot, M. Crabeel, P. Halleux, and L. Thiry. 1976. Repression of Escherichia coli carbamoylphosphate synthase: relationships with enzyme synthesis in the arginine and pyrimidine pathways. J. Bacteriol. 127291-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Piette, J., H. Nyunoya, C. J. Lusty, R. Cunin, G. Weyens, M. Crabeel, D. Charlier, N. Glansdorff, and A. Piérard. 1984. DNA sequence of the carA gene and the control region of carAB: tandem promoters, respectively controlled by arginine and the pyrimidines, regulate the synthesis of carbamoyl-phosphate synthetase in Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 814134-4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Poulsen, P., F. Bonekamp, and K. F. Jensen. 1984. Structure of the Escherichia coli pyrE operon and control of pyrE expression by a UTP modulated intercistronic attenuation. EMBO J. 31783-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Poulsen, P., and K. F. Jensen. 1987. Effect of UTP and GTP pools on attenuation at the pyrE gene of Escherichia coli. Mol. Gen. Genet. 208152-158. [DOI] [PubMed] [Google Scholar]
- 136.Poulsen, P., K. F. Jensen, P. Valentin-Hansen, P. Carlsson, and L. G. Lundberg. 1983. Nucleotide sequence of the Escherichia coli pyrE gene and of the DNA in front of the protein-coding region. Eur. J. Biochem. 135223-229. [DOI] [PubMed] [Google Scholar]
- 137.Qi, F., and C. L. Turnbough, Jr. 1995. Regulation of codBA operon expression in Escherichia coli by UTP-dependent reiterative transcription and UTP-sensitive transcriptional start site switching. J. Mol. Biol. 254552-565. [DOI] [PubMed] [Google Scholar]
- 138.Quinn, C. L., B. T. Stephenson, and R. L. Switzer. 1991. Functional organization and nucleotide sequence of the Bacillus subtilis pyrimidine biosynthetic operon. J. Biol. Chem. 2669113-9127. [PubMed] [Google Scholar]
- 139.Roland, K. L., C. G. Liu, and C. L. Turnbough, Jr. 1988. Role of the ribosome in suppressing transcriptional termination at the pyrBI attenuator of Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 857149-7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Roland, K. L., F. E. Powell, and C. L. Turnbough, Jr. 1985. Role of translation and attenuation in the control of pyrBI operon expression in Escherichia coli K-12. J. Bacteriol. 163991-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Roof, W. D., K. F. Foltermann, and J. R. Wild. 1982. The organization and regulation of the pyrBI operon in E. coli includes a rho-independent attenuator sequence. Mol. Gen. Genet. 187391-400. [DOI] [PubMed] [Google Scholar]
- 142.Sadler, W. C., and R. L. Switzer. 1977. Regulation of Salmonella phosphoribosylpyrophosphate synthetase activity in vivo. Deductions from pool measurements. J. Biol. Chem. 2528504-8511. [PubMed] [Google Scholar]
- 143.Savacool, H. K., and R. L. Switzer. 2002. Characterization of the interaction of Bacillus subtilis PyrR with pyr mRNA by site-directed mutagenesis of the protein. J. Bacteriol. 1842521-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Schneider, D. A., W. Ross, and R. L. Gourse. 2003. Control of rRNA expression in Escherichia coli. Curr. Opin. Microbiol. 6151-156. [DOI] [PubMed] [Google Scholar]
- 145.Schwartz, M., and J. Neuhard. 1975. Control of expression of the pyr genes in Salmonella typhimurium: effects of variations in uridine and cytidine nucleotide pools. J. Bacteriol. 121814-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Shimada, T., K. Hirao, A. Kori, K. Yamamoto, and A. Ishihama. 2007. RutR is the uracil/thymine-sensing master regulator of a set of genes for synthesis and degradation of pyrimidines. Mol. Microbiol. 66744-757. [DOI] [PubMed] [Google Scholar]
