Abstract
Biapenem is a parenteral carbapenem antibiotic that exhibits wide-ranging antibacterial activity, remarkable chemical stability, and extensive stability against human renal dehydropeptidase-I. Tebipenem is the active form of tebipenem pivoxil, a novel oral carbapenem antibiotic that has a high level of bioavailability in humans, in addition to the above-mentioned features. β-lactam antibiotics, including carbapenems, target penicillin-binding proteins (PBPs), which are membrane-associated enzymes that play essential roles in peptidoglycan biosynthesis. To envisage the binding of carbapenems to PBPs, we determined the crystal structures of the trypsin-digested forms of both PBP 2X and PBP 1A from Streptococcus pneumoniae strain R6, each complexed with biapenem or tebipenem. The structures of the complexes revealed that the carbapenem C-2 side chains form hydrophobic interactions with Trp374 and Thr526 of PBP 2X and with Trp411 and Thr543 of PBP 1A. The Trp and Thr residues are conserved in PBP 2B. These results suggest that interactions between the C-2 side chains of carbapenems and the conserved Trp and Thr residues in PBPs play important roles in the binding of carbapenems to PBPs.
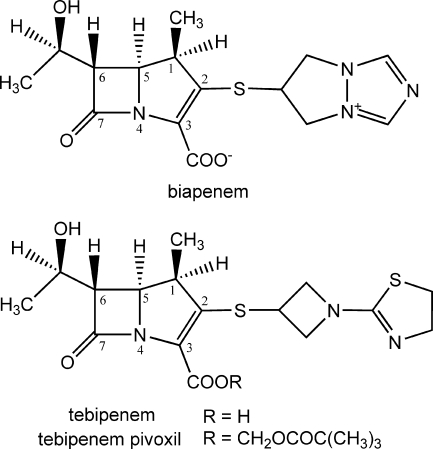
Carbapenems are widely recognized to have broad antibacterial activities. Six parenteral carbapenem antibiotics, imipenem (34), panipenem (11), meropenem (25), ertapenem (19), biapenem (33), and doripenem (1), are used clinically as chemotherapeutic agents to treat severe bacterial infections, and tebipenem pivoxil, a new oral carbapenem, has been developed as the prodrug ester of tebipenem. Biapenem has a σ-symmetric (6,7-dihydro-5H-pyrazolo[1,2-a][1,2,4]triazolium-6-yl)thio group as its C-2 side chain (Fig. 1) (30). Biapenem exhibits a wide range of antibacterial activity encompassing many gram-negative and gram-positive aerobic and anaerobic bacteria, including species producing various β-lactamases, with the exception of class B β-lactamases (4). Biapenem also exhibits remarkable chemical stability and extensive stability against human renal dehydropeptidase-I (36). Tebipenem is the active form of tebipenem pivoxil, and it has the [1-(1,3-thiazolin-2-yl)azetidin-3-yl]thio group at the C-2 position (Fig. 1). Tebipenem pivoxil has a high level of bioavailability in humans, in addition to the above-mentioned features (14, 16, 20).
FIG. 1.
Chemical structures of biapenem and tebipenem, with the atomic numbering scheme of the carbapenem skeleton.
