Abstract
Summary: The phenomenon of peptidoglycan recycling is reviewed. Gram-negative bacteria such as Escherichia coli break down and reuse over 60% of the peptidoglycan of their side wall each generation. Recycling of newly made peptidoglycan during septum synthesis occurs at an even faster rate. Nine enzymes, one permease, and one periplasmic binding protein in E. coli that appear to have as their sole function the recovery of degradation products from peptidoglycan, thereby making them available for the cell to resynthesize more peptidoglycan or to use as an energy source, have been identified. It is shown that all of the amino acids and amino sugars of peptidoglycan are recycled. The discovery and properties of the individual proteins and the pathways involved are presented. In addition, the possible role of various peptidoglycan degradation products in the induction of β-lactamase is discussed.
INTRODUCTION
Many studies of bacterial cell walls have been concerned with their structures and mechanisms of synthesis. Much less studied and hidden from view is the story of how gram-negative bacteria such as Escherichia coli break down and reutilize all components of their cell wall peptidoglycan (PG). Our present knowledge of PG recycling is actually quite limited and fragmented. This review brings together these diverse observations and assesses their significance.
Overview of PG Recycling
Nine enzymes, one permease, and one periplasmic binding protein that appear to have the recovery of degradation products from PG as their sole function, thereby making them available for the cell to resynthesize more PG or to use as an energy source, have been identified in E. coli. Since PG represents only 2% of the cell mass, it does not represent a large source of nutrients. However, faced with a sudden loss of carbon, recycling may provide the necessary building blocks to complete a round of cell division and enter stationary phase. Alternatively, in the competition for food in the large intestine, the ability to recycle PG may have survival value. As will be discussed below, gram-negative bacteria living in the colon tend to have a complete repertoire of enzymes for recycling, whereas other organisms have a more limited but nevertheless purposeful arsenal.
What is known is that recycling is not essential under laboratory conditions. E. coli cells lacking ampG permease (essential for recycling) grow well in minimal media, although cells freshly transduced to ΔampG initially grow poorly for unknown reasons. With one exception, mutations in the genes known to be involved in recycling have no obvious phenotype. Thus, the genes do not appear to be required for viability, normal growth rate, normal morphology, or normal PG content. All of the enzymes are produced and are active during logarithmic growth. All are active on substrates that are present at concentrations in the range of 10−5 to 10−6 M. A loss of one of the enzymes usually results in the accumulation of millimolar amounts of its substrate. This may be taken to be an indirect indication of their normal activity.
Table 1 lists the proteins associated with PG recycling, their genes, their locations in the cell, and references.
TABLE 1.
Murein recycling enzymes
| Gene | Activity | Location | Reference(s) |
|---|---|---|---|
| ampG | GlcNAc-anhMurNAc permease | Inner membrane | 21,57 |
| ampD | anhMurNAc-l-Ala amidase | Cytoplasm | 48,57 |
| mpl (yjfG) | UDP-MurNAc:l-Ala-γ-d-Glu-Dap ligase | Cytoplasm | 46,83 |
| ldcA (f304 gene) | ld-Carboxypeptidase | Cytoplasm | 115,124 |
| mpaA (ycjI) | γ-d-Glu-Dap amidase | Cytoplasm | 120 |
| ycjG | l-Ala-d/l-Glu epimerase | Cytoplasm | 105 |
| nagZ (ycfO) | β-N-Acetylglucosaminidase | Cytoplasm | 20 |
| nagA | GlcNAc-6-P deacetylase | Cytoplasm | 130 |
| nagK (ycfX) | GlcNAc kinase | Cytoplasm | 121 |
| anmK (ydhH) | anhMurNAc kinase | Cytoplasm | 123 |
| murQ (yfeU) | MurNAc-6-P etherase | Cytoplasm | 59,122 |
| amiD (ybjR) | anhMurNAc-l-Ala amidase | Outer membrane | 118 |
| mppA | murein peptide binding protein | Periplasm | 94 |
Biological Effects of PG Recycling Intermediates
A discussion of the known biological effects of PG recycling intermediates is beyond the scope of this review. Activities such as cytotoxicity, pyrogenicity, arthritogenicity, adjuvanticity, and sleep promotion are well documented and have been reviewed by Adam and Lederer (1). Recent work has focused on the role of PG recycling in pathogenicity and the role of PG fragments in stimulating the innate immune response. Excellent reviews of these subjects by Boneca (11), Chaput and Boneca (19), and Cloud-Hansen et al. (23) have recently been published.
PG Structure and Composition
The term PG was originally defined as a class of polymers composed of approximately equal amounts of amino acids and sugars, and the term murein was reserved for the PG present in the cell envelopes of bacteria. However, as the only PG polymer presently recognized in nature is that found in bacterial cell walls and since the term murein is easily confused with murine, PG has been used increasingly in place of murein as a name for the polymer. The term murein sacculus survives to describe the rigid, shape-forming component of the cell envelope of bacteria that is composed of PG. In this review, PG and murein will be used interchangeably to refer to the polymer, and cell wall and murein sacculus to refer to the rigid structure free of inner and outer membranes, which together with the cell wall make up the cell envelope.
As shown in Fig. 1, PG consists of glycan strands cross-linked by short peptides referred to as stem peptides. The glycan strands consist of alternating units of N-acetylglucosamine (GlcNAc)- and N-acetylmuramic acid (MurNAc)-linked β-1,4. MurNAc is a derivative of GlcNAc with d-lactic acid attached to position 3 by an ether linkage. In gram-negative bacteria, the stem peptide attached to the carboxyl group of each muramic acid usually consists of l-Ala-γ-d-Glu-(l)-meso-diaminopimelic acid (Dap)-d-Ala, although the stem peptide often lacks d-Ala or, more rarely, terminates in d-Ala-d-Ala. About one-half of the stem peptides are involved in cross-links between neighboring glycan strands. The glycan strand length varies widely, with an average length of 30 disaccharide-peptide units (47). A comprehensive review of PG composition in a wide variety of bacteria was done by Schleifer and Kandler (104).
FIG. 1.
Basic structure of PG of E. coli.
Turnover of PG and Discovery of the Recycling of Murein Tripeptide
Turnover refers to the actual loss of PG components from the cell. However, the products of turnover are normally captured and reutilized by the cell by a process termed PG recycling. The PG recycling phenomenon was accidentally discovered by Goodell and Schwarz in 1985 during an investigation of cell wall turnover (40). At the time, there was evidence of substantial loss, i.e., turnover, of the cell wall murein of gram-positive bacilli such as Bacillus subtilis (77) and Lactobacillus acidophilus (12) and of the gram-negative organism Neisseria gonorrhoeae (38, 43). The rate of loss ranged from 25% to 50% per generation.
In marked contrast, turnover of E. coli murein ranged from 0% (77) to perhaps 5% per generation (18). When Goodell and Schwarz reinvestigated this phenomenon (40), they observed a 6 to 8% loss of Dap from the sacculus per generation. The Dap-containing materials released into the medium were identified as being l-Ala-γ-d-Glu-meso-Dap-d-Ala, l-Ala-γ-d-Glu-meso-Dap, and Dap-d-Ala. All of these results were obtained from pulse-chase experiments employing radioactive Dap. The small differences in turnover between these three reports were presumably due to strain variation or cultural conditions. However, as an internal control, no turnover of protein was detected in any of these studies.
Goodell and Schwarz then went on to do a pulse-chase experiment in which the Dap-containing compounds present in the cytoplasm were studied, and this led to the discovery that much of the Dap present in the wall was being recycled (40). An E. coli lys dap mutant was labeled with [3H]Dap for two generations and then chased. The pulse-chase experiment using [3H]Dap described by Goodell and Schwarz is shown in Fig. 2. During the first 2 h of chase, the amount of radioactive Dap in the sacculi actually increased as the cytoplasmic pool of Dap was utilized to synthesize PG. During this initial 2-h period, the free [3H]Dap content of the cytoplasm dropped almost 100-fold. In sharp contrast, the amount of radioactive Dap in the UDP-MurNAc-l-Ala-γ-d-Glu-meso-Dap-d-Ala-d-Ala (UDP-MurNAc-pentapeptide) pool decreased only twofold, although one would expect it to decrease more rapidly than the Dap pool because of its smaller size. UDP-MurNAc-pentapeptide, a precursor for the synthesis of PG, is present in relatively high concentrations in the cytoplasm (approximately 0.27 mM in rich medium) (79) (see Fig. 3 for a diagram of the PG biosynthetic pathway). Radioactive Dap to replenish the UDP-MurNAc-pentapeptide pool could have come from PG only, and hence, recycling of cell wall material was occurring. From the data in that experiment, Goodell calculated that about 45% of the PG was being recycled per generation (37).
FIG. 2.
Kinetics of labeling of intracellular [3H]Dap-labeled compounds during a chase. (Reprinted from reference 40 with permission.)
FIG. 3.
Cytoplasmic reactions leading to the formation of the PG monomer and transfer to the periplasmic space.
From data indicating that l-Ala-γ-d-Glu-[3H]Dap was taken up and used without degradation and that oligopeptide permease (Opp) was required for its uptake, Goodell and colleagues speculated that recycling involved the cleavage of PG to release the tripeptide into the periplasm, followed by uptake via Opp, and the direct utilization of the tripeptide in the biosynthetic pathway (37, 39). As described below, only the last of these three predictions proved correct. Eight years after the discovery of recycling, it was demonstrated that the deletion of genes for Opp permease did not prevent recycling by E. coli (92). Instead of turning over 45% of their cell wall per generation, as predicted if cells required Opp for the uptake of compounds containing Dap, it was demonstrated that several opp-negative strains turned over only 0 to 8% of their Dap per generation, depending on the strain studied (92). Hence, contrary to the original prediction (37), Opp played no detectable role in recycling. Obviously, there must be another permease for the Dap-labeled compound(s). When this result was published, S. Normark (personal communication) suggested that AmpG was a possible permease candidate. AmpG is a membrane protein required for the induction of AmpC β-lactamase in several gram-negative bacteria such as Citrobacter freundii and Enterobacter cloacae, hence the name AmpG (67, 76). It was considered to function either as a sensor or as a permease for the activating molecule. When an ampG mutant was tested for turnover of cell wall Dap, a turnover rate of 40% per generation was observed (57). Thus, AmpG was the permease required for the recycling of cell wall degradation products containing Dap. Note that E. coli does not have a β-lactamase-inducible system, but most studies of β-lactamase induction utilize plasmids in E. coli that carry the ampC and ampR genes from an inducible bacterium. Another protein has been named AmpD because the deletion or mutation of E. coli ampD led to the constitutive expression of ampC β-lactamase from a plasmid carrying the C. freundii ampR and ampC genes. AmpD was thus considered to be a negative regulator involved in β-lactamase induction (75). AmpD also proved to be required for recycling. In its absence, the turnover rate was comparable although slightly less than that in the ampG mutant (57). As demonstrated by molecular sieve chromatography, the cytoplasm of ampD cells labeled with Dap contained huge amounts of a compound or compounds with molecular weights (MWs) 600 to 1,000 that were not present in wild-type cells. The material did not accumulate in ampG cells, indicating that the material was derived from PG. An ampG ampD double mutant also contained only trace amounts of this material, which indicates that AmpG functions before AmpD.
The compound that accumulated was identified as being 1,6-anhydro-N-acetylmuramyl-l-Ala-γ-d-Glu-Dap (anhMurNAc-tripeptide) (57). This at once suggested that anhMurNAc-tripeptide was the inducer of β-lactamase in ampD cells and that AmpD served as a “negative regulator” of β-lactamase synthesis by preventing its accumulation. Since E. coli does not have an inducible β-lactamase operon, participation in PG recycling is probably the normal function of AmpG and AmpD. The relationship of recycling to β-lactamase induction will be discussed below.