- 147.Shultzaberger, R. K., Z. Chen, K. A. Lewis, and T. D. Schneider. 2007. Anatomy of Escherichia coli sigma-70 promoters. Nucleic Acids Res. 35771-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sinha, S. C., J. Krahn, B. S. Shin, D. R. Tomchick, H. Zalkin, and J. L. Smith. 2003. The purine repressor of Bacillus subtilis: a novel combination of domains adapted for transcription regulation. J. Bacteriol. 1854087-4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sinha, S. C., and J. L. Smith. 2001. The PRT protein family. Curr. Opin. Struct. Biol. 11733-739. [DOI] [PubMed] [Google Scholar]
- 150.Sipos, K., R. Szigeti, X. Dong, and C. L. Turnbough, Jr. 2007. Systematic mutagenesis of the thymidine tract of the pyrBI attenuator and its effects on intrinsic transcription termination in Escherichia coli. Mol. Microbiol. 66127-138. [DOI] [PubMed] [Google Scholar]
- 151.Sørensen, K. I. 1994. Conformational heterogeneity in the Salmonella typhimurium pyrC and pyrD leader mRNAs produced in vivo. Nucleic Acids Res. 22625-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sørensen, K. I., K. E. Baker, R. A. Kelln, and J. Neuhard. 1993. Nucleotide pool-sensitive selection of the transcriptional start site in vivo at the Salmonella typhimurium pyrC and pyrD promoters. J. Bacteriol. 1754137-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Sørensen, K. I., and J. Neuhard. 1991. Dual transcriptional initiation sites from the pyrC promoter control expression of the gene in Salmonella typhimurium. Mol. Gen. Genet. 225249-256. [DOI] [PubMed] [Google Scholar]
- 154.Steitz, J. A., and K. Jakes. 1975. How ribosomes select initiator regions in mRNA: base pair formation between the 3′ terminus of 16S rRNA and the mRNA during initiation of protein synthesis in Escherichia coli. Proc. Natl. Acad. Sci. USA 724734-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Switzer, R. L., R. J. Turner, and Y. Lu. 1999. Regulation of the Bacillus subtilis pyrimidine biosynthetic operon by transcriptional attenuation: control of gene expression by an mRNA-binding protein. Prog. Nucleic Acids Res. Mol. Biol. 62329-367. [DOI] [PubMed] [Google Scholar]
- 156.Tomchick, D. R., R. J. Turner, R. L. Switzer, and J. L. Smith. 1998. Adaptation of an enzyme to regulatory function: structure of Bacillus subtilis PyrR, a bifunctional pyr RNA-binding attenuation protein and uracil phosphoribosyltransferase. Structure 6337-350. [DOI] [PubMed] [Google Scholar]
- 157.Tu, A. H., and C. L. Turnbough, Jr. 1997. Regulation of upp expression in Escherichia coli by UTP-sensitive selection of transcriptional start sites coupled with UTP-dependent reiterative transcription. J. Bacteriol. 1796665-6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Turnbough, C. L., Jr. 1983. Regulation of Escherichia coli aspartate transcarbamylase synthesis by guanosine tetraphosphate and pyrimidine ribonucleoside triphosphates. J. Bacteriol. 153998-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Turnbough, C. L., Jr., K. L. Hicks, and J. P. Donahue. 1983. Attenuation control of pyrBI operon expression in Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 80368-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Turner, R. J., E. R. Bonner, G. K. Grabner, and R. L. Switzer. 1998. Purification and characterization of Bacillus subtilis PyrR, a bifunctional pyr mRNA-binding attenuation protein/uracil phosphoribosyltransferase. J. Biol. Chem. 2735932-5938. [DOI] [PubMed] [Google Scholar]
- 161.Turner, R. J., Y. Lu, and R. L. Switzer. 1994. Regulation of the Bacillus subtilis pyrimidine biosynthetic (pyr) gene cluster by an autogenous transcriptional attenuation mechanism. J. Bacteriol. 1763708-3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Van de Casteele, M., P. Chen, M. Roovers, C. Legrain, and N. Glansdorff. 1997. Structure and expression of a pyrimidine gene cluster from the extreme thermophile Thermus strain ZO5. J. Bacteriol. 1793470-3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Vial, T. C., K. E. Baker, and R. A. Kelln. 1993. Dual control by purines and pyrimidines of the expression of the pyrD gene of Salmonella typhimurium. FEMS Microbiol. Lett. 111309-314. [DOI] [PubMed] [Google Scholar]