Penicillin-binding proteins (PBPs) are enzymes that catalyze the polymerization and cross-linking of peptidoglycan precursors during bacterial cell wall biosynthesis (12, 26). The PBPs have been divided into three classes. High-molecular-weight (HMW) class A PBPs are bifunctional enzymes with transglycosylase and transpeptidase activities. HMW class B PBPs act only as transpeptidases. The low-molecular-weight PBPs generally act as dd-carboxypeptidases. Streptococcus pneumoniae contains six PBPs: the HMW class A PBPs 1A, 1B, and 2A; the HMW class B PBPs 2B and 2X; and low-molecular-weight PBP 3. In the cross-linking reaction, the transpeptidase or dd-carboxypeptidase must bind to a first peptidoglycan substrate, called the donor strand. The active site Ser residue then attacks the carbonyl carbon atom of the C-terminal d-Ala-d-Ala peptide bond, leading to an acyl-enzyme complex, with subsequent release of the C-terminal d-Ala. The transient acyl-enzyme then has two possible fates: hydrolysis, which releases the shortened peptidoglycan strand (dd-carboxypeptidation), or cross-link formation with an acceptor strand from a neighboring peptidoglycan polymer (transpeptidation) (28). The β-lactam antibiotics inhibit transpeptidase and dd-carboxypeptidase activities by acylating the active-site Ser of PBPs. The active site of PBPs is bordered by three conserved motifs: Ser-X-X-Lys (SXXK), which includes the catalytic Ser; Ser-X-Asn (SXN); and Lys-Thr/Ser-Gly (KT/SG) (12). Amino acid alterations of the PBPs can reduce their affinities for β-lactam antibiotics, resulting in drug resistance. It has been documented that in S. pneumoniae, PBPs 2X, 2B, and 1A are frequently associated with β-lactam resistance (2, 3, 8, 35).
S. pneumoniae R6 PBP 2X (750 residues) is composed of a short cytoplasmic region, a transmembrane region, and a periplasmic unit containing three domains: the N-terminal, transpeptidase, and C-terminal domains. A soluble form of PBP 2X lacking the cytoplasmic and transmembrane regions, as well as a trypsin-digested form of PBP 2X containing the N-terminal, transpeptidase, and C-terminal domains, has been crystallized (13, 32, 38). S. pneumoniae R6 PBP 1A (719 residues) is composed of a short cytoplasmic region, a transmembrane region, and a periplasmic unit containing transglycosylase and transpeptidase domains and a Ser-rich C-terminal tail. A trypsin-digested form of PBP 1A containing a short peptide derived from the transglycosylase domain, the linker region between the transglycosylase and transpeptidase domains, the transpeptidase domain, and the C-terminal tail has also been crystallized (5). However, the crystal structure of a PBP complexed with a carbapenem has not yet been determined; hence, structural information regarding interactions between carbapenems and PBPs to facilitate rational design of new β-lactam antibiotics has remained elusive to date. Here, we present the crystal structures of biapenem and tebipenem complexed with the trypsin-digested forms of PBPs 2X and 1A from the S. pneumoniae R6 strain. All four of the complex structures, which are products of inactivation by carbapenems, exhibit hydrophobic interactions between the C-2 side chains of the carbapenems and the Trp and Thr residues in the active sites of the PBPs. Our results suggest that these hydrophobic interactions play important roles in the binding of carbapenems to PBPs.
MATERIALS AND METHODS
Protein expression and purification.
The soluble PBP 2X (amino acids 49 to 750) from S. pneumoniae strain R6 was expressed, and its trypsin-digested form (amino acids 71 to 238, 241 to 625, and 626 to 750) was purified as described previously (38). The soluble PBP 1A (amino acids 37 to 719 with the Arg545Gln mutation) from S. pneumoniae R6 was expressed, and its trypsin-digested form (amino acids 47 to 70 and 264 to 653) was purified according to previously described methods, with slight modifications (5, 17). Briefly, the gene for the soluble PBP 1A protein was cloned into the pGEX-4T-1 vector (GE Healthcare Biosciences, Piscataway, NJ). The soluble PBP 1A with a glutathione S-transferase tag was expressed in Escherichia coli BL21(DE3) (Novagen, Madison, WI). The cells were disrupted by sonication, and the protein was purified by a glutathione Sepharose 4 fast flow column (GE Healthcare Biosciences). Pooled fractions were dialyzed against 20 mM Tris-HCl (pH 7.9) and 1 mM EDTA. The dialyzed protein was then digested with 0.13 mg/ml trypsin (Sigma-Aldrich, St. Louis, MO) for 1 h at room temperature. The protein was subsequently purified by using MonoQ, Macro-Prep ceramic hydroxyapatite type I, and Superdex 75 columns with methods similar to those described previously for PBP 2X (38) and then concentrated to a final protein concentration of 5 mg/ml.