Lytic Enzymes of E. coli and Their Role in PG Recycling
We shall now turn to the question of what compounds are being imported into the cytoplasm by the AmpG permease. The compounds must be derived from PG since they contain Dap. The PG degradation products released into the periplasm and, hence, available for uptake by AmpG permease were determined by the relative activities of the various lytic enzymes during the growth of E. coli. E. coli has three classes of enzymes that cleave bonds in PG. Class 1 includes endopeptidases such as penicillin-binding protein 4 (PBP4), PBP7, and MepA that cleave the cross-link between d-Ala of one glycan strand and Dap of a neighboring strand. Obviously, the endopeptidases acting alone would not release PG fragments. Class 2 includes the MurNAc-l-Ala amidases AmiA, AmiB, and AmiC, which are present in the periplasm and have been shown to participate in cell separation, that cleave the bond between MurNAc and the stem peptide of intact PG (44, 99). Another amidase, AmiD, which is present in the outer membrane, was recently identified (118). The amidases thus release murein tri-, tetra-, and pentapeptides into the periplasm from which they diffuse out of the cell, although a small fraction may enter the cytoplasm via the MppA-OppBCDF permease. Class 3 includes the lytic transglycosylases, most of which are exoenzymes, that cleave the glycan strand between carbon 1 of MurNAc and carbon 4 of GlcNAc and simultaneously form the 1,6-anhydro bond of anhMurNAc (49). The principal lytic transglycosylase responsible for the turnover of PG is SltY, a soluble lytic transglycosylase found in the periplasm. Figure 4 shows the release of GlcNAc-anhMurNAc-tetrapeptide from the end of a glycan strand by the exoenzyme SltY. In addition, there are multiple membrane-bound lytic transglycosylases (reviewed in reference 47). Some of these membrane-bound enzymes that cleave glycan strands are thought to serve specialized functions, such as providing space for a flagellum (31, 66, 102, 134). Surprisingly, mutants lacking three lytic transglycosylases (SltY, MltA, and MltB) grow normally, although the rate of turnover is reduced (69).
FIG. 4.
Formation of GlcNAc-1,6-anhydro-MurNAc-peptides during cleavage of PG by lytic transglycosylases.
In addition to the lytic enzymes, PBP4, PBP5, and PBP6 have d,d-carboxypeptidase activity that converts stem pentapeptides to stem tetrapeptides (47). Mutations in some of the PG hydrolases affect septum cleavage (45) and also cell shape (78, 98, 100, 125, 133).
Thus, the turnover of PG is initiated by lytic transglycosylases and endopeptidases to produce anhydromuropeptide (anh-muropeptide) monomers (GlcNAc-anhMurNAc-peptides). As reflected in the composition of the murein sacculus, the predominant muropeptide released by lytic transglycosylases is GlcNAc-anhMurNAc-l-Ala-γ-d-Glu-meso-Dap-d-Ala, i.e., GlcNAc-anhMurNAc-tetrapeptide, with lesser amounts of GlcNAc-anhMurNAc-tripeptide and a trace amount of GlcNAc-anhMurNAc-pentapeptide (35).
All of the lytic enzymes are active during growth and can potentially cause lysis. Hence, they must be carefully controlled both during growth and during stationary phase. For more detailed information on murein hydrolases, see a review by Shockman and Höltje (106).
AmpG: THE KEY PERMEASE INVOLVED IN RECYCLING
As noted above, in the absence of Opp permease, the rate of PG turnover remained at only a few percent per generation (92), but in the absence of AmpG, cells lost 40% of their PG Dap per generation, proving that Opp permease was not involved in PG turnover (57). However, loss from the side walls may be much greater than 40%. This is because the PG in the poles of the cell is stable; i.e., it does not turn over and thus appears to be eternal. This was originally clearly demonstrated in 1983 by Burman et al., who observed that [3H]Dap-labeled cells lost no label from the poles in three generations of chase, whereas the concentration of label in the side walls decreased greatly during this period (16). In 1997, de Pedro et al. confirmed that the polar PG was stable by using an improved technique in which d-cysteine was incorporated into PG and later tagged with gold particles so that the fate of the preformed murein could be monitored during a chase (29). Thus, since the side walls contain approximately two-thirds of the cell's PG and the stable poles contain one-third of the PG, the side walls must be turning over and recycling about 60% of their murein per generation.
As noted above, AmpG was originally identified as being a protein required for the induction of AmpC β-lactamase in Enterobacter cloacae (67). It was shown to be a membrane protein predicted to have 10 transmembrane segments (76). Strains with the ampG mutation G151D, G268D, or G373D were shown to be noninducible (76). Recently, the topology of AmpG was investigated (17) using a β-lactamase method described by Broome-Smith et al. (13). Ten membrane-spanning segments were identified, and four other hydrophobic segments remained in the cytoplasm: two of these were too short to span the membrane, and the other two contained a midsegment proline (17). Interestingly, two of the three mutations previously reported to result in a loss of function (76) are in the cytoplasmic hydrophobic domains, which suggests a role for these regions in transport (17).
It is now known that AmpG is the permease involved in the transport of anh-muropeptides (57). Nearly all of the Dap from PG that is turned over and recycled passes through the AmpG permease. The ampG gene and yajG, coded upstream of ampG, form an operon. However, it has been shown that yajG, a putative lipoprotein, was not involved in either β-lactamase induction or the expression of AmpG (76).
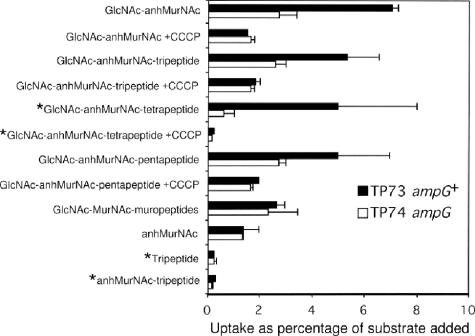
AmpG, which belongs to the major facilitator superfamily, is an interesting permease in that it transports relatively large molecules (MW 992). Because freeze-thawed E. coli cells were known to be permeable to these large-MW muropeptides, a simplified method for measuring the uptake of radioactive muropeptides using freeze-thawed cells was developed (21). Although the freeze-thawed cells exhibited high background (TP74 ampG) (Fig. 5), it was possible to demonstrate clearly that AmpG was specific for the free GlcNAc-anhMurNAc disaccharide and the disaccharide carrying stem peptides and that proton motive force was required (Fig. 5) (21). AmpG did not transport anhMurNAc-tripeptide; hence, GlcNAc-anhMurNAc-peptides, the primary products of the action of lytic transglycosylases on PG, are the compounds imported by AmpG.
FIG. 5.
Uptake of various ligands by freeze-thawed cells of wild-type and ampG strains. *, labeled with [3H]Dap. All other ligands were labeled with [3H]GlcN. (Reprinted from reference 21.)
DISCOVERY AND CHARACTERIZATION OF ENZYMES INVOLVED IN RECYCLING PG AMINO ACIDS
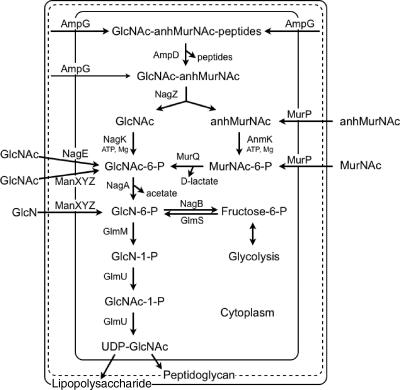
In order to study the fate of the PG amino acids, PGs labeled with [3H]Dap and various [3H]Dap-labeled substrates were prepared from the labeled murein or from extracts of mutant cells known to accumulate a given substrate of interest (21, 58, 118, 120). All the strains used in these studies lacked LysA to prevent the conversion of Dap to l-Lys. The following seven enzymes appear to have evolved to allow cells to recycle the amino acids from their murein (Fig. 6).
FIG. 6.
Pathway for recycling murein tripeptide and amino acids.
AmpD: Anhydro-N-Acetylmuramyl-l-Ala Amidase
As noted above, an E. coli strain lacking AmpD contained large amounts of anhMurNAc-tripeptide in its cytoplasm. This suggested that anhMurNAc-tripeptide might be a substrate for AmpD. When AmpD was cloned and a highly purified sample was obtained, it was shown to cleave the anhMurNAc-l-Ala bond. Interestingly, it proved to be highly specific for the anhMurNAc-l-Ala bond (58). The rate of cleavage of MurNAc-peptides and UDP-MurNAc-pentapeptide was at least 10,000 times slower than that of anhMurNAc-tripeptide (58). Thus, AmpD functioning in the cytoplasm rapidly cleaves the anhMurNAc-peptides without destroying the UDP-MurNAc-pentapeptide needed for the synthesis of PG. Höltje et al. also demonstrated that AmpD was specific for the anhMurNAc-l-Ala bond by showing that it cleaved the anhMurNAc-l-Ala bond from the anhMurNAc-peptides at the ends of PG strands but not from the MurNAc-l-Ala bonds in the remainder of the glycan strands (48).
The three-dimensional structure of AmpD from Citrobacter freundii has been solved by nuclear magnetic resonance (72). It contained a zinc ion that was shown to be essential for activity (33, 72). Replacement of each of the presumed zinc triad amino acids (His34, His154, and Asp164) with Ala resulted in a lack of a zinc ion in the active site and loss of amidase activity (33). Tyr63 and Lys162 were proposed to be involved in the binding of the substrate by site-directed mutagenesis (33).
Mpl: UDP-N-Acetylmuramate:l-Alanyl-γ-d-Glutamyl- meso-Diaminopimelate Ligase
In Goodell's study of PG recycling, it was demonstrated that the murein tripeptide l-Ala-γ-d-Glu-meso-Dap was reused directly without first being degraded to Dap (37). When l-Ala-γ-d-Glu-[3H]Dap was added to growing E. coli cells, radioactive UDP-MurNAc-pentapeptide and PG were formed even in the presence of a huge excess of nonradioactive Dap (37). One candidate for an enzyme to add murein tripeptide to UDP-MurNAc was MurC, the ligase that adds l-Ala to UDP-MurNAc (Fig. 3). However, purified MurC was shown to totally lack activity for the addition of the tripeptide to UDP-MurNAc (73).
A search of the E. coli chromosome database revealed an open reading frame, yjfG, or o457, with 25.8% amino acid sequence identity to MurC (15). Since paralogs often have similar functions, it was speculated that yjfG was the gene encoding UDP-MurNAc:l-Ala-γ-d-Glu-meso-Dap ligase (83). This proved to be the case, and yjfG was renamed mpl for murein peptide ligase (83). Strains overexpressing the yjfG gene product overproduced Mpl activity, and strains in which the mpl gene was disrupted by the insertion of the 1.28-kb kanamycin resistance cartridge lacked Mpl activity (83). One of the mpl::Km strains was studied further and was shown to have normal morphology and PG content and had a 12-fold-higher pool of murein tripeptide in its cytoplasm than the wild type. This was in contrast to an ampD mutant, which accumulated well over a 100-fold-higher pool of its substrate in the cytoplasm. Interestingly, the cytoplasm was also found to contain an increased level of l-Ala-d-Glu, indicating the presence of an enzyme capable of cleaving Dap from the tripeptide.
The properties and specificity of the Mpl ligase have been studied recently. Mpl was shown to utilize tripeptide, tetrapeptide, and pentapeptide with similar efficiencies (46). Mpl also accepts peptides containing lysine in place of Dap (5). During entry into stationary phase, the expression level of Mpl increases under the control of sigma S (114).
LdcA: ld-Carboxypeptidase
The principal compound to accumulate in an E. coli strain lacking ampD was anhMurNAc-l-Ala-γ-d-Glu-Dap (57). It lacked the GlcNAc and d-Ala normally present in the principal anh-muropeptide imported by AmpG. This indicated that a β-N-acetylglucosaminidase that removes GlcNAc and a peptidase that removes d-Ala are likely to be present in the cytoplasm. Indeed, a peptidase named LdcA was found to be present in the cytoplasm (115). The crystal structure of LdcA from Pseudomonas aeruginosa shows that LdcA is a serine protease with a Ser-His-Glu catalytic triad (68). LdcA was shown to cleave d-Ala from l-Ala-γ-d-Glu-meso-Dap-d-Ala (tetrapeptide), disaccharide-tetrapeptide, anh-disaccharide-tetrapeptide, MurNAc-tetrapeptide, and UDP-MurNAc-tetrapeptide at similar rates, although free tetrapeptide was cleaved most efficiently (115). Surprisingly, the cross-linked dimers bis-disaccharide-tetrapeptide and bis-anhydrodisaccharide-tetrapeptide as well as murein sacculi were not substrates. It was shown previously that UDP-MurNAc-pentapeptide was not a substrate (124).
Among the 11 genes involved in recycling, a mutation in the ldcA gene is the only known mutation that produces a phenotype. The ldcA mutant forms thick, oval cells in late log phase, and many lyse (115). Baum et al. identified a dithiazoline inhibitor of LdcA and showed that the drug causes cell lysis in stationary phase, which is similar to the phenotype of an ldcA mutant (6). Those authors' explanation for this phenotype is that in the absence of LdcA, tetrapeptide accumulates and is used by Mpl to form UDP-MurNAc-tetrapeptide (115). In fact, as noted above, the tetrapeptide is a good substrate for Mpl (46). The MurNAc-tetrapeptide is incorporated into the murein sacculus instead of MurNAc-pentapeptide. Hence, a shortage of pentapeptide, which is required for cross-linking the glycan chains, leads to a deficiency of cross-links and a weakened wall, resulting in bulging and lysis. The fact that the phenotype develops during the onset of stationary phase suggests that recycling may be of critical importance during this period.
MpaA: γ-d-Glutamyl-meso-Diaminopimelate Amidase
There were two clues that a γ-d-Glu-Dap amidase may be present in the cytoplasm of E. coli: (i) Dap-d-Ala was identified in the cytoplasm by Goodell and Schwarz (40), and (ii) l-Ala-d-Glu was identified in the cytoplasm of an mpl mutant (83). These dipeptides could be derived from murein tetrapeptide, and l-Ala-d-Glu could also be produced from murein tripeptide, by cleavage of the bond between d-Glu and Dap.
Bacillus sphaericus contains a sporulation-related γ-d-Glu-Dap amidase named ENP1 (2, 32). The catalytic domain of ENP1 contains a zinc binding site, suggesting that a zinc ion is essential for its activity (51). In a search of the E. coli protein database, regions of the YcjI amino acid sequence displayed significant homology with the catalytic domain and putative zinc-binding triad of ENP1, although the overall identity between the two sequences was only 13% (120). In particular, the putative zinc-binding triad His162-Glu165-His307 of ENP1 corresponds to His69-Glu72-His177 of YcjI in the alignment of the two amino acid sequences (120). YcjI proved to be the gene for an enzyme that cleaved the γ-d-Glu-Dap bond and was named mpaA for murein peptide amidase A. An mpl mpaA (ycjI::Cm) double mutant was constructed and was shown to contain over 10 times as much tripeptide as the mpl mutant and over 100 times as much tripeptide as the wild-type strain in its cytoplasm (120). This demonstrated that the enzyme was active in vivo, as expected. It was difficult to detect activity in vitro. When cloned and overproduced, MpaA formed huge inclusion bodies in the cytoplasm, and purification of MpaA was unsuccessful. However, extracts from cells expressing mpaA from a plasmid were shown to cleave the tripeptide with the release of Dap. More information on the substrate specificity of MpaA and on a possible requirement for zinc awaits overproduction and purification of the enzyme.
YcjG: l-Alanyl-d/l-Glutamate Epimerase
Schmidt et al. showed that YcjG, divergently coded downstream of mpaA, is l-Ala-d/l-Glu epimerase, which converts l-Ala-d-Glu to l-Ala-l-Glu (105). l-Ala-l-Glu is a substrate for PepD, a dipeptidase of E. coli with a broad specificity (107). The epimerase belongs to the enolase superfamily (42), and the reaction mechanism of the enzyme has been studied (42, 64).
Thus, E. coli can release all of the amino acids from the tripeptide. However, this is a minor pathway because most of the tripeptide is utilized by Mpl to form the PG biosynthesis intermediate UDP-MurNAc-tripeptide.
AmiD: an Outer Membrane Anhydro-N-Acetylmuramyl- l-Ala Amidase
A nagZ ampD mutant accumulated GlcNAc-anhMurNAc-tripeptide in its cytoplasm, as expected (20). However, a significant amount of the disaccharide GlcNAc-anhMurNAc was also present (20). This suggested that in addition to AmpD, E. coli may have a second enzyme with anhMurNAc-l-Ala amidase activity. A search of the genome revealed a paralog, YbjR, with 40% identity to amino acids 65 to 171 of AmpD. The paralog was named AmiD to distinguish it from AmiA, AmiB, and AmiC, the three MurNAc-l-Ala amidases found in the periplasm. Deletion of amiD from the nagZ ampD double mutant produced a strain that totally lacked anhMurNAc-l-Ala amidase activity, demonstrating that AmiD and AmpD were the only enzymes in E. coli able to cleave the anhMurNAc-l-Ala bond (118).
Substrate specificity of AmiD.
AmiD was shown to cleave the anhMurNAc-l-Ala bond in GlcNAc-anhMurNAc-tripeptide much more rapidly than that in anhMurNAc-tripeptide. Surprisingly, muropeptides lacking the anhydro ring were good substrates, and in fact, PG itself was cleaved by the enzyme. Thus, AmiD has a relatively broad specificity. AmiD, like AmpD (33), was shown to require zinc for activity (118).
AmiD is a lipoprotein.
The amiD gene product has a short signal sequence and a lipobox motif (L14AGC), followed by Ala, which suggested that AmiD is a lipoprotein destined for the outer membrane (62, 117). Indeed, fractionation of membranes of ampD cells in which AmiD was the only anhMurNAc-l-Ala amidase present demonstrated that the AmiD amidase activity was in the outer membrane (118).
Why GlcNAc-anhMurNAc is present in the cytoplasm of the amiD ampD double mutant even though the strain totally lacks anhMurNAc-l-Ala amidase activity.
The amiD ampD nagZ mutant still accumulated disaccharide but only one-fourth as much GlcNAc-anhMurNAc as the ampD nagZ mutant. This indicated that 75% of the GlcNAc-anhMurNAc present in the cytoplasm of the triple mutant was produced by AmiD from anh-muropeptides in the periplasm, followed by the uptake of the disaccharide by AmpG. This demonstrates that significant AmiD activity occurs in growing cells. The activity was somewhat surprising in view of the fact that the amiD gene has tandem rare arginine codons, AGA_AGA, as its second and third codons, suggesting that relatively few AmiD molecules are produced. The remaining 25% of the GlcNAc-anhMurNAc present in the amiD ampD nagZ mutant must have been produced by the action of periplasmic amidases, AmiA, AmiB, and/or AmiC, that cleave the stem peptides from PG, followed by digestion of PG by lytic transglycosylases to release GlcNAc-anhMurNAc and uptake of the disaccharide by AmpG.
Interestingly, an amiD ampG mutant, which one would expect to accumulate GlcNAc-anhMurNAc-peptides in the periplasm, because their MW exceeds 850 and diffusion through the outer membrane would therefore be expected to be slow (85), actually released these large-MW compounds into the medium (118). Not incidentally, the main compound released into the medium, GlcNAc-anhMurNAc-tetrapeptide, is identical to tracheal cytotoxin (TCT) of Bordetella pertussis (101).
MppA, a Periplasmic Murein Peptide-Binding Protein, and OppBCDF and Their Role in Uptake of Murein Tripeptide
When the probable existence of a UDP-MurNAc:l-Ala-γ-d-Glu-meso-Dap ligase was first predicted (37) (see Fig. 3 for its role in PG synthesis) and the role of AmpG in recycling the tripeptide became known (57), efforts to identify the ligase gene were undertaken. Based on Goodell's observation that a Dap-requiring strain of E. coli could utilize l-Ala-γ-d-Glu-meso-Dap for PG synthesis and that the intact tripeptide was apparently used directly (37), mutants that required murein tripeptide as a source of Dap for growth of a Dap auxotroph were sought. E. coli TP981 (opp+ ampG::Km lysA+ dapD2) was mutagenized by transposon mutagenesis (94). Mutants of the Dap-requiring strain that did not form colonies on plates containing low levels of the murein tripeptide l-Ala-γ-d-Glu-Dap as the sole source of Dap were selected by the replica plating technique. Four mutants were obtained from about 27,000 colonies replicated (94). Two of the mutants carried the transposon in a previously unidentified gene whose product was about 46% identical to the OppA amino acid sequence. This gene product has been named MppA, for murein peptide permease, because of its functional and structural similarity to OppA, the periplasmic binding protein required for the uptake of oligopeptides. MppA was shown to be a periplasmic binding protein required for the uptake of the murein tripeptide (94). However, OppB, OppC, OppD, and OppF, components of an ATP-dependent oligopeptide transporter (97), were also required, suggesting that MppA transported the tripeptide to the membrane components of Opp for transport into the cytoplasm. In support of this notion, one of the four mutants obtained was found to be in oppB (94). Curiously, the fourth mutant obtained in this screen proved to be in the promoter region of groESL, suggesting that groESL was required to produce sufficient MppA-OppBCDF permease and/or the hypothetical UDP-MurNAc:l-Ala-γ-d-Glu-Dap ligase to allow growth on plates with limited murein tripeptide as the source of Dap.
MppA was found to be a negative regulator of multiple-antibiotic resistance in E. coli strain TP980 (71). An insertional mutation in mppA produced a typical multidrug resistance (MDR) phenotype including the overproduction of the transcriptional activator MarA and of the membrane-bound AcrAB proteins that function as a drug efflux pump. Surprisingly, this phenotype was shown to be restricted to a single strain of E. coli. When the mutation was transduced into another strain, the MDR phenotype disappeared (10). Nevertheless, it is of interest that when the original mutagenized culture was used to directly select multidrug-resistant mutants, strains with mutations in mppA and marA were obtained, as anticipated, showing a relationship of these genes to MDR. In addition, strains with mutations in trkA, gshA, sapC, sapD, sapF, and ompR were isolated (H. Li, personal communication). It was suggested previously that these genes may directly or indirectly affect K+ transport (96). By inference, the K+ concentration may affect AcrAB efflux expression. The role of the sap operon (sapABCDF) in E. coli remains unknown. The sap operon (sensitivity to antimicrobial peptides) in Salmonella enterica serovar Typhimurium was discovered when sap mutants were found to be more resistant to protamine (41).
Recently, it was reported that MppA could be used to transport heme into the cell via dipeptide permease (70). A recombinant E. coli strain expressing a foreign heme outer membrane receptor was able to use exogenous heme as an iron source. The uptake of heme required the dipeptide inner membrane transporter (DppBCDF) and either MppA or DppA, the dipeptide binding protein (70).
Pathways for Utilization of PG Amino Acids
Figure 6 illustrates the pathways for the utilization of cell wall amino acids. The peptides are released from anh-muropeptides by AmpD amidase. LdcA carboxypeptidase cleaves d-Ala from the tetrapeptide. The tripeptide is efficiently used by Mpl ligase to form UDP-MurNAc-tripeptide, which is a normal intermediate in the pathway for PG synthesis (Fig. 3). It is surprising that the cell also maintains a second pathway utilizing MpaA amidase and l-Ala-d/l-Glu epimerase (YcjG) in its arsenal, which together allow the cell to recover all the individual amino acids from the muropeptides.
DISCOVERY AND CHARACTERIZATION OF ENZYMES INVOLVED IN RECYCLING PG AMINO SUGARS
Since E. coli can recycle amino acids from the cell wall, it was natural to consider whether the cells can also utilize the amino sugars. In order to study the fate of PG amino sugars, the PG amino sugars were specifically labeled with [3H]GlcN. This was possible by using strains that carried the nagB mutation, which prevented the conversion of GlcN-6-phosphate (GlcN-6-P) to fructose-6-P (Fig. 7). Thus, essentially all of the label was restricted to GlcNAc and MurNAc of PG and the GlcNAc of lipopolysaccharide. As reported below, it was found that both free GlcNAc and anhMurNAc derived from PG were converted to GlcNAc-6-P and were thus available for reuse. The following five enzymes were found to be involved in recycling of the PG amino sugars.
FIG. 7.
Pathway for recycling PG amino sugars.
NagA: N-Acetylglucosamine-6-P Deacetylase
NagA, an N-acetylglucosamine-6-P deacetylase (129), is an essential enzyme for the metabolism of amino sugars and for recycling GlcNAc and anhMurNAc. As indicated in Fig. 7, conversion of GlcNAc-6-P to UDP-GlcNAc requires that it first be deacetylated by NagA and then converted to GlcN-1-P by GlmM (80), followed by reacetylation and reaction with UTP by GlmU to form UDP-GlcNAc (81, 82). Alternatively, GlcN-6-P can be converted to fructose-6-P by NagB (129). As indicated, the reverse reaction to synthesize Glc-N-6-P is catalyzed by GlmS (reviewed in reference 116).
NagZ: β-N-Acetylglucosaminidase
Since AmpG permease transported only compounds containing GlcNAc-anhMurNAc, whereas anhMurNAc-tripeptide rather than GlcNAc-anhMurNAc-tripeptide accumulated in ampD cells, it was apparent that GlcNAc must be cleaved from the incoming disaccharide-peptides. A β-N-acetylglucosaminidase from E. coli was characterized by Yem and Wu in 1976 (132). It was shown to be present in the cytoplasm and to cleave p-nitrophenyl-β-N-acetyl-d-glucosaminide and muropeptides obtained from PG by digestion with lysozyme. Those authors obtained mutants deficient in enzyme activity but did not map the mutations (131). Hrebenda also reported the isolation of a mutant deficient in enzyme activity and found that the mutation cotransduced with the tryptophan operon at a low frequency (52). As these mutants were no longer available, Cheng et al. isolated a mutant deficient in β-N-acetylglucosaminidase from E. coli cells treated with nitrosoguanidine (20). Individual colonies were screened for the ability to hydrolyze p-nitrophenyl-β-N-acetyl-d-glucosaminide, and one with reduced activity, TP75, was chosen for further studies. The mutated gene mapped 3.2 min counterclockwise from the tryptophan operon and contained a point mutation. This identified an open reading frame, ycfO, which was renamed nagZ. A deletion mutant of nagZ totally lacked β-N-acetylglucosaminidase activity and contained large amounts of the disaccharide GlcNAc-anhMurNAc in its cytoplasm (20). Thus, NagZ is the only β-N-acetylglucosaminidase expressed in E. coli. As one would expect, a nagZ ampD double mutant accumulated GlcNAc-anhMurNAc-tripeptide in its cytoplasm (20). The enzyme was active against muropeptides as well as anh-muropeptides. The substrate specificity of NagZ was also studied by Vötsch and Templin (128). NagZ was inhibited by GlcNAc, the product of the reaction, and, interestingly, by bulgecin, which is known to inhibit the soluble lytic transglycosylase SltY (128). The NagZ enzyme has a substrate specificity similar to that of ExoII (22), an unusual β-N-acetylglucosaminidase from Vibrio furnissii whose amino acid sequence is 57% identical to that of NagZ.
It was reported previously that a strain lacking NagZ can be induced to synthesize only one-fourth as much β-lactamase as normal from the ampC-ampR system of E. cloacae (128). Small-molecule inhibitors of NagZ from Vibrio cholerae were synthesized and were shown to form a complex with NagZ by crystallographic methods (112). One of these compounds decreased β-lactamase induction by fourfold (112), which is consistent with the result obtained with a nagZ mutant (128).
NagK: N-Acetylglucosamine Kinase
The cleavage of anh-muropeptides by the cytoplasmic β-N-acetylglucosaminidase NagZ leads to the release of GlcNAc in the cytoplasm. The free sugar was shown to be recycled into PG and lipopolysaccharide, but exactly how the cell was able to utilize the free GlcNAc was not determined (90). Recycling of this GlcNAc would be readily achieved if it could be converted to the phosphorylated form (Fig. 7). A kinase that phosphorylated GlcNAc was purified from E. coli, and its properties were studied in 1966 (3). Building on this purification procedure, a GlcNAc kinase from E. coli was purified, and the gene was identified by determining the N-terminal sequence of the purified enzyme (121). The gene, ycfX, was named nagK for N-acetylglucosamine kinase. A null mutant exhibited very low kinase activity. The nagK gene was cloned, and the NagK protein was overexpressed and purified. The kinase was shown to be highly specific for GlcNAc, although at extremely high concentrations, glucose was also phosphorylated (121).
A nagEBACD mutant fed [3H]GlcN was shown to lack radioactive UDP-GlcNAc in its cytoplasm and accumulated large amounts of radioactive GlcNAc-P and GlcNAc because of the absence of NagA deacetylase (90). In contrast, a nagK derivative of the nagEBACD mutant contained just the opposite, i.e., large amounts of UDP-[3H]GlcNAc but relatively little [3H] GlcNAc and no [3H]GlcNAc-P (121). This suggested the intriguing possibility that a pathway that is able to utilize GlcNAc without first phosphorylating it had been activated (121). This hypothetical pathway remains to be investigated.
As judged by its amino acid sequence (which includes the conserved domain of the ROK family), NagK is a member of the ROK family, which includes repressors, open reading frames, and kinases. A crystal structure of Mlc, which is a transcriptional regulator belonging to the ROK family, revealed that the conserved domain of the ROK family coordinates a zinc ion, which is essential for its function (103). However, whether zinc is required for NagK activity remains unknown.
AnmK: Anhydro-N-Acetylmuramic Acid Kinase
As noted above, preventing the recycling of PG amino sugars by the inactivation of nagZ caused an accumulation of GlcNAc-anhMurNAc. Likewise, the deletion of nagEBACD resulted in high levels of GlcNAc and GlcNAc-P in the cytoplasm, since NagA is required for the utilization of GlcNAc-P. Surprisingly, the nag operon deletion strain completely lacked anhMurNAc. This result strongly suggested that anhMurNAc was metabolized, since during PG recycling, equal amounts of each amino sugar are released into the cytoplasm, and anhMurNAc was not found in the cytoplasm or in the spent medium.
Incubation of [3H]anhMurNAc with soluble cell extract from 1 ml of an E. coli culture converted only a trace amount to a compound or compounds that were separable from anhMurNAc by high-performance liquid chromatography or thin-layer chromatography (90). This was surprising, since in the case of the other enzymes involved in recycling, this amount of cell extract was sufficient to convert essentially all of the substrate to product in less than 2 h. However, when ATP and Mg2+ were included in the incubation mixture, the reaction proceeded rapidly. With thin-layer chromatography as a guide, the enzymes(s) was partially purified and used to digest anhMurNAc. Analysis of the digest mixture by mass spectrometry unexpectedly identified both GlcNAc-P and MurNAc-P as products (123). Further purification on a MonoQ column separated an activity that converted anhMurNAc to MurNAc-P from an activity converting MurNAc-P to GlcNAc-P (123). Analysis of a tryptic digest of the fraction containing the partially purified anhMurNAc kinase by mass spectrometry identified three known proteins and one unknown open reading frame, ydhH, which proved to be the gene for anhMurNAc kinase, renamed AnmK (123). Thus, the first step in the conversion of anhMurNAc to a metabolizable form was phosphorylation. The reaction is unusual in that it involves the cleavage of the 1,6-anhydro bond in addition to phosphorylation. A comparable reaction converts 1,6-anhydroglucose (levoglucosan) to glucose-6-P (135). An anmK mutant did not accumulate anhMurNAc in its cytoplasm. This is in contrast to other enzymes in the recycling pathway in which the loss of an enzyme resulted in the accumulation of the substrate for said enzyme in the cytoplasm. In the case of AnmK, the substrate anhMurNAc was found in the spent medium, suggesting that an efflux pump for anhMurNAc is present in E. coli that lacks AnmK (123).
MurQ: N-Acetylmuramic Acid-6-P Etherase
In 1983, Parquet et al. (95) reported the surprising finding that E. coli cells could grow on MurNAc as the sole source of carbon and energy. Dahl et al. (26) recently demonstrated that a special phosphotransferase system, MurP (YfeV), was required for growth on MurNAc and that MurNAc was phosphorylated during uptake. It was further demonstrated that cells lacking GlcNAc-6-P deacetylase (ΔnagA) or glucosamine-6-P deaminase (ΔnagB) could not grow on MurNAc. This suggested that GlcNAc-6-P was an intermediate involved in the utilization of MurNAc and that MurNAc-6-P was converted to GlcNAc-6-P. This was followed by the identification of a gene, murQ (yfeU), for the hypothetical “etherase” (59). Jaeger et al. showed that murQ was required for growth on MurNAc; that in the absence of murQ, cells fed MurNAc accumulated MurNAc-P; and that purified MurQ converted MurNAc-6-P to GlcNAc-6-P with the release of d-lactate (59). Interestingly, murQ proved to be in an operon together with murP and a gene for PBP4B (126). MurQ was subsequently shown to be the only MurNAc-P etherase in E. coli, and it was shown that both MurQ and AnmK were required for the utilization of anhMurNAc derived from PG (122). Data indicating that anhMurNAc was taken up by cells provided that MurP was present and that AnmK was required for phosphorylation were also presented (122). Thus, the pathway for recycling anhMurNAc and the pathway that allows the growth of E. coli on MurNAc (and possibly anhMurNAc) merge at MurNAc-6-P. Growth on anhMurNAc was not adequately tested due to the lack of sufficient substrate, although in a preliminary experiment, E. coli cells failed to grow on anhMurNAc as a sole carbon source.
Pathway for Utilization of PG Amino Sugars
As shown in Fig. 7, the amino sugars from PG enter the cell via AmpG permease in a bound form as GlcNAc-anhMurNAc-peptides. The peptides are removed by anhMurNAc-l-Ala-amidase (AmpD), and the disaccharide is cleaved by β-N-acetylglucosaminidase (NagZ), thus freeing GlcNAc and anhMurNAc. NagK, a highly specific GlcNAc kinase, converts the free GlcNAc derived from PG to the phosphorylated form. GlcNAc is normally phosphorylated upon uptake by a phosphotransferase system, and GlcNAc-P is readily metabolized (Fig. 7). anhMurNAc is also phosphorylated by a specific kinase (AnmK) and then converted to GlcNAc-P by MurNAc-P etherase (MurQ). Hence, both amino sugars are converted to GlcNAc-P and can enter the normal metabolic pathways for the synthesis of UDP-GlcNAc and for glycolysis.
OCCURRENCE OF ORTHOLOGS OF PG RECYCLING ENZYMES IN OTHER GENERA
Table 2 lists orthologs of PG recycling enzymes present in selected bacteria. A more extensive list can be found in Table S1 in the supplemental material. With the reservation that the presence of an ortholog does not guarantee the expected activity, one can see that many familiar bacteria have the four genes (ampG, ampD, ldcA, and mpl) involved in the recycling of the murein tripeptide. Several bacteria lack LdcA, which may not be needed if the tetrapeptide content of their murein is low or if their murein peptide ligase does not use the tetrapeptide efficiently. It is interesting that many bacteria may possess the outer membrane anhMurNAc-l-Ala amidase (AmiD) even though it is not directly involved in recycling. The degradation of the tripeptide by MpaA and YcjG appears to be employed by fewer organisms.
TABLE 2.
Orthologs of proteins involved in PB recyclinga
| Organism | Presence of ortholog of:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AmpG | AmpD | Mpl | LdcA | AmiD | NagZ | NagK | AnmK | MurQ | MpaA | YcjG | |
| Escherichia coli K-12 | + | + | + | + | +* | + | + | + | + | + | + |
| Escherichia coli CFT073 | + | + | + | + | +* | + | + | + | + | + | |
| Salmonella enterica | + | + | + | + | +* | + | + | + | + | + | + |
| Yersinia pestis | + | + | + | +* | + | + | + | + | + | ||
| Shigella flexneri | + | + | + | + | +* | + | + | + | + | + | + |
| Haemophilus influenzae | + | + | + | + | + | + | + | ||||
| Pseudomonas aeruginosa | ++ | ++ | + | + | +* | + | + | + | + | ||
| Bordetella pertussis | + | + | + | + | +* | + | + | + | |||
| Neisseria gonorrhoeae | + | + | + | + | + | + | |||||
| Vibrio cholerae | + | + | + | + | + | + | + | ++ | + | ||
| Legionella pneumophila | + | +* | + | +* | + | + | |||||
| Caulobacter crescentus | + | +* | + | + | + | + | |||||
| Helicobacter pylori | |||||||||||
| Borrelia burgdorferi | |||||||||||
| Bacteroides fragilis | + | + | + | + | + | + | |||||
| Bacillus subtilis | + | +* | + | + | + | ||||||
This table lists selected organisms listed in Table S1 in the supplemental material. Organisms with two orthologs of a gene are indicated as ++. * indicates orthologs containing a putative signal sequence.
With regard to the proteins involved in the utilization of PG amino sugars (AmpG, NagZ, NagK, AnmK, and MurQ), it appears that many of the listed bacteria may have all five proteins. Remarkably, about one-half of the bacteria listed, as well as Xanthomonas, Shewanella, Legionella, and Ralstonia (see Table S1 in the supplemental material), have an anmK ortholog but lack a murQ ortholog. The product of anmK, MurNAc-P, presumably cannot be utilized unless converted to GlcNAc-P by MurQ. This suggests that a structurally unrelated protein serves the MurQ etherase function or that AnmK serves a different function.
Helicobacter pylori and Borrelia burgdorferi lack AmpG and enzymes to cleave PG degradation products. Hence, these compounds, when cleaved from PG by lytic transglycosylases, must be released into their surroundings or injected into the cytoplasm of the host (unless their SltY ortholog is inactive). It has been demonstrated that H. pylori injects Dap-containing material into the host cytoplasm via a bacterial type IV secretion system (127).
It is surprising that Bordetella pertussis and Neisseria gonorrhoeae possess orthologs of AmpG and most of the recycling enzymes because Bordetella pertussis secretes huge amounts of the intact TCT (anh-disaccharide-tetrapeptide) (24) and Neisseria gonorrhoeae also releases TCT (109). This suggests that AmpG proteins of B. pertussis and N. gonorrhoeae are either not expressed or not active. A strain of B. pertussis which overexpresses AmpG and lacks a gene encoding dermonecrotic toxin is attenuated (84), suggesting that TCT, which is normally released into the medium, is taken up by AmpG permease.
Remarkably, mammals have a membrane protein with significant homology to AmpG (see Table S1 in the supplemental material). If active, the protein may play a role in the importation of PG fragments into the cytoplasm, allowing their detection by Nod1 or Nod2.
From the possession of orthologs, we can speculate further how certain other bacteria may recycle their PG degradation products. Legionella pneumophila possesses an ortholog of AmpD. However, AmpD of L. pneumophila appears to have a signal sequence that would export the protein into the periplasm. Thus, GlcNAc-anhMurNAc-peptides may be cleaved in the periplasm to release GlcNAc-anhMurNAc and peptides. GlcNAc-anhMurNAc would be imported by AmpG into the cytoplasm, which contains orthologs of the enzymes needed for the degradation and reutilization of amino sugars. Free peptides could be taken up by the MppA-OppBCDF system and reused by Mpl.
In E. coli, murQ and murP are in the same operon. Vibrio cholerae is unique in that it possesses two MurQ orthologs, both of which are in chromosome 1. One MurQ is in an operon with murP, and the other MurQ is coded just downstream of anmK. This seems to indicate the horizontal transfer of one of these operons. Interestingly, NagZ of V. cholerae is coded divergently to anmK.
Pseudomonas aeruginosa has three ampD orthologs, one of which is amiD and all of which are involved in β-lactamase induction (61). It may also have two ampG genes.
Although the gram-positive bacterium Bacillus subtilis lacks AmpG permease, it has orthologs of NagZ (YbbD), MurQ (YbbI), MurP (YbbF), LdcA (YkfA), MpaA (YqgT), and YcjG (YkfB). B. subtilis cleaves PG primarily by a muramidase and an amidase to produce GlcNAc-MurNAc and peptides. GlcNAc-MurNAc could be cleaved by NagZ, which seems to be exported by a putative signal sequence of the protein. GlcNAc is imported into the cytoplasm by a specific phosphotransferase system, NagP (YflF), and metabolized by NagA and NagB. MurNAc is phosphorylated and imported into the cytoplasm by MurP to produce MurNAc-6-P, which is converted into GlcNAc-6-P by MurQ. Interestingly, NagZ, MurQ, and MurP are encoded in a putative operon that also includes a gene coding for a putative β-lactamase. Free peptides are imported by an unknown permease and degraded by YkfA, a putative γ-d-Glu-Dap amidase (YkfC), and YcjG epimerase (YkfB). YkfA, YkfB, and YkfC are encoded downstream of the dpp operon, which is regulated by the transcriptional regulator CodY and is expressed in the early stationary phase (108, 110). Thus, B. subtilis may have the potential to utilize PG degradation products when carbon and energy sources become scarce.
RELATED TOPICS
Some conclusions from early work die hard. For instance, although it has been known for many years that the Opp permease plays no detectable role in recycling PG amino acids (92) and that the uptake of murein tripeptide via Opp cannot occur unless MppA is present (94), both of these notions persist. The same might be said for the hypothesis that the cell elongates the sacculus by inserting three glycan strands at a time to replace one existing strand. This idea was a good attempt to explain cell wall turnover, but the concept is incompatible with the fact that single strands are inserted one at a time during elongation (25, 27, 89).
β-Lactamase Revisited: Relation of Recycling Intermediates to β-Lactamase Induction
In 1994, anhMurNAc-tripeptide accumulation in an ampD mutant of E. coli was shown to correlate with the constitutive expression of C. freundii ampC β-lactamase carried on a plasmid (57). This was followed by the demonstration that AmpR was an activator of ampC expression and that the concentration of UDP-MurNAc-pentapeptide normally present in E. coli acted as a corepressor to repress ampC expression (56). The amount of anhMurNAc-tripeptide present in the ampD mutant was sufficient to restore expression (56). This explained the constitutive expression of the C. freundii ampC β-lactamase in the ampD mutant but did not explain the induction of ampC β-lactamase expression caused by the addition of cefoxitin or imipenem to growing E. coli cells. As reported in the original publication, anhMurNAc-tripeptide did not accumulate under these conditions, although a significant amount of a smaller-MW compound did accumulate (57). It was recently shown that the compound that accumulates in this fraction is the pentapeptide l-Ala-γ-d-Glu-Dap-d-Ala-d-Ala and that a smaller amount of anhMurNAc-pentapeptide also accumulates (T. Uehara, K. Suefuji, and J. T. Park, unpublished data). Dietz et al. (30) showed that cefoxitin and imipenem, which are good inducers of β-lactamase, caused cells to accumulate anh-disaccharide-pentapeptide in their periplasm, whereas four other β-lactam antibiotics that caused minimal induction failed to accumulate anh-disaccharide-pentapeptide. Those authors concluded that anhMurNAc-pentapeptide is the active inducer, although the above-described result suggests that free pentapeptide may be a more likely candidate. In vitro studies of ampC mRNA synthesis are required to identify the true inducer.
However, other observations suggest that UDP-MurNAc-pentapeptide may not be the corepressor of AmpR. For instance, both a temperature-sensitive murG mutant at the restrictive temperature (MurG is required for lipid 2 synthesis) (Fig. 3) and envA cells carrying C. freundii ampR ampC and treated with ramoplanin (which binds to lipid 2) have been shown to express β-lactamase (113). envA cells have a leaky outer membrane, allowing the entry of ramoplanin and other antibiotics (87). In addition to ramoplanin, other PG synthesis inhibitors including vancomycin, moenomycin, fosfomycin, and cycloserine also induce β-lactamase but to a lesser extent (113). All these conditions may result in the reduced availability of lipid 2. It has been shown that a crude preparation of lipid 2 from 1 ml of E. coli culture is sufficient to block AmpR-driven ampC β-lactamase mRNA synthesis in a cell-free system (K. Suefuji, T. Uehara, and J. T. Park, unpublished data). We are left with several observations that need to be explained before the induction of β-lactamase by β-lactam antibiotics can be fully understood: (i) the fact that AmpG is required for β-lactamase induction by cefoxitin and imipenem implicates PG recycling intermediates other than anhMurNAc-tripeptide in the process; (ii) the fact that cefoxitin and imipenem, which are good inducers, are the only β-lactams tested that caused the accumulation of anhMurNAc-pentapeptide implicates this compound in the induction process (30); (iii) the fact that pentapeptide is the principal compound to accumulate in cells treated with cefoxitin implicates pentapeptide as being the more likely inducer than anhMurNAc-pentapeptide; and (iv) reports that both ramoplanin treatment and a murG(Ts) mutant at the restricted temperature cause β-lactamase induction seems to implicate lipid 2 as a corepressor. Unfortunately, a clear connection between lipid 2, recycling intermediates containing pentapeptide, and UDP-MurNAc-pentapeptide remains to be established.
More questions about β-lactamase induction concerning the possible role of AmpE, a membrane protein coded just downstream of ampD, have arisen. AmpE is involved in the regulation of β-lactamase expression (50, 75), but the mechanism is unknown. Recently, AmpE in Pseudomonas aeruginosa was shown to play an indirect role in resistance to β-lactams (60). While an ampE mutation did not change the amounts of PG recycling intermediates in the cytoplasm of E. coli, treatment of the ampE mutant with cefoxitin increased the amount of free pentapeptide present in the cytoplasm over 100-fold compared to that of the wild type (T. Uehara, unpublished data), indicating that AmpE is involved in PG recycling when the peptide moiety contains d-Ala-d-Ala. It is very surprising that pentapeptide accumulates in the cytoplasm of the ampE mutant since Mpl protein ligates pentapeptide as well as tripeptide to UDP-MurNAc, and this should act against the accumulation of pentapeptide (46).
Recycling intermediates and gene regulation.
Due to the fact that recycling intermediates triggered the induction of β-lactamase, it was suggested that changes in the cytoplasmic levels of an intermediate might be sensed and cause a response to cell wall damage (57, 93). However, no such relationship has emerged in E. coli. On the other hand, in Pseudomonas aeruginosa, the AmpR transcriptional regulator regulates pyocyanin production and the expression of LasA protease and LasB elastase in addition to β-lactamase (65). It has also been reported that the treatment of P. aeruginosa biofilms with imipenem, a good inducer of β-lactamase, changes global gene expression and alginate production as well as β-lactamase expression (4). Another piece of evidence that supports the linkage of alginate production to PG recycling is that algD transcription is increased by a mutation in ampD in Azotobacter vinelandii (88). Encystment and the cell shape of A. vinelandii are also affected by ampD and ampE (88). Lastly, in Acinetobacter baylyi, mutants in ampD and mpl are hypersensitive to β-lactam antibiotics (36), whereas ampD mutants of E. cloacae and C. freundii are hyperresistant to β-lactams by overproducing AmpC β-lactamase (74). In A. baylyi, PG degradation products might negatively regulate an efflux pump involved in the acquisition of resistance. Thus, recycling intermediates may play a regulatory role in some bacteria.
Turnover of New PG Formed during Septation
The recent discovery that the process of cell separation may involve the removal of the equivalent of one or perhaps two layers of murein from between the two newly formed cell poles dramatizes the hidden nature of turnover (119). One of us, T. Uehara, realized that the amount of anhMurNAc-peptides that accumulated in ampD mutants could be used as a measure of turnover of the cell wall. This led to a series of experiments measuring the amount of synthesis and degradation occurring during elongation and septation of the cell, with surprising results (119).
When cells were treated with A22, a compound known to inhibit MreB function (34, 63), elongation was inhibited, septation continued, and the cells gradually changed their shape, becoming round (55). Thus, by the exposure of E. coli MG1655 lysA ampD mutant cells to A22 to prevent the PG synthesis associated with elongation, followed by pulse-labeling of the cells with [3H]Dap (or lysA ampD nagB nagZ cells with [3H]GlcN), it was possible to determine the rate of murein turnover occurring during septation (119). Surprisingly, even though the poles of the cell are known to be stable and do not turn over (16, 28, 29), at least 30% of the murein made during septation was rapidly turned over during a 10-min labeling period (119). This indicates that a large fraction of the PG made during septation is removed as the two new poles are being formed and the daughter cells separate. One interpretation of this result is that at least three layers of wall are laid down simultaneously as the division machinery forms the septum, and most of the middle layer(s) is rapidly removed by lytic transglycosylases. An early indication that material from between the two new poles may be removed during septation can be seen from the electron-transparent space between the new poles revealed in thin-section micrographs of septating E. coli B cells shown in Fig. 8 (14). Similar open spaces have been seen in thin-section micrographs of the chain-forming envA (86) and envC (91) mutants. Presumably, the space between the poles contains proteins or other polymers, including partially cross-linked PG, that keep the poles separated. Interestingly, if the loss was only one-third of the new polar material made during septation, it would be consistent with the three-for-one (actually, three formed and one removed) model for septation proposed previously by Höltje (47). However, there is a major caveat. The turnover of murein observed is a minimum value because it does not take into account the unknown amount of PG made and not yet turned over, the amount turned over and not trapped by the ampD cells, or the undercount of label due to a lower specific activity in the anh-muropeptides than in the sacculi. If only 67% of the unstable septal murein was degraded during the 10-min experimental period, it would indicate that four layers of PG are formed during septum formation. Another reason to consider four layers as a possibility rather than three is the report that PBP1B, which is believed to form the glycan chains during septation, is reported to be active when in the form of a dimer (8). If both enzymes of the PBP1B dimer are forming glycan chains, this would produce an even number of glycan layers. The results could not help distinguish between three layers or four layers of PG in the growing septa (119).
FIG. 8.
Thin section of the septum of E. coli. cm, cytoplasmic membrane; om, outer membrane; mp, peptidoglycan. (Reprinted from reference 14 with permission.)
Amidase activity during septation.
The amidases present in the periplasm, AmiA, AmiB, and AmiC, have been shown to facilitate cell separation (44, 99). Without the amidases, many cells remain attached to each other in chains, indicating that separation is delayed but not completely prevented. However, in terms of the release of Dap from cells during septation, only one-sixth of the septal PG cleaved by the lytic transglycosylases had been cleaved by the amidases, indicating that lytic transglycosylases are primarily responsible for septal PG degradation (119). The murein tri- and tetrapeptides detected in spent media by Goodell and Schwarz (40) are products of amidase activity. Amidase activity seems to be greater during septation than during elongation because it was shown previously that asynchronously growing cells turn over more PG than elongating cells (92). Treatment of septating cells (i.e., A22-treated cells) with furazlocillin, which prevents cross-linking by PBP3 during cell division, resulted in turnover rates of over 75% (our unpublished data). Thus, when cross-linking was blocked, the nascent poles were rapidly degraded, along with the PG between the new poles.
Turnover of New PG Formed during Elongation
When cells overexpress sulA for a short period of time, septation ceases, cell elongation continues, and cells become filamentous (9). It is well established that during elongation, single new glycan strands are inserted between two existing strands (25, 27, 89). Under these elongation conditions, during a 10-min period of labeling of cells, about 7% of their PG turned over, as measured by the entrapment of the anhMurNAc-peptides released by lytic transglycosylases. As noted above, since the PG in the poles of the cell is not turned over during a chase, over 60% of the side wall is presumably turned over each generation for at least the four generations studied (92). The turnover of about 7% of the murein formed during 10 min of elongation by cells with a 50-min generation time is actually less than the rate at which the preexisting side wall turns over (119). Thus, the turnover does not appear to be related to the elongation process.
It is believed that amdinocillin (mecillinam) inhibits the transpeptidation reaction by PBP2 that is required for cross-linking the new glycan strands to old strands during elongation (53, 54, 111). The treatment of elongating cells with amdinocillin resulted in an apparent 78% inhibition of murein synthesis and recovery of the missing material as anhMurNAc-peptides in the cytoplasm of ampD cells (119). Thus, as in the case of furazlocillin-treated septating cells described above, un-cross-linked murein was being made and rapidly degraded by lytic transglycoslyases. A very different result was obtained when elongating cells were inhibited by compound A22. At the concentration used (30 μg/ml), cell wall synthesis was inhibited by 60%, but a relatively small amount of anhMurNAc-peptides accumulated, indicating that glycan strands were not being formed (119). One interpretation of these results is that A22 prevented the utilization of the lipid 2 intermediate so that glycan strands could not be formed, whereas in the presence of amdinocillin, un-cross-linked glycan strands were synthesized and rapidly cleaved by lytic transglycosylases.
CONCLUDING REMARKS
Unraveling the process of cell wall recycling has been a pioneering effort. We have reported here how the individual enzymes were discovered one at a time, how their genes were identified and cloned, and how the proteins were purified and initially characterized. One can see now that E. coli and many other colon-dwelling bacteria may have three different recycling pathways. The major one is dependent on Mpl for the direct utilization of murein tripeptide, a second one hydrolyzes the tripeptide to make the individual amino acids available for reuse, and the third, also a major pathway, enables cells to reutilize anhMurNAc and GlcNAc. As speculated in the discussion of orthologs, some bacteria may have only one or two of these pathways.
Lytic transglycosylase and AmpG permease are required for the pathways in E. coli and related enteric bacteria. AmpG is a remarkable protein and plays a key role in recycling. Without it, there would be no recycling. AmpG, driven by proton motive force, can transport hydrophilic molecules as large as GlcNAc-anhMurNAc-pentapeptide MW 992 across the inner membrane. AmpG permease is specific for compounds containing GlcNAc-anhMurNAc. It might be possible to transport biologically active molecules into the cell linked to GlcNAc-anhMurNAc since the disaccharide can carry a peptide of MW 532.
Two other proteins that are required for the recycling of anhMurNAc are no less remarkable. anhMurNAc kinase carries out two reactions: cleavage of the 1,6-anhydro bond and phosphorylation of the sugar to yield MurNAc-6-P. MurQ, the enzyme that converts MurNAc-P to GlcNAc-P with the release of d-lactic acid, is remarkable for the fact that it cleaves an ether bond. Few such enzymes have been identified in nature. Interestingly, E. coli is also equipped to capture such PG products as murein tripeptide (94), Dap (7), MurNAc (26, 95), and anhMurNAc (122) from the environment.
Although murein turnover and recycling are largely hidden from view, they play a major role in the metabolism of the murein sacculus. Much work remains to be done in the areas of regulation and structural studies. Beyond the question of why turnover and recycling of PG occur, the greatest unsolved mysteries are why over 60% of the side wall is turned over each generation and why the poles of the cell wall are completely resistant to turnover.
Supplementary Material
Acknowledgments
We thank Michael Malamy (Tufts University) for critical comments.
Footnotes
Supplemental material for this article may be found at http://mmbr.asm.org/.
REFERENCES
- 1.Adam, A., and E. Lederer. 1984. Muramyl peptides: immunomodulators, sleep factors, and vitamins. Med. Res. Rev. 4111-152. [DOI] [PubMed] [Google Scholar]
- 2.Arminjon, F., M. Guinand, M. J. Vacheron, and G. Michel. 1977. Specificity profiles of the membrane-bound γ-D-glutamyl-(L)meso-diaminopimelateendopeptidase and LD-carboxypeptidase from Bacillus sphaericus 9602. Eur. J. Biochem. 73557-565. [DOI] [PubMed] [Google Scholar]
- 3.Asensio, C., and M. Ruiz-Amil. 1966. N-Acetyl-D-glucosamine kinase. II. Escherichia coli. Methods Enzymol. 9421-425. [Google Scholar]
- 4.Bagge, N., M. Schuster, M. Hentzer, O. Ciofu, M. Givskov, E. P. Greenberg, and N. Høiby. 2004. Pseudomonas aeruginosa biofilms exposed to imipenem exhibit changes in global gene expression and β-lactamase and alginate production. Antimicrob. Agents Chemother. 481175-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baum, E. Z., S. M. Crespo-Carbone, D. Abbanat, B. Foleno, A. Maden, R. Goldschmidt, and K. Bush. 2006. Utility of muropeptide ligase for identification of inhibitors of the cell wall biosynthesis enzyme MurF. Antimicrob. Agents Chemother. 50230-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baum, E. Z., S. M. Crespo-Carbone, B. Foleno, S. Peng, J. J. Hilliard, D. Abbanat, R. Goldschmidt, and K. Bush. 2005. Identification of a dithiazoline inhibitor of Escherichia coli L,D-carboxypeptidase A. Antimicrob. Agents Chemother. 494500-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger, E. A., and L. A. Heppel. 1972. A binding protein involved in the transport of cystine and diaminopimelic acid in Escherichia coli. J. Biol. Chem. 2477684-7694. [PubMed] [Google Scholar]
- 8.Bertsche, U., E. Breukink, T. Kast, and W. Vollmer. 2005. In vitro murein peptidoglycan synthesis by dimers of the bifunctional transglycosylase-transpeptidase PBP1B from Escherichia coli. J. Biol. Chem. 28038096-38101. [DOI] [PubMed] [Google Scholar]
- 9.Bi, E., and J. Lutkenhaus. 1993. Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J. Bacteriol. 1751118-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bina, X., V. Perreten, and S. B. Levy. 2003. The periplasmic protein MppA requires an additional mutated locus to repress marA expression in Escherichia coli. J. Bacteriol. 1851465-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boneca, I. G. 2005. The role of peptidoglycan in pathogenesis. Curr. Opin. Microbiol. 846-53. [DOI] [PubMed] [Google Scholar]
- 12.Boothby, D., L. Daneo-Moore, M. L. Higgins, J. Coyette, and G. D. Shockman. 1973. Turnover of bacterial cell wall peptidoglycans. J. Biol. Chem. 2482161-2169. [PubMed] [Google Scholar]
- 13.Broome-Smith, J. K., M. Tadayyon, and Y. Zhang. 1990. β-Lactamase as a probe of membrane protein assembly and protein export. Mol. Microbiol. 41637-1644. [DOI] [PubMed] [Google Scholar]
- 14.Burdett, I. D., and R. G. Murray. 1974. Electron microscope study of septum formation in Escherichia coli strains B and B-r during synchronous growth. J. Bacteriol. 1191039-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burland, V., G. Plunkett III, H. J. Sofia, D. L. Daniels, and F. R. Blattner. 1995. Analysis of the Escherichia coli genome VI: DNA sequence of the region from 92.8 through 100 minutes. Nucleic Acids Res. 232105-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burman, L. G., J. Raichler, and J. T. Park. 1983. Evidence for diffuse growth of the cylindrical portion of the Escherichia coli murein sacculus. J. Bacteriol. 155983-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chahboune, A., M. Decaffmeyer, R. Brasseur, and B. Joris. 2005. Membrane topology of the Escherichia coli AmpG permease required for recycling of cell wall anhydromuropeptides and AmpC β-lactamase induction. Antimicrob. Agents Chemother. 491145-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaloupka, J., and M. Strnadová. 1972. Turnover of murein in a diaminopimelic acid dependent mutant of Escherichia coli. Folia Microbiol. (Prague) 17446-455. [DOI] [PubMed] [Google Scholar]
- 19.Chaput, C., and I. G. Boneca. 2007. Peptidoglycan detection by mammals and flies. Microbes Infect. 9637-647. [DOI] [PubMed] [Google Scholar]
- 20.Cheng, Q., H. Li, K. Merdek, and J. T. Park. 2000. Molecular characterization of the β-N-acetylglucosaminidase of Escherichia coli and its role in cell wall recycling. J. Bacteriol. 1824836-4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng, Q., and J. T. Park. 2002. Substrate specificity of the AmpG permease required for recycling of cell wall anhydro-muropeptides. J. Bacteriol. 1846434-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chitlaru, E., and S. Roseman. 1996. Molecular cloning and characterization of a novel β-N-acetyl-D-glucosaminidase from Vibrio furnissii. J. Biol. Chem. 27133433-33439. [DOI] [PubMed] [Google Scholar]
- 23.Cloud-Hansen, K. A., S. B. Peterson, E. V. Stabb, W. E. Goldman, M. J. McFall-Ngai, and J. Handelsman. 2006. Breaching the great wall: peptidoglycan and microbial interactions. Nat. Rev. Microbiol. 4710-716. [DOI] [PubMed] [Google Scholar]
- 24.Cookson, B. T., H. L. Cho, L. A. Herwaldt, and W. E. Goldman. 1989. Biological activities and chemical composition of purified tracheal cytotoxin of Bordetella pertussis. Infect. Immun. 572223-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cooper, S., M. L. Hsieh, and B. Guenther. 1988. Mode of peptidoglycan synthesis in Salmonella typhimurium: single-strand insertion. J. Bacteriol. 1703509-3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dahl, U., T. Jaeger, B. T. Nguyen, J. M. Sattler, and C. Mayer. 2004. Identification of a phosphotransferase system of Escherichia coli required for growth on N-acetylmuramic acid. J. Bacteriol. 1862385-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Jonge, B. L., F. B. Wientjes, I. Jurida, F. Driehuis, J. T. Wouters, and N. Nanninga. 1989. Peptidoglycan synthesis during the cell cycle of Escherichia coli: composition and mode of insertion. J. Bacteriol. 1715783-5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Pedro, M. A., W. D. Donachie, J. V. Höltje, and H. Schwarz. 2001. Constitutive septal murein synthesis in Escherichia coli with impaired activity of the morphogenetic proteins RodA and penicillin-binding protein 2. J. Bacteriol. 1834115-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Pedro, M. A., J. C. Quintela, J. V. Höltje, and H. Schwarz. 1997. Murein segregation in Escherichia coli. J. Bacteriol. 1792823-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dietz, H., D. Pfeifle, and B. Wiedemann. 1997. The signal molecule for β-lactamase induction in Enterobacter cloacae is the anhydromuramyl-pentapeptide. Antimicrob. Agents Chemother. 412113-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dijkstra, A. J., and W. Keck. 1996. Peptidoglycan as a barrier to transenvelope transport. J. Bacteriol. 1785555-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garnier, M., M. J. Vacheron, M. Guinand, and G. Michel. 1985. Purification and partial characterization of the extracellular γ-D-glutamyl-(L)meso-diaminopimelate endopeptidase I, from Bacillus sphaericus NCTC 9602. Eur. J. Biochem. 148539-543. [DOI] [PubMed] [Google Scholar]
- 33.Généreux, C., D. Dehareng, B. Devreese, J. Van Beeumen, J. M. Frère, and B. Joris. 2004. Mutational analysis of the catalytic centre of the Citrobacter freundii AmpD N-acetylmuramyl-L-alanine amidase. Biochem. J. 377111-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gitai, Z., N. A. Dye, A. Reisenauer, M. Wachi, and L. Shapiro. 2005. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell 120329-341. [DOI] [PubMed] [Google Scholar]
- 35.Glauner, B. 1988. Separation and quantification of muropeptides with high-performance liquid chromatography. Anal. Biochem. 172451-464. [DOI] [PubMed] [Google Scholar]
- 36.Gomez, M. J., and A. A. Neyfakh. 2006. Genes involved in intrinsic antibiotic resistance of Acinetobacter baylyi. Antimicrob. Agents Chemother. 503562-3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goodell, E. W. 1985. Recycling of murein by Escherichia coli. J. Bacteriol. 163305-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goodell, E. W., M. Fazio, and A. Tomasz. 1978. Effect of benzylpenicillin on the synthesis and structure of the cell envelope of Neisseria gonorrhoeae. Antimicrob. Agents Chemother. 13514-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goodell, E. W., and C. F. Higgins. 1987. Uptake of cell wall peptides by Salmonella typhimurium and Escherichia coli. J. Bacteriol. 1693861-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goodell, E. W., and U. Schwarz. 1985. Release of cell wall peptides into culture medium by exponentially growing Escherichia coli. J. Bacteriol. 162391-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Groisman, E. A., C. Parra-Lopez, M. Salcedo, C. J. Lipps, and F. Heffron. 1992. Resistance to host antimicrobial peptides is necessary for Salmonella virulence. Proc. Natl. Acad. Sci. USA 8911939-11943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gulick, A. M., D. M. Schmidt, J. A. Gerlt, and I. Rayment. 2001. Evolution of enzymatic activities in the enolase superfamily: crystal structures of the L-Ala-D/L-Glu epimerases from Escherichia coli and Bacillus subtilis. Biochemistry 4015716-15724. [DOI] [PubMed] [Google Scholar]
- 43.Hebeler, B. H., and F. E. Young. 1976. Chemical composition and turnover of peptidoglycan in Neisseria gonorrhoeae. J. Bacteriol. 1261180-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heidrich, C., M. F. Templin, A. Ursinus, M. Merdanovic, J. Berger, H. Schwarz, M. A. de Pedro, and J. V. Höltje. 2001. Involvement of N-acetylmuramyl-L-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol. Microbiol. 41167-178. [DOI] [PubMed] [Google Scholar]
- 45.Heidrich, C., A. Ursinus, J. Berger, H. Schwarz, and J. V. Höltje. 2002. Effects of multiple deletions of murein hydrolases on viability, septum cleavage, and sensitivity to large toxic molecules in Escherichia coli. J. Bacteriol. 1846093-6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hervé, M., A. Boniface, S. Gobec, D. Blanot, and D. Mengin-Lecreulx. 2007. Biochemical characterization and physiological properties of Escherichia coli UDP-N-acetylmuramate:L-alanyl-γ-D-glutamyl-meso-diaminopimelate ligase. J. Bacteriol. 1893987-3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Höltje, J. V. 1998. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 62181-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Höltje, J. V., U. Kopp, A. Ursinus, and B. Wiedemann. 1994. The negative regulator of β-lactamase induction AmpD is a N-acetyl-anhydromuramyl-L-alanine amidase. FEMS Microbiol. Lett. 122159-164. [DOI] [PubMed] [Google Scholar]
- 49.Höltje, J. V., D. Mirelman, N. Sharon, and U. Schwarz. 1975. Novel type of murein transglycosylase in Escherichia coli. J. Bacteriol. 1241067-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Honoré, N., M. H. Nicolas, and S. T. Cole. 1989. Regulation of enterobacterial cephalosporinase production: the role of a membrane-bound sensory transducer. Mol. Microbiol. 31121-1130. [DOI] [PubMed] [Google Scholar]
- 51.Hourdou, M. L., M. Guinand, M. J. Vacheron, G. Michel, L. Denoroy, C. Duez, S. Englebert, B. Joris, G. Weber, and J. M. Ghuysen. 1993. Characterization of the sporulation-related γ-D-glutamyl-(L)meso-diaminopimelic-acid-hydrolysing peptidase I of Bacillus sphaericus NCTC 9602 as a member of the metallo(zinc) carboxypeptidase A family. Modular design of the protein. Biochem. J. 292563-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hrebenda, J. 1979. Mutants of Escherichia coli with altered level of β-N-acetylglucosaminidase activities. Acta Microbiol. Pol. 2853-62. [PubMed] [Google Scholar]
- 53.Ishino, F., W. Park, S. Tomioka, S. Tamaki, I. Takase, K. Kunugita, H. Matsuzawa, S. Asoh, T. Ohta, B. G. Spratt, and M. Matsuhashi. 1986. Peptidoglycan synthetic activities in membranes of Escherichia coli caused by overproduction of penicillin-binding protein 2 and RodA protein. J. Biol. Chem. 2617024-7031. [PubMed] [Google Scholar]
- 54.Ishino, F., S. Tamaki, B. G. Spratt, and M. Matsuhashi. 1982. A mecillinam-sensitive peptidoglycan crosslinking reaction in Escherichia coli. Biochem. Biophys. Res. Commun. 109689-696. [DOI] [PubMed] [Google Scholar]
- 55.Iwai, N., K. Nagai, and M. Wachi. 2002. Novel S-benzylisothiourea compound that induces spherical cells in Escherichia coli probably by acting on a rod-shape-determining protein(s) other than penicillin-binding protein 2. Biosci. Biotechnol. Biochem. 662658-2662. [DOI] [PubMed] [Google Scholar]
- 56.Jacobs, C., J. M. Frère, and S. Normark. 1997. Cytosolic intermediates for cell wall biosynthesis and degradation control inducible β-lactam resistance in gram-negative bacteria. Cell 88823-832. [DOI] [PubMed] [Google Scholar]
- 57.Jacobs, C., L. J. Huang, E. Bartowsky, S. Normark, and J. T. Park. 1994. Bacterial cell wall recycling provides cytosolic muropeptides as effectors for β-lactamase induction. EMBO J. 134684-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jacobs, C., B. Joris, M. Jamin, K. Klarsov, J. Van Beeumen, D. Mengin-Lecreulx, J. van Heijenoort, J. T. Park, S. Normark, and J. M. Frère. 1995. AmpD, essential for both β-lactamase regulation and cell wall recycling, is a novel cytosolic N-acetylmuramyl-L-alanine amidase. Mol. Microbiol. 15553-559. [DOI] [PubMed] [Google Scholar]
- 59.Jaeger, T., M. Arsic, and C. Mayer. 2005. Scission of the lactyl ether bond of N-acetylmuramic acid by Escherichia coli “etherase”. J. Biol. Chem. 28030100-30106. [DOI] [PubMed] [Google Scholar]
- 60.Juan, C., M. D. Maciá, O. Gutierrez, C. Vidal, J. L. Pérez, and A. Oliver. 2005. Molecular mechanisms of β-lactam resistance mediated by AmpC hyperproduction in Pseudomonas aeruginosa clinical strains. Antimicrob. Agents Chemother. 494733-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Juan, C., B. Moya, J. L. Pérez, and A. Oliver. 2006. Stepwise upregulation of the Pseudomonas aeruginosa chromosomal cephalosporinase conferring high-level β-lactam resistance involves three AmpD homologues. Antimicrob. Agents Chemother. 501780-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Juncker, A. S., H. Willenbrock, G. Von Heijne, S. Brunak, H. Nielsen, and A. Krogh. 2003. Prediction of lipoprotein signal peptides in gram-negative bacteria. Protein Sci. 121652-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Karczmarek, A., R. M. Baselga, S. Alexeeva, F. G. Hansen, M. Vicente, N. Nanninga, and T. den Blaauwen. 2007. DNA and origin region segregation are not affected by the transition from rod to sphere after inhibition of Escherichia coli MreB by A22. Mol. Microbiol. 6551-63. [DOI] [PubMed] [Google Scholar]
- 64.Klenchin, V. A., D. M. Schmidt, J. A. Gerlt, and I. Rayment. 2004. Evolution of enzymatic activities in the enolase superfamily: structure of a substrate-liganded complex of the L-Ala-D/L-Glu epimerase from Bacillus subtilis. Biochemistry 4310370-10378. [DOI] [PubMed] [Google Scholar]
- 65.Kong, K. F., S. R. Jayawardena, S. D. Indulkar, A. Del Puerto, C. L. Koh, N. Høiby, and K. Mathee. 2005. Pseudomonas aeruginosa AmpR is a global transcriptional factor that regulates expression of AmpC and PoxB β-lactamases, proteases, quorum sensing, and other virulence factors. Antimicrob. Agents Chemother. 494567-4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Koraimann, G. 2003. Lytic transglycosylases in macromolecular transport systems of gram-negative bacteria. Cell. Mol. Life Sci. 602371-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Korfmann, G., and C. C. Sanders. 1989. ampG is essential for high-level expression of AmpC β-lactamase in Enterobacter cloacae. Antimicrob. Agents Chemother. 331946-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Korza, H. J., and M. Bochtler. 2005. Pseudomonas aeruginosa LD-carboxypeptidase, a serine peptidase with a Ser-His-Glu triad and a nucleophilic elbow. J. Biol. Chem. 28040802-40812. [DOI] [PubMed] [Google Scholar]
- 69.Kraft, A. R., J. Prabhu, A. Ursinus, and J. V. Höltje. 1999. Interference with murein turnover has no effect on growth but reduces β-lactamase induction in Escherichia coli. J. Bacteriol. 1817192-7198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Létoffé, S., P. Delepelaire, and C. Wandersman. 2006. The housekeeping dipeptide permease is the Escherichia coli heme transporter and functions with two optional peptide binding proteins. Proc. Natl. Acad. Sci. USA 10312891-12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li, H., and J. T. Park. 1999. The periplasmic murein peptide-binding protein MppA is a negative regulator of multiple antibiotic resistance in Escherichia coli. J. Bacteriol. 1814842-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liepinsh, E., C. Généreux, D. Dehareng, B. Joris, and G. Otting. 2003. NMR structure of Citrobacter freundii AmpD, comparison with bacteriophage T7 lysozyme and homology with PGRP domains. J. Mol. Biol. 327833-842. [DOI] [PubMed] [Google Scholar]
- 73.Liger, D., A. Masson, D. Blanot, J. van Heijenoort, and C. Parquet. 1995. Over-production, purification and properties of the uridine-diphosphate-N-acetylmuramate:L-alanine ligase from Escherichia coli. Eur. J. Biochem. 23080-87. [DOI] [PubMed] [Google Scholar]
- 74.Lindberg, F., S. Lindquist, and S. Normark. 1987. Inactivation of the ampD gene causes semiconstitutive overproduction of the inducible Citrobacter freundii β-lactamase. J. Bacteriol. 1691923-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lindquist, S., M. Galleni, F. Lindberg, and S. Normark. 1989. Signalling proteins in enterobacterial AmpC β-lactamase regulation. Mol. Microbiol. 31091-1102. [DOI] [PubMed] [Google Scholar]
- 76.Lindquist, S., K. Weston-Hafer, H. Schmidt, C. Pul, G. Korfmann, J. Erickson, C. Sanders, H. H. Martin, and S. Normark. 1993. AmpG, a signal transducer in chromosomal β-lactamase induction. Mol. Microbiol. 9703-715. [DOI] [PubMed] [Google Scholar]
- 77.Mauck, J., L. Chan, and L. Glaser. 1971. Turnover of the cell wall of gram-positive bacteria. J. Biol. Chem. 2461820-1827. [PubMed] [Google Scholar]
- 78.Meberg, B. M., A. L. Paulson, R. Priyadarshini, and K. D. Young. 2004. Endopeptidase penicillin-binding proteins 4 and 7 play auxiliary roles in determining uniform morphology of Escherichia coli. J. Bacteriol. 1868326-8336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mengin-Lecreulx, D., B. Flouret, and J. van Heijenoort. 1982. Cytoplasmic steps of peptidoglycan synthesis in Escherichia coli. J. Bacteriol. 1511109-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mengin-Lecreulx, D., and J. van Heijenoort. 1996. Characterization of the essential gene glmM encoding phosphoglucosamine mutase in Escherichia coli. J. Biol. Chem. 27132-39. [DOI] [PubMed] [Google Scholar]
- 81.Mengin-Lecreulx, D., and J. van Heijenoort. 1994. Copurification of glucosamine-1-phosphate acetyltransferase and N-acetylglucosamine-1-phosphate uridyltransferase activities of Escherichia coli: characterization of the glmU gene product as a bifunctional enzyme catalyzing two subsequent steps in the pathway for UDP-N-acetylglucosamine synthesis. J. Bacteriol. 1765788-5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mengin-Lecreulx, D., and J. van Heijenoort. 1993. Identification of the glmU gene encoding N-acetylglucosamine-1-phosphate uridyltransferase in Escherichia coli. J. Bacteriol. 1756150-6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mengin-Lecreulx, D., J. van Heijenoort, and J. T. Park. 1996. Identification of the mpl gene encoding UDP-N-acetylmuramate:L-alanyl-γ-D-glutamyl-meso-diaminopimelate ligase in Escherichia coli and its role in recycling of cell wall peptidoglycan. J. Bacteriol. 1785347-5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mielcarek, N., A. S. Debrie, D. Raze, J. Quatannens, J. Engle, W. E. Goldman, and C. Locht. 2006. Attenuated Bordetella pertussis: new live vaccines for intranasal immunisation. Vaccine 24S2-54-55. [DOI] [PubMed] [Google Scholar]
- 85.Nikaido, H. 1996. Outer membrane, p.29-47. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed., vol. 2. American Society for Microbiology, Washington, DC. [Google Scholar]
- 86.Normark, S., H. G. Boman, and G. D. Bloom. 1971. Cell division in a chain-forming envA mutant of Escherichia coli K12. Fine structure of division sites and effects of EDTA, lysozyme and ampicillin. Acta Pathol. Microbiol. Scand. B Microbiol. Immunol. 79651-664. [DOI] [PubMed] [Google Scholar]
- 87.Normark, S., H. G. Boman, and E. Matsson. 1969. Mutant of Escherichia coli with anomalous cell division and ability to decrease episomally and chromosomally mediated resistance to ampicillin and several other antibiotics. J. Bacteriol. 971334-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Núñez, C., S. Moreno, L. Cárdenas, G. Sóberon-Chávez, and G. Espín. 2000. Inactivation of the ampDE operon increases transcription of algD and affects morphology and encystment of Azotobacter vinelandii. J. Bacteriol. 1824829-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Park, J. T. 1993. An overview of the assembly, turnover, and recycling of the murein sacculus, p. 119-126. In M. A. de Pedro et al. (ed.), Bacterial growth and lysis. Plenum Press, New York, NY.
- 90.Park, J. T. 2001. Identification of a dedicated recycling pathway for anhydro-N-acetylmuramic acid and N-acetylglucosamine derived from Escherichia coli cell wall murein. J. Bacteriol. 1833842-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Park, J. T. 1996. The murein sacculus, p. 48-57. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed., vol. 2. American Society for Microbiology, Washington, DC. [Google Scholar]
- 92.Park, J. T. 1993. Turnover and recycling of the murein sacculus in oligopeptide permease-negative strains of Escherichia coli: indirect evidence for an alternative permease system and for a monolayered sacculus. J. Bacteriol. 1757-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Park, J. T. 1995. Why does Escherichia coli recycle its cell wall peptides? Mol. Microbiol. 17421-426. [DOI] [PubMed] [Google Scholar]
- 94.Park, J. T., D. Raychaudhuri, H. Li, S. Normark, and D. Mengin-Lecreulx. 1998. MppA, a periplasmic binding protein essential for import of the bacterial cell wall peptide L-alanyl-γ-D-glutamyl-meso-diaminopimelate. J. Bacteriol. 1801215-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Parquet, C., B. Flouret, M. Leduc, Y. Hirota, and J. van Heijenoort. 1983. N-Acetylmuramoyl-L-alanine amidase of Escherichia coli K12. Possible physiological functions. Eur. J. Biochem. 133371-377. [DOI] [PubMed] [Google Scholar]
- 96.Parra-Lopez, C., R. Lin, A. Aspedon, and E. A. Groisman. 1994. A Salmonella protein that is required for resistance to antimicrobial peptides and transport of potassium. EMBO J. 133964-3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pearce, S. R., M. L. Mimmack, M. P. Gallagher, U. Gileadi, S. C. Hyde, and C. F. Higgins. 1992. Membrane topology of the integral membrane components, OppB and OppC, of the oligopeptide permease of Salmonella typhimurium. Mol. Microbiol. 647-57. [DOI] [PubMed] [Google Scholar]
- 98.Popham, D. L., and K. D. Young. 2003. Role of penicillin-binding proteins in bacterial cell morphogenesis. Curr. Opin. Microbiol. 6594-599. [DOI] [PubMed] [Google Scholar]
- 99.Priyadarshini, R., M. A. de Pedro, and K. D. Young. 2007. Role of peptidoglycan amidases in the development and morphology of the division septum in Escherichia coli. J. Bacteriol. 1895334-5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Priyadarshini, R., D. L. Popham, and K. D. Young. 2006. Daughter cell separation by penicillin-binding proteins and peptidoglycan amidases in Escherichia coli. J. Bacteriol. 1885345-5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rosenthal, R. S., W. Nogami, B. T. Cookson, W. E. Goldman, and W. J. Folkening. 1987. Major fragment of soluble peptidoglycan released from growing Bordetella pertussis is tracheal cytotoxin. Infect. Immun. 552117-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Scheurwater, E., C. W. Reid, and A. J. Clarke. 30 March 2007, posting date. Lytic transglycosylases: bacterial space-making autolysins. Int. J. Biochem. Cell Biol. doi: 10.1016/j.biocel.2007.03.018. [DOI] [PubMed]
- 103.Schiefner, A., K. Gerber, S. Seitz, W. Welte, K. Diederichs, and W. Boos. 2005. The crystal structure of Mlc, a global regulator of sugar metabolism in Escherichia coli. J. Biol. Chem. 28029073-29079. [DOI] [PubMed] [Google Scholar]
- 104.Schleifer, K. H., and O. Kandler. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36407-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schmidt, D. M., B. K. Hubbard, and J. A. Gerlt. 2001. Evolution of enzymatic activities in the enolase superfamily: functional assignment of unknown proteins in Bacillus subtilis and Escherichia coli as L-Ala-D/L-Glu epimerases. Biochemistry 4015707-15715. [DOI] [PubMed] [Google Scholar]
- 106.Shockman, G. D., and J. V. Höltje. 1994. Microbial peptidoglycan (murein) hydrolases, p. 131-166. In J.-M. Ghuysen and R. Hakenbeck (ed.), Bacterial cell wall, vol. 27. Elsevier Science B.V., Amsterdam, The Netherlands. [Google Scholar]
- 107.Schroeder, U., B. Henrich, J. Fink, and R. Plapp. 1994. Peptidase D of Escherichia coli K-12, a metallopeptidase of low substrate specificity. FEMS Microbiol. Lett. 123153-159. [DOI] [PubMed] [Google Scholar]
- 108.Serror, P., and A. L. Sonenshein. 1996. Interaction of CodY, a novel Bacillus subtilis DNA-binding protein, with the dpp promoter region. Mol. Microbiol. 20843-852. [DOI] [PubMed] [Google Scholar]
- 109.Sinha, R. K., and R. S. Rosenthal. 1980. Release of soluble peptidoglycan from growing conococci: demonstration of anhydro-muramyl-containing fragments. Infect. Immun. 29914-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Slack, F. J., P. Serror, E. Joyce, and A. L. Sonenshein. 1995. A gene required for nutritional repression of the Bacillus subtilis dipeptide permease operon. Mol. Microbiol. 15689-702. [DOI] [PubMed] [Google Scholar]
- 111.Spratt, B. G., and A. B. Pardee. 1975. Penicillin-binding proteins and cell shape in E. coli. Nature 254516-517. [DOI] [PubMed] [Google Scholar]
- 112.Stubbs, K. A., M. Balcewich, B. L. Mark, and D. J. Vocadlo. 2007. Small molecule inhibitors of a glycoside hydrolase attenuate inducible AmpC mediated β-lactam resistance. J. Biol. Chem. 28221382-21391. [DOI] [PubMed] [Google Scholar]
- 113.Sun, D., S. Cohen, N. Mani, C. Murphy, and D. M. Rothstein. 2002. A pathway-specific cell based screening system to detect bacterial cell wall inhibitors. J. Antibiot. (Tokyo) 55279-287. [DOI] [PubMed] [Google Scholar]
- 114.Talukder, A. A., S. Yanai, T. Nitta, A. Kato, and M. Yamada. 1996. RpoS-dependent regulation of genes expressed at late stationary phase in Escherichia coli. FEBS Lett. 386177-180. [DOI] [PubMed] [Google Scholar]
- 115.Templin, M. F., A. Ursinus, and J. V. Höltje. 1999. A defect in cell wall recycling triggers autolysis during the stationary growth phase of Escherichia coli. EMBO J. 184108-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Teplyakov, A., C. Leriche, G. Obmolova, B. Badet, and M. A. Badet-Denisot. 2002. From Lobry de Bruyn to enzyme-catalyzed ammonia channelling: molecular studies of D-glucosamine-6P synthase. Nat. Prod. Rep. 1960-69. [DOI] [PubMed] [Google Scholar]
- 117.Tokuda, H., and S. Matsuyama. 2004. Sorting of lipoproteins to the outer membrane in E. coli. Biochim. Biophys. Acta 16935-13. [DOI] [PubMed] [Google Scholar]
- 118.Uehara, T., and J. T. Park. 2007. An anhydro-N-acetylmuramyl-L-alanine amidase with broad specificity tethered to the outer membrane of Escherichia coli. J. Bacteriol. 1895634-5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Uehara, T., and J. T. Park. 2008. Growth of Escherichia coli: significance of peptidoglycan degradation during elongation and septation. J. Bacteriol. 1903914-3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Uehara, T., and J. T. Park. 2003. Identification of MpaA, an amidase in Escherichia coli that hydrolyzes the γ-D-glutamyl-meso-diaminopimelate bond in murein peptides. J. Bacteriol. 185679-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Uehara, T., and J. T. Park. 2004. The N-acetyl-D-glucosamine kinase of Escherichia coli and its role in murein recycling. J. Bacteriol. 1867273-7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Uehara, T., K. Suefuji, T. Jaeger, C. Mayer, and J. T. Park. 2006. MurQ etherase is required by Escherichia coli in order to metabolize anhydro-N-acetylmuramic acid obtained either from the environment or from its own cell wall. J. Bacteriol. 1881660-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Uehara, T., K. Suefuji, N. Valbuena, B. Meehan, M. Donegan, and J. T. Park. 2005. Recycling of the anhydro-N-acetylmuramic acid derived from cell wall murein involves a two-step conversion to N-acetylglucosamine-phosphate. J. Bacteriol. 1873643-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ursinus, A., H. Steinhaus, and J. V. Höltje. 1992. Purification of a nocardicin A-sensitive ld-carboxypeptidase from Escherichia coli by affinity chromatography. J. Bacteriol. 174441-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Varma, A., and K. D. Young. 2004. FtsZ collaborates with penicillin binding proteins to generate bacterial cell shape in Escherichia coli. J. Bacteriol. 1866768-6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Vega, D., and J. A. Ayala. 2006. The DD-carboxypeptidase activity encoded by pbp4B is not essential for the cell growth of Escherichia coli. Arch. Microbiol. 18523-27. [DOI] [PubMed] [Google Scholar]
- 127.Viala, J., C. Chaput, I. G. Boneca, A. Cardona, S. E. Girardin, A. P. Moran, R. Athman, S. Memet, M. R. Huerre, A. J. Coyle, P. S. DiStefano, P. J. Sansonetti, A. Labigne, J. Bertin, D. J. Philpott, and R. L. Ferrero. 2004. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat. Immunol. 51166-1174. [DOI] [PubMed] [Google Scholar]
- 128.Vötsch, W., and M. F. Templin. 2000. Characterization of a β-N-acetylglucosaminidase of Escherichia coli and elucidation of its role in muropeptide recycling and β-lactamase induction. J. Biol. Chem. 27539032-39038. [DOI] [PubMed] [Google Scholar]
- 129.White, R. J. 1968. Control of amino sugar metabolism in Escherichia coli and isolation of mutants unable to degrade amino sugars. Biochem. J. 106847-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.White, R. J., and C. A. Pasternak. 1967. The purification and properties of N-acetylglucosamine 6-phosphate deacetylase from Escherichia coli. Biochem. J. 105121-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yem, D. W., and H. C. Wu. 1976. Isolation of Escherichia coli K-12 mutants with altered level of β-N-acetylglucosaminidase. J. Bacteriol. 125372-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yem, D. W., and H. C. Wu. 1976. Purification and properties of β-N-acetylglucosaminidase from Escherichia coli. J. Bacteriol. 125324-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Young, K. D. 2003. Bacterial shape. Mol. Microbiol. 49571-580. [DOI] [PubMed] [Google Scholar]
- 134.Zahrl, D., M. Wagner, K. Bischof, M. Bayer, B. Zavecz, A. Beranek, C. Ruckenstuhl, G. E. Zarfel, and G. Koraimann. 2005. Peptidoglycan degradation by specialized lytic transglycosylases associated with type III and type IV secretion systems. Microbiology 1513455-3467. [DOI] [PubMed] [Google Scholar]
- 135.Zhuang, X., and H. Zhang. 2002. Identification, characterization of levoglucosan kinase, and cloning and expression of levoglucosan kinase cDNA from Aspergillus niger CBX-209 in Escherichia coli. Protein Expr. Purif. 2671-81. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.