- 164.Wagner, L. A., R. B. Weiss, R. Driscoll, D. S. Dunn, and R. F. Gesteland. 1990. Transcriptional slippage occurs during elongation at runs of adenine or thymine in Escherichia coli. Nucleic Acids Res. 183529-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Walker, K. A., P. Mallik, T. S. Pratt, and R. Osuna. 2004. The Escherichia coli fis promoter is regulated by changes in the levels of its transcription initiation nucleotide CTP. J. Biol. Chem. 27950818-50828. [DOI] [PubMed] [Google Scholar]
- 166.Walker, K. A., and R. Osuna. 2002. Factors affecting start site selection at the Escherichia coli fis promoter. J. Bacteriol. 1844783-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wang, H., N. Glansdorff, and D. Charlier. 1998. The arginine repressor of Escherichia coli K-12 makes direct contacts to minor and major groove determinants of the operators. J. Mol. Biol. 277805-824. [DOI] [PubMed] [Google Scholar]
- 168.Wanner, B. L. 1996. Phosphorus assimilation and control of the phosphate regulon, p. 1357-1381. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. American Society for Microbiology, Washington, DC. [Google Scholar]
- 169.Wilson, H. R., C. D. Archer, J. Liu, and C. L. Turnbough, Jr. 1992. Translational control of pyrC expression mediated by nucleotide-sensitive selection of transcriptional start sites in Escherichia coli. J. Bacteriol. 174514-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wilson, H. R., P. T. Chan, and C. L. Turnbough, Jr. 1987. Nucleotide sequence and expression of the pyrC gene of Escherichia coli K-12. J. Bacteriol. 1693051-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wilson, H. R., and C. L. Turnbough, Jr. 1990. Role of the purine repressor in the regulation of pyrimidine gene expression in Escherichia coli K-12. J. Bacteriol. 1723208-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Winkler, W. C. 2005. Metabolic monitoring by bacterial mRNAs. Arch. Microbiol. 183151-159. [DOI] [PubMed] [Google Scholar]
- 173.Winkler, W. C., and R. R. Breaker. 2005. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 59487-517. [DOI] [PubMed] [Google Scholar]
- 174.Xiong, X. F., and W. S. Reznikoff. 1993. Transcriptional slippage during the transcription initiation process at a mutant lac promoter in vivo. J. Mol. Biol. 231569-580. [DOI] [PubMed] [Google Scholar]
- 175.Yanofsky, C. 1981. Attenuation in the control of expression of bacterial operons. Nature 289751-758. [DOI] [PubMed] [Google Scholar]
- 176.Yanofsky, C., R. L. Kelley, and V. Horn. 1984. Repression is relieved before attenuation in the trp operon of Escherichia coli as tryptophan starvation becomes increasingly severe. J. Bacteriol. 1581018-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhang, H., C. M. Jørgensen, and R. L. Switzer. 2005. Mutations affecting transcription pausing in the Bacillus subtilis pyr operon. Arch. Microbiol. 184101-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhang, H., and R. L. Switzer. 2003. Transcriptional pausing in the Bacillus subtilis pyr operon in vitro: a role in transcriptional attenuation? J. Bacteriol. 1854764-4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Zubay, G., D. Schwartz, and J. Beckwith. 1970. Mechanism of activation of catabolite-sensitive genes: a positive control system. Proc. Natl. Acad. Sci. USA 66104-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Zuker, M., D. H. Mathews, and D. H. Turner. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide, p. 11-43. In J. Barciszewski and B. F. C. Clark (ed.), RNA biochemistry and biotechnology. Kluwer Academic Publishers, Dordrecht, The Netherlands.