Crystallization and data collection.
Crystals of PBP 2X-biapenem and -tebipenem complexes were prepared by soaking trypsin-digested PBP 2X crystals for 6 h in a solution containing 5 mg/ml biapenem (Wyeth K.K., Tokyo, Japan) and for 1 h in a solution containing 10 mg/ml tebipenem (Wyeth K. K.), respectively, and diffraction data were collected on the BL32B2 beam line at the SPring-8 synchrotron facility by methods similar to those described previously (38). Crystals of trypsin-digested PBP 1A were grown by the hanging-drop vapor-diffusion method at 293 K. The protein solution (1 μl) was mixed with 3 μl of reservoir solution containing 4 to 6 mM zinc sulfate and 50 mM MES (4-morpholineethanesulfonic acid; pH 6.8) and equilibrated against 500 μl of reservoir solution with 250 μl Al's oil (Hampton Research, Laguna Niguel, CA) as a barrier. Crystals of PBP 1A-biapenem and -tebipenem complexes were prepared by soaking crystals of uncomplexed PBP 1A for 4.5 h in a solution containing 5 mg/ml biapenem and for 4 h in a solution containing 5 mg/ml tebipenem, respectively. The resulting crystals were cryoprotected by using 30% (vol/vol) ethylene glycol. Data were collected at 100 K at a wavelength of 1.0 Å on the BL41XU beam line at the SPring-8 synchrotron facility. All diffraction data were integrated and scaled using the CrystalClear software package (Rigaku Corporation, Tokyo, Japan).
Structure determination and refinement.
The crystals of PBP 2X-biapenem and -tebipenem complexes were isomorphous with those of the uncomplexed PBP 2X (38). The model phases of both complexes were improved by rigid body refinement, using the program Refmac5 (29) with the structure of the uncomplexed PBP 2X (Protein Data Bank code 2Z2L) as a starting model. Model building was performed using the programs Coot (9) and DS Modeling (Accelrys, San Diego, CA). Crystallographic refinement was performed using the programs Refmac5 and CNX (Accelrys). Two PBP 2X molecules (designated molecules 1 and 2) per asymmetric unit were present. The final model of the PBP 2X-biapenem complex includes 1,286 residues (74 to 231, 254 to 619, and 626 to 750 [molecule 1]; 73 to 102, 113 to 169, 175 to 232, 254 to 620, and 626 to 750 [molecule 2]), two products derived from biapenem, one sulfate ion, and 82 water molecules. The final model of the PBP 2X-tebipenem complex includes 1,265 residues (74 to 231, 254 to 619, and 626 to 750 [molecule 1]; 73 to101, 133 to 169, 173 to 232, 254 to 377, 380 to 620, and 626 to 750 [molecule 2]), two products derived from tebipenem, one sulfate ion, and 33 water molecules. The structure of the PBP 1A-tebipenem complex was solved by molecular replacement, using the program Molrep (37) with the structure of the uncomplexed PBP 1A as a search model (Protein Data Bank code 2C6W) (5). The crystal of the PBP 1A-biapenem complex was isomorphous with the crystal of the PBP 1A-tebipenem complex. Two PBP 1A molecules (designated molecules 1 and 2) per asymmetric unit were present. The final model of each complex includes 800 residues (51 to 66 and 267 to 650 in both molecules 1 and 2), two products derived from carbapenems, 8 zinc ions, and 6 and 40 water molecules in the biapenem and tebipenem complexes, respectively. All structures were validated by using the PROCHECK program (23). A summary of statistics from data collection and refinement is given in Table 1.
TABLE 1.
Data collection and refinement statistics
| Data set | Value for indicated complex:f
|
|||
|---|---|---|---|---|
| PBP 2X-biapenem | PBP 2X-tebipenem | PBP 1A-biapenem | PBP 1A-tebipenem | |
| Unit cell parameters | ||||
| a (Å) | 106.79 | 107.31 | 185.70 | 185.75 |
| b (Å) | 171.91 | 172.06 | 50.67 | 50.28 |
| c (Å) | 88.67 | 88.80 | 110.78 | 109.54 |
| Data collection statisticsa | ||||
| Resolution (Å) | 2.50 (2.58-2.50) | 2.80 (2.90-2.80) | 3.00 (3.11-3.00) | 2.70 (2.80-2.70) |
| Total no. of reflections | 282,689 | 231,874 | 73,665 | 131,215 |
| No. of unique reflections | 53,714 | 41,099 | 19,127 | 29,074 |
| Completeness (%) | 92.7 (90.1) | 99.7 (99.1) | 88.1 (91.5) | 100.0 (99.6) |
| Redundancy | 5.3 (4.8) | 5.6 (5.3) | 3.9 (3.6) | 4.5 (4.3) |
| Rmergeb (%) | 5.9 (22.8) | 7.0 (24.0) | 13.0 (35.0) | 12.3 (36.4) |
| <I/σ(I)>c | 18.9 (5.5) | 14.4 (4.9) | 6.2 (2.8) | 7.2 (2.7) |
| Refinement statistics | ||||
| Total no. of atoms | 10,016 | 9,786 | 6,362 | 6,398 |
| Rcrystd (%) | 23.5 | 22.5 | 22.2 | 22.8 |
| Rfreee (%) | 28.1 | 27.4 | 27.6 | 27.1 |
| RMSD from ideal | ||||
| Bond length (Å) | 0.007 | 0.007 | 0.007 | 0.007 |
| Bond angle (°) | 1.0 | 1.0 | 1.0 | 1.0 |
| Avg B factor (Å2) | 48.0 | 60.0 | 50.5 | 42.0 |
| Ramachandran plot | ||||
| Most favored (%) | 88.4 | 87.4 | 87.1 | 89.9 |
| Additional allowed (%) | 11.0 | 12.1 | 12.5 | 9.5 |
| Generously allowed (%) | 0.2 | 0.5 | 0.4 | 0.6 |
| Disallowed (%) | 0.4 | 0.0 | 0.0 | 0.0 |
Values in parentheses are for the shell with the highest resolution.
Rmerge = ΣhklΣi|Ihkl,i − <Ihkl>|/ΣhklΣi<Ihkl>, where Ihkl,i represents the ith measurement of the intensity of the hkl reflection and its symmetry equivalent and <Ihkl> is the average intensity of the hkl reflection.
Averages of the diffraction intensities divided by their standard deviations.
Rcryst = Σhkl‖Fo| − |Fc‖/Σhkl|Fo|, where |Fo| and |Fc| are the observed and calculated structure factor amplitudes, respectively, for reflection hkl.
Rfree is the same as Rcryst for a random 5% of reflections excluded from refinement.
All complexes are in space group P21212.
Protein structure accession numbers.
Atomic coordinates have been deposited in the Protein Data Bank with accession codes 2ZC3 (PBP 2X-biapenem complex), 2ZC4 (PBP 2X-tebipenem complex), 2ZC5 (PBP 1A-biapenem complex), and 2ZC6 (PBP 1A-tebipenem complex).
RESULTS
PBP 2X-carbapenem complex structures.
There were two trypsin-digested PBP 2X molecules (designated molecules 1 and 2) per asymmetric unit in each biapenem and tebipenem complex structure. The electron density within each active site revealed the carbapenem molecule covalently bound to Ser337 (Fig. 2A and B). Although the loop regions (residues 376 to 386) in the transpeptidase domains of molecules 1 and 2 adopted distinct conformations, presumably due to different crystal-packing interactions with the neighboring molecule, the ligands in the active site of each carbapenem complex bound in a substantially similar mode. Thus, for clarity we will refer to molecule 1 of each complex in all further discussion.
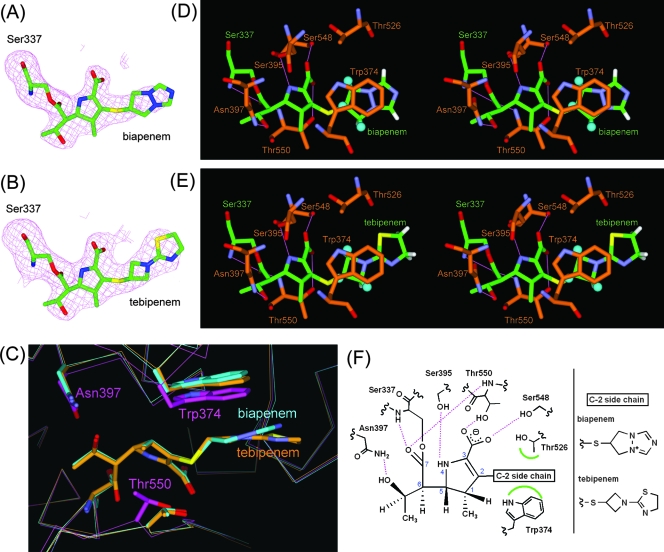
FIG. 2.
PBP 2X-carbapenem complex structures. (A and B) Fo − Fc omit electron density maps calculated for the structures omitting the carbapenem moiety covalently attached to Ser337 in molecule 1 of PBP 2X in complex with biapenem and tebipenem. The maps, depicted as magenta meshes, are contoured at 2.8 σ. Carbapenem-acylated Ser residues are shown as stick models, with carbon atoms colored green, oxygen atoms red, nitrogen atoms blue, and sulfur atoms yellow. (C) Superpositioned active-site regions of the uncomplexed (magenta) and biapenem (light blue)- and tebipenem (orange)-complexed structures. The carbapenems and side chains of selected residues are shown as thick sticks. Atoms are colored as described for panel A, except that the carbon atoms of the carbapenems and selected residues in the uncomplexed and complexed structures are magenta, light blue, and orange, respectively. (D and E) Stereo view of the PBP 2X active site complexed with biapenem and tebipenem, respectively. Atoms are colored as described for panel A, except that the carbon atoms of PBP 2X are orange. The positions of hydrogen atoms of the C-2 side chain were calculated by DS Modeling (Accelrys). The hydrogen atoms that form CH/π interactions are shown as balls (light blue), and other hydrogen atoms are shown as thick sticks (white). Hydrogen bonds are represented by thin lines (magenta). (F) Schematic diagram of the carbapenem binding mode. Hydrogen bonds are represented by broken lines (magenta), and hydrophobic contacts are shown as arcs (green). The atomic numbering scheme of the carbapenem skeleton is also shown.
In each biapenem and tebipenem complex structure, the formation of the complex was accompanied by an induced-fit conformational change at the pocket of the enzyme, and the C-2 side chain of each carbapenem formed characteristic hydrophobic interactions. The Cα atoms of Thr550 in both the biapenem and tebipenem complexes were displaced 1.1 Å toward the opening of the active pocket compared to their positions in the uncomplexed structure (38), and the Cα atoms of Trp374 in the biapenem and tebipenem complexes were displaced 1.0 and 1.1 Å, respectively, toward the opening of the active pockets compared to their positions in the uncomplexed structure (Fig. 2C). The C-2 side chain of each carbapenem participated in hydrophobic interactions with Trp374 and Thr526 (Fig. 2D and E). Three hydrogen atoms in the C-2 side chain of each carbapenem were involved in CH/π interactions (31) with Trp374. The side of the ring in the C-2 side chain formed hydrophobic interactions with Thr526. Concerning the interactions of the carbapenem skeleton, the NH atom at position 4 in the ring formed a hydrogen bond to the Ser395 side chain, and the oxygen atoms in the carboxylate moiety at the third position in the ring formed hydrogen bonds to the side chains of Ser548 and Thr550. The hydroxyethyl group of the C-6 side chain formed a hydrogen bond to Asn397, which corresponded to the hydrogen bond seen in PBPs complexed with cephalosporins (5, 27, 38), in which the amido oxygen in the C-7 side chain of the cephalosporin binds to a conserved Asn residue. A schematic representation of the carbapenem binding mode is shown in Fig. 2F.
PBP 1A-carbapenem complex structures.
There were two trypsin-digested PBP 1A molecules (designated molecules 1 and 2) per asymmetric unit in each biapenem and tebipenem complex structure. Although our crystals were not isomorphous with the previously reported PBP 1A crystals, in which there was only one molecule per asymmetric unit (5), all complexed structures in both asymmetric units of our crystals showed close structural similarity to the earlier uncomplexed structure, with Cα root mean square deviations of 0.59 and 0.49 Å for molecules 1 and 2, respectively, of the biapenem complex, and 0.54 and 0.50 Å for molecules 1 and 2, respectively, of the tebipenem complex. The electron density within each active site revealed the carbapenem molecule covalently bound to Ser370 (Fig. 3A and B). Because the ligands in each active site bind in a substantially similar mode, we will refer only to molecule 1 in all further discussion.
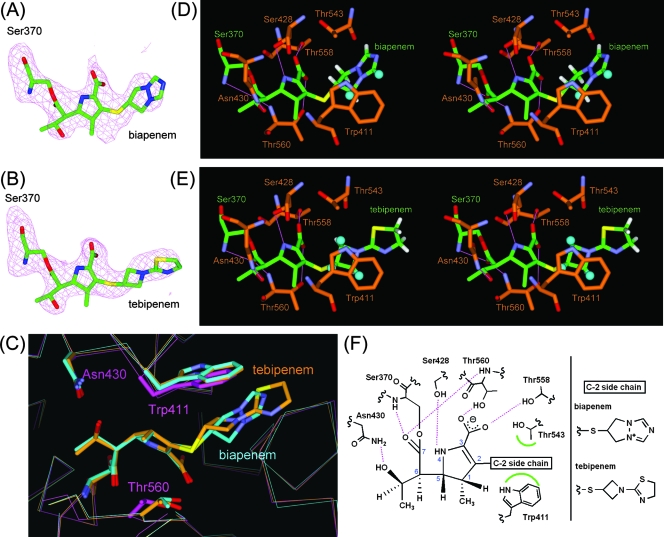
FIG. 3.
PBP1A-carbapenem complex structures. (A and B) Fo − Fc omit electron density maps calculated for the structures omitting the carbapenem moiety covalently attached to Ser370 in molecule 1 of PBP 1A in complex with biapenem and tebipenem. The maps and models are displayed in the same manner as for Fig. 2A and B. (C) Superpositioned active-site regions of the uncomplexed (magenta) and biapenem (light blue)- and tebipenem (orange)-complexed structures. The carbapenems and side chains of selected residues are shown as thick sticks. Atoms are colored as described for Fig. 2C. (D and E) Stereo view of the active sites of PBP 1A in complex with biapenem and tebipenem, respectively. Atoms are colored as described for Fig. 2A and B, except that the carbon atoms of PBP 1A are orange. Hydrogen atoms of the C-2 side chain and hydrogen bonds are displayed in the same manner as for Fig. 2D and E. (F) Schematic diagram of the carbapenem binding mode. Hydrogen bonds are represented by broken lines (magenta), and hydrophobic contacts are shown as arcs (green). The atomic numbering scheme of the carbapenem skeleton is also shown.
In each biapenem and tebipenem complex structure, the formation of the complex was accompanied by an induced-fit conformational change in the pocket of the enzyme, and the C-2 side chain of each carbapenem formed characteristic hydrophobic interactions similar to those noted in the PBP 2X-carbapenem complexes, with the exception of interactions between the C-2 side chain of biapenem and PBP 1A. At the interaction sites for the C-2 side chain of each carbapenem, the Cα atoms of Trp411 were displaced 0.8 and 0.7 Å, respectively, toward the opening of the active pockets compared to their positions in the uncomplexed structure (Fig. 3C), and the C-2 side chains of each carbapenem formed hydrophobic interactions with Trp411 and Thr543 (Fig. 3D and E). However, CH/π interactions with Trp411 were different in each carbapenem complex. In the biapenem complex, the ring in the C-2 side chain was positioned almost perpendicular to the indole ring in Trp411. Two hydrogen atoms on one side of the ring in the C-2 side chain were involved in CH/π interactions with Trp411, and the other side of the ring formed hydrophobic interactions with Thr543. On the other hand, in the tebipenem complex, the four-membered ring in the C-2 side chain was parallel to the indole ring in Trp411, and three hydrogen atoms were involved in CH/π interactions, as seen in the PBP 2X-tebipenem complex. A schematic representation of these interactions is shown in Fig. 3F.
Comparison of the PBP 2X and PBP 1A complexes.
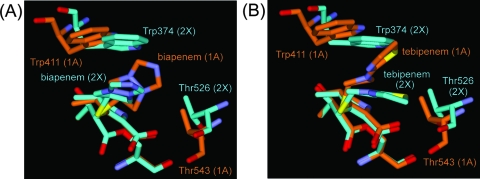
Superpositioning of the Cα atoms of the conserved motifs in the PBP 2X- and PBP 1A-carbapenem complexes (337STMK, 395SSN, and 547KSG in PBP 2X; 370STMK, 428SRN, and 557KTG in PBP 1A) revealed that the carbapenem skeletons overlapped well. However, the C-2 side chains adopted different conformations to accommodate the different positions of the Trp and Thr residues by changing their torsion angles around the sulfur atoms that bound directly to the C-2 carbon atoms in the carbapenem skeletons (Fig. 4A and B). After the atoms were superposited, the Cα distances between Trp374 in PBP 2X and Trp411 in PBP 1A were 1.4 and 1.5 Å in the biapenem and tebipenem complexes, respectively. The Cα distances between Thr526 in PBP 2X and Thr543 in PBP 1A were 1.7 and 1.2 Å in the biapenem and tebipenem complexes, respectively. Consequently, the space between Trp411 and Thr543 in the PBP 1A-carbapenem complexes was slightly wider than that between Trp374 and Thr526 in the PBP 2X-carbapenem complexes. Thus, there is enough space in the active site of PBP 1A, but not 2X, for the C-2 side chain of the carbapenems to sample both conformations: perpendicular and parallel to the indole ring in Trp411. Owing to the different chemical structures of the C-2 side chains, the optimal mode to enhance the binding of the C-2 side chain of biapenem to PBP 1A is probably different from that for tebipenem.
FIG. 4.
Comparison of the PBP 2X and PBP 1A complexes. (A) Superpositioned active-site regions of PBP 2X (light blue)- and PBP 1A (orange)-biapenem complex structures. Biapenem and the side chains of the Trp and Thr residues in the active sites are shown as thick sticks. Atoms are colored as described for Fig. 2A, except that the carbon atoms in PBP 2X- and PBP 1A-carbapenem complex structures are light blue and orange, respectively. (B) Superpositioned active-site regions of PBP 2X (light blue)- and PBP 1A (orange)-tebipenem complex structures. The models are displayed in the same manner as for panel A.
DISCUSSION
We have shown here that the C-2 side chains of biapenem and tebipenem formed hydrophobic interactions with Trp374 and Thr526 in PBP 2X and with Trp411 and Thr543 in PBP 1A after acylation. Because the crystal structures of PBP 2X-meropenem and -imipenem complexes also exhibit similar interactions (M. Yamada and T. Watanabe, unpublished data), these interactions are supposed to be common features of carbapenems. Previous studies have indicated that one of the features of carbapenems is the high chemical reactivity of the carbapenem skeleton (6). On the other hand, other studies have indicated that the C-2 side chains of carbapenems contribute antimicrobial activity, because the carbapenem with only a hydrogen atom as its C-2 side chain has less antimicrobial activity than does imipenem (15). Although no kinetics data regarding the binding of carbapenems to S. pneumoniae PBPs 1A, 2X, and 2B have been published to date, kinetics data for S. pneumoniae PBP 2A are available (39). Imipenem shows a better affinity for PBP 2A than do cephalosporins and penicillins. The k2/K (acylation efficiency) values of PBP 2A for imipenem, cefuroxime, cephalothin, penicillin G, and piperacillin are 13,800, 12,000, 3,200, 640, and 230 (M−1 s−1), respectively. In addition, Davies and colleagues carried out competition assays using a labeled penicillin G (7). They reported that the binding affinities of carbapenems, doripenem, imipenem, and meropenem for PBPs 1A, 2X, and 2B from S. pneumoniae were similar to that of ceftriaxone. Doripenem, imipenem, and meropenem showed a strong affinity for PBPs 1A (50% inhibitory concentrations [IC50s] are 0.007, 0.015, and 0.03, respectively), 2X (IC50s are 0.015, 0.013, and 0.013, respectively), and 2B (IC50s are 0.012, 0.008, and 0.03, respectively) of a penicillin-susceptible S. pneumonia strain. Ceftriaxone bound tightly to PBPs 1A (IC50 = 0.02) and 2X (IC50 = 0.03) not but to PBP 2B (IC50 > 1), as expected, as PBP 2B is not a primary target for cephalosporins. A similar tendency was observed for a penicillin-resistant S. pneumoniae (PRSP) strain, although the binding affinities of all β-lactams were reduced. Although the structures studied here were products of inactivation reactions by carbapenems, hydrophobic interactions between the C-2 side chains of the carbapenems and Trp and Thr residues in the active sites of PBPs 2X and 1A are likely to play a role in drug binding upon acylation.
The crystal structure of PBP 2B from S. pneumoniae revealed that Trp429 and Thr605 occupy positions similar to those of the Trp and Thr residues in the active sites of PBPs 2X and 1A (M. Yamada and T. Watanabe, unpublished data). Moreover, no amino acids besides Trp and Thr have been observed at these positions in PBPs 2X, 1A, and 2B from 40 S. pneumoniae clinical isolates, including PRSP, with the exception of a Thr543Ile mutation in PBP 1A from five strains (35). These results suggest that the C-2 side chains of the carbapenems possibly interact with the Trp and Thr residues in the active sites of PBPs 2X, 1A, and 2B from PRSP.
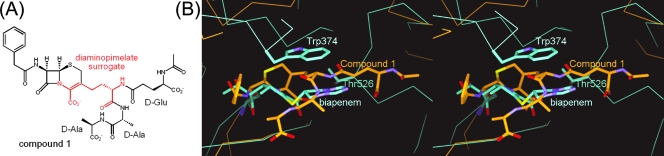
In order to investigate a possible role for the Trp and Thr residues of S. pneumoniae PBPs 2X and 1A in the recognition of the acceptor strand, the structure of the S. pneumoniae PBP 2X-biapenem complex was superimposed on the structure of a Streptomyces sp. strain R61 dd-peptidase/transpeptidase complexed with a cephalosporin that mimics the acceptor strand of a Streptomyces peptidoglycan (Fig. 5A) (24). The superimposed structures showed that part of the diaminopimelate surrogate of the peptidoglycan-mimetic compound was located near Trp374 and Thr 526 of PBP 2X (Fig. 5B). These Trp and Thr residues are highly conserved in PBPs 2X and 1A from S. pneumoniae clinical isolates, as described above. However, the shapes of the binding pockets of S. pneumoniae PBPs 2X and 1A differ from that of the Streptomyces dd-peptidase/transpeptidase. Furthermore, the approaching nucleophile of S. pneumoniae, which is frequently the terminal amino group of an Ala-Ser or Ala-Ala dipeptide on the ɛ-amino group of the stem peptide Lys residue, differs from that of Streptomyces, which is the terminal amino group of a Gly residue on the ɛ-amino group of the stem peptide diaminopimelate (10, 22). Thus, a full understanding of the role of the Trp and Thr residues in the active sites of S. pneumoniae PBPs 2X and 1A in the recognition of the acceptor strand must await further crystallographic and biