Abstract
A detailed biochemical characterization of the Pseudomonas aeruginosa VIM-11 metallo-β-lactamase (MβL) is reported. The only substitution differentiating VIM-11 from VIM-2 (N165S) promoted a slightly improved catalytic efficiency of the former on 3 out of 12 substrates, notably the bulky cephalosporins. Thus, MβL-mediated resistance also may be modulated by remote mutations.
Acquired metallo-β-lactamases (MβLs) are emerging resistance determinants in numerous bacterial species of clinical relevance (3, 17). These enzymes pose particular threats, including their capacity to confer broad-spectrum β-lactam resistance and the unavailability of clinically useful inhibitors. In addition, their potential for rapid and generalized dissemination represents a great concern (2, 17). In recent years, two major groups of MβLs, corresponding to IMP and VIM types, increasingly have been identified in gram-negative pathogens (2). Recently, a novel variant of the VIM family encoded by blaVIM-11 was reported from a Pseudomonas aeruginosa clinical strain that differs from the blaVIM-2 sequence in a unique nonsynonymous mutation (N165S) (13). Despite their clinical relevance, studies addressing the impact of blaVIM allelic mutations on kinetic performance still are scarce. We report here the cloning, expression, purification, and biochemical characterization of VIM-11 as well as a comparison of the properties of VIM-11 to those of VIM-2. We also studied the contribution of each of these enzymes to in vitro resistance to different β-lactams when produced in Escherichia coli.
A VIM-11-producing P. aeruginosa clinical strain, M5109, recovered from Hospital de Niños Ricardo Gutierrez, Buenos Aires, Argentina (13), and P. aeruginosa COL-1, a VIM-2-producing strain (14), were used as sources of genomic DNA for the isolation of the cognate genes. Bacterial DNA was obtained essentially as described previously (15).
The contribution of VIM-11 or VIM-2 to bacterial β-lactam resistance was investigated by testing the susceptibility of E. coli DH5α harboring plasmids directing the production of these enzymes to various β-lactams. For this purpose, we designed the primers CIM-F (5′-GCCGGATCCCATAGTTAAGTAGCACTCACC-3′) and CIM-R (5′-ATCGGATCCCTACTCAACGACTGAGC-3′), both containing BamHI restriction sites, which allow the amplification of the complete VIM-2 or VIM-11 coding sequences, including the corresponding signal peptides. PCR products then were cloned into the corresponding sites of the broad-host-range expression plasmid pαΩ (5). As a control, we also employed E. coli DH5α cells carrying the empty vector. MICs were determined by the standard agar macrodilution method according to the guidelines of the Clinical and Laboratory Standards Institute (formerly NCCLS) (1). As shown in Table 1, the production of VIM-11 or VIM-2 in E. coli DH5α decreased the susceptibility of the host cells to all β-lactam substrates tested, with the expected exception of aztreonam (14). These results demonstrate that either enzyme can confer broad-spectrum β-lactam resistance to the bacterial host. Moreover, the inhibition of imipenem hydrolysis by EDTA using a microbiological assay confirmed the presence of MβL activity (9).
TABLE 1.
MICs of β-lactams for E. coli DH5α harboring recombinant plasmidsa
| β-Lactam | MIC (μg/ml) for:
|
||
|---|---|---|---|
| E. coli DH5α pαΩ | E. coli DH5α pαΩ blaVIM-11 | E. coli DH5α pαΩ blaVIM-2 | |
| Benzylpenicillin | 32 | 128 | 128 |
| Ampicillin | 8 | 64 | 256 |
| Cephalothin | 4 | 64 | 64 |
| Cefuroxime | 4 | 64 | 128 |
| Cefoxitin | 4 | 32 | 64 |
| Ceftazidime | 0.125 | 8 | 8 |
| Cefotaxime | 0.03 | 4 | 8 |
| Cefepime | 0.03 | 0.5 | 0.5 |
| Cefpirome | 0.015 | 0.5 | 0.5 |
| Aztreonam | 0.25 | 0.25 | 0.25 |
| Imipenem | 0.125 | 0.5 | 1 |
| Meropenem | 0.03 | 0.06 | 0.125 |
E. coli DH5α pαΩ, E. coli DH5α containing pαΩ (as a control); E. coli DH5α pαΩ blaVIM-11 or blaVIM-2, E. coli DH5α harboring recombinant plasmid pαΩ blaVIM-11 or pαΩ blaVIM-2, respectively.
Primers VIM-C (5′-CTCGCTGGATCCGTAGATTCTAGCGGTGAG-3′) and VIM-N (5′-GCTCAGTCGTTGAGTAGAAGCTTGATGCG-3′) were designed after the corresponding VIM gene sequences (11, 13). These primers contain an additional BamHI and HindIII restriction site, respectively, and were used to amplify and clone the DNA regions encoding the corresponding mature VIM sequences into the corresponding sites of the pET-Term expression vector (12). The resulting pET-TermVIM-2 and pET-TermVIM-11 plasmids direct the production of the corresponding enzymes as C-terminal fusions to glutathione S-transferase (12). By using these plasmids, the corresponding fusion proteins were overproduced in E. coli BL21(DE3) pLysS and purified to homogeneity essentially as described previously (12), with the following modifications: (i) expression was induced at 20°C during 6 h, and (ii) the purified fusion proteins were dialyzed against 100 mM Tris-HCl (pH 8.0) to remove glutathione, digested with thrombin, and finally loaded onto a glutathione-agarose column (GE) equilibrated with 100 mM Tris-HCl (pH 8.0) to separate the cleaved proteins from glutathione S-transferase. The eluted proteins were more than 95% pure as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (not shown) and were used for the further comparative studies detailed below.
Metal content analysis of the purified proteins, as determined by using 4-(2-pyridylazo)-resorcinol (PAR) (16), revealed that both enzymes contain two bound Zn(II) ions.
To gain insight into the active sites of VIM-11 and VIM-2, we produced the corresponding dicobalt(II) derivatives as described elsewhere (12). Both Co(II)-substituted VIM-11 and VIM-2 displayed four ligand field bands in the visible region (see Fig. S1 in the supplemental material) that were similar to those reported for Co(II)-BcII from Bacillus cereus (12) and other MβLs (3). This shows that the Co1 sites are structurally similar among different Co(II) derivatives. On the contrary, a charge transfer band attributed to a Cys-Co(II) moiety, a probe of the Co2 site, is found on both VIM-11 and VIM-2 at 383 and 380 nm, respectively (see Fig. S1 in the supplemental material). A comparison of these spectral features with the charge transfer band of Co(II)-substituted BcII (340 nm) reveals a slightly different Co2 site (8a, 12).
Kinetic parameters with several representative β-lactam compounds, including penicillins, cephalosporins, carbapenems, and aztreonam (Table 2), were determined as previously reported (10, 16). β-Lactamase activity was determined by monitoring the changes in absorbance in a Jasco V-550 spectrophotometer. Δɛlmax values for the different substrates were detailed previously (8, 10, 16). As seen in Table 2, all tested β-lactams were hydrolyzed by these two enzymes, with the exception of aztreonam. The highest catalytic efficiencies for both enzymes were observed for benzylpenicillin, cephalothin, cefuroxime, cefotaxime, imipenem, and nitrocefin, while cefepime was a rather poor substrate. Between the two carbapenem substrates tested, VIM-11 showed greater hydrolytic efficiency on imipenem (Table 2), a result also reported for VIM-2 by other authors (4). As seen in Table 2, VIM-11 displayed a slightly better catalytic efficiency on 3 of the 12 hydrolyzed substrates compared to that of VIM-2 when tested under similar experimental conditions. Remarkably, these substrates (i.e., ceftazidime, cefepime, and cefpirome) are cephalosporins with bulky substituents at the C-3 position. The differences for these substrates are reflected in fourfold (ceftazidime), threefold (cefepime), and twofold (cefpirome) increases in kcat/Km values of VIM-11 relative to those of VIM-2 (Table 2).
TABLE 2.
Kinetic parameters for the hydrolysis of different β-lactam substrates of VIM-11 and VIM-2 enzymes
| β-Lactam | VIM-11a
|
VIM-2a
|
kcat/Km (s−1 M−1) for VIM-11/VIM-2 ratio | ||||
|---|---|---|---|---|---|---|---|
| Km (μM) | kcat (s−1) | kcat/Km (s−1 M−1) | Km (μM) | kcat (s−1) | kcat/Km (s−1 M−1) | ||
| Benzylpenicillin | 15.3 | 113 | 7.4 × 106 | 19.1 | 141 | 7.4 × 106 | 1.0 |
| Ampicillin | 109 | 190 | 1.8 × 106 | 136 | 302 | 2.2 × 106 | 0.81 |
| Cephalothin | 4.5 | 81 | 1.8 × 107 | 6.2 | 96.4 | 1.6 × 107 | 1.12 |
| Cefuroxime | 5.5 | 27 | 4.9 × 106 | 5.5 | 30.5 | 5.5 × 106 | 0.89 |
| Cefoxitin | 7.4 | 6.4 | 8.6 × 105 | 9.9 | 10 | 1.0 × 106 | 0.86 |
| Ceftazidime | 110 | 14 | 1.3 × 105 | 88.2 | 2.8 | 3.2 × 104 | 4.06 |
| Cefotaxime | 11.4 | 41.3 | 3.6 × 106 | 13.4 | 61 | 4.6 × 106 | 0.78 |
| Cefepime | >1,200 | >100 | 8.3 × 104 | >1,200 | >33 | 2.8 × 104 | 2.9 |
| Cefpirome | 183 | 102 | 5.6 × 105 | 171 | 48.1 | 2.8 × 105 | 2.0 |
| Nitrocefin | 8.2 | 129 | 1.6 × 107 | 13.8 | 206 | 1.5 × 107 | 1.06 |
| Imipenem | 9.4 | 20 | 2.1 × 106 | 9.9 | 30.2 | 3.1 × 106 | 0.67 |
| Meropenem | 22.7 | 3.2 | 1.4 × 105 | 24.5 | 3.5 | 1.4 × 105 | 1.0 |
| Aztreonam | NHb | NH | |||||
The Km and kcat values represent the means of two independent measurements. Standard deviation values were less than 5%.
NH, no hydrolysis was detected.
These minor differences in catalytic efficiency for the purified enzymes were not reflected in the MICs for ceftazidime, cefepime, or cefpirome measured for the E. coli host producing the corresponding recombinant enzymes (Table 1). This also could result from differential intrinsic properties between these enzymes, such as different levels of stability in vivo. In this context, the measurements of MβL-specific activity on periplasmic extracts (using a saturated concentration of substrates) indicated that E. coli cells producing VIM-11 contained severalfold lower levels of activity than those producing VIM-2 on all β-lactam substrates tested (e.g., 0.9 versus 5.5 μmol ceftazidime, 5.4 versus 141 μmol cephalothin, or 1.3 versus 27 μmol imipenem hydrolyzed/min/mg total periplasmic protein for VIM-11- and VIM-2-producing bacteria, respectively).
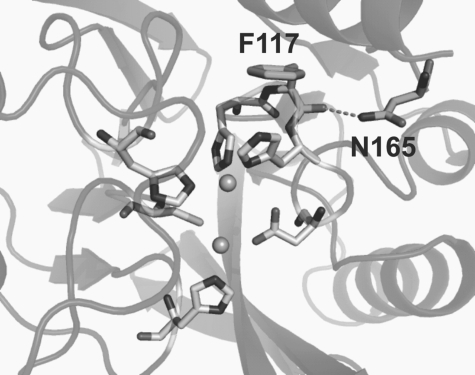
The crystal structure of dizinc(II) VIM-2 (6) revealed that the Asn165 side chain forms an H bond with the peptide carbonyl of Phe117, flanked in the protein sequence by two Zn ligands (His116 and His118) (Fig. 1). The N165S mutation is expected to remove the interaction between loops L7 and L8, possibly enhancing loop flexibility and resulting in a relatively wider active-site groove, a situation that would better accommodate cephalosporins with bulkier substituents at C-3. In line with these observations, a number of pieces of evidence account for the impact of second-shell ligands in enzymatic activity (7, 16), suggesting that mutations outside the active site also contribute to tuning the MβL catalytic performance.
FIG. 1.
Upper view of the active site of VIM-2 (Protein Data Bank entry 1ko3) (6). The protein backbone is shown in the cartoons. Residues of the active site and residues F117 and N165 (involved in the mentioned H-bond interaction) are indicated by sticks. Zn(II) ions are shown as spheres, and the H bond is indicated with dashed lines.
Supplementary Material
Acknowledgments
We are indebted to P. Nordmann for kindly providing P. aeruginosa strain COL-1. We also are grateful to M. Galas for the generous gift of P. aeruginosa strain M5109 (Malbrán Institute collection, Buenos Aires, Argentina).
This work was supported by grants from the Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT, Argentina), Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Howard Hughes Medical Institute (HHMI), and Departamento de Salud Pública, Municipalidad de Rosario.
A.M.V. and A.J.V. are staff members of CONICET, and M.A.M. and P.E.T. are fellows of this institution. A.J.V. also is an international scholar of HHMI. P.M. and A.L. are researchers of the National University of Rosario. F.P. is a researcher of Malbrán Institute.
Footnotes
Published ahead of print on 24 March 2008.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.CLSI. 2007. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M100-S17. CLSI, Wayne, PA.
- 2.Cornaglia, G., M. Akova, G. Amicosante, R. Cantón, R. Cauda, J.-D. Docquier, M. Edelstein, J.-M. Frére, M. Fuzi, M. Galleni, H. Giamarellou, M. Gniadkowski, R. Koncan, B. Libisch, F. Luzzaro, V. Miriagou, F. Navarro, P. Nordmann, L. Pagani, L. Peixe, L. Poirel, M. Souli, E. Tacconelli, A. Vatopoulos, G. M. Rossolini, et al. 2007. Metallo-β-lactamases as emerging resistance determinants in gram-negative pathogens: open issues. Int. J. Antimicrob. Agents 29:380-388. [DOI] [PubMed] [Google Scholar]
- 3.Crowder, M. W., J. Spencer, and A. J. Vila. 2006. Metallo-β-lactamases: novel weaponry for antibiotic resistance in bacteria. Acc. Chem. Res. 39:721-728. [DOI] [PubMed] [Google Scholar]
- 4.Docquier, J. D., J. Lamotte-Brasseur, M. Galleni, G. Amicosante, J. M. Frère, and G. M. Rossolini. 2003. On functional and structural heterogeneity of VIM-type metallo-β-lactamases. J. Antimicrob. Chemother. 51:257-266. [DOI] [PubMed] [Google Scholar]
- 5.Frey, J., E. Mudd, and H. Krisch. 1988. A bacteriophage T4 expression cassette that functions efficiently in a wide range of gram-negative bacteria. Gene 62:237-247. [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Saez, I., J. D. Docquier, G. M. Rossolini, and O. Dideberg. 2008. The three-dimensional structure of VIM-2, a Zn-β-lactamase from Pseudomonas aeruginosa in its reduced and oxidised form. J. Mol. Biol. 375:604-611. [DOI] [PubMed] [Google Scholar]
- 7.Iyobe, S., H. Kusadokoro, J. Ozaki, N. Matsumura, S. Minami, S. Haruta, T. Sawai, and K. O'Hara. 2000. Amino acid substitutions in a variant of IMP-1 metallo-β-lactamase. Antimicrob. Agents Chemother. 44:2023-2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laraki, N., N. Franceschini, G. M. Rossolini, P. Santucci, C. Meunier, E. de Pauw, G. Amicosante, J. M. Frère, and M. Galleni. 1999. Biochemical characterization of the Pseudomonas aeruginosa 101/1477 metallo-β-lactamase IMP-1 produced by Escherichia coli. Antimicrob. Agents Chemother. 43:902-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8a.Llarrull, L. L., M. F. Tioni, J. Kowalski, B. Bennett, and A. J. Vila. 2007. Evidence for a dinuclear active site in the metallo-β-lactamase BCII with substoichiometric Co(II): a new model for metal uptake. J. Biol. Chem. 282:30586-30595. [DOI] [PubMed] [Google Scholar]
- 9.Marchiaro, P., M. A. Mussi, V. Ballerini, F. Pasteran, A. M. Viale, A. J. Vila, and A. S. Limansky. 2005. Sensitive EDTA-based microbiological assays for the detection of metallo-β-lactamases in nonfermentative gram-negative bacteria. J. Clin. Microbiol. 43:5648-5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matagne, A., A. M. Misselyn-Bauduin, B. Joris, T. Erpicum, B. Granier, and J. M. Frere. 1990. The diversity of the catalytic properties of class A beta-lactamases. Biochem. J. 265:131-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nordmann, P., and L. Poirel. 2002. Emerging carbapenemases in gram-negative aerobes. Clin. Microbiol. Infect. 8:321-331. [DOI] [PubMed] [Google Scholar]
- 12.Orellano, E. G., J. E. Girardini, J. A. Cricco, E. A. Ceccarelli, and A. J. Vila. 1998. Spectroscopic characterization of a binuclear metal site in Bacillus cereus beta-lactamase II. Biochemistry 37:10173-10180. [DOI] [PubMed] [Google Scholar]
- 13.Pasteran, F., D. Faccone, A. Petroni, M. Rapoport, M. Galas, M. Vazquez, and A. Procopio. 2005. Novel variant (blaVIM-11) of the metallo-β-lactamase blaVIM family in a GES-1 extended-spectrum-β-lactamase-producing Pseudomonas aeruginosa clinical isolate in Argentina. Antimicrob. Agents Chemother. 49:474-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poirel, L., T. Naas, D. Nicolas, L. Collet, S. Bellais, J. D. Cavallo, and P. Nordmann. 2000. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob. Agents Chemother. 44:891-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 16.Tomatis, P. E., R. M. Rasia, L. Segovia, and A. J. Vila. 2005. Mimicking natural evolution in metallo-β-lactamases through second-shell ligand mutations. Proc. Natl. Acad. Sci. USA 102:13761-13766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walsh, T. R., M. A. Toleman, L. Poirel, and P. Nordmann. 2005. Metallo-beta-lactamases: the quiet before the storm? Clin. Microbiol. Rev. 18:306-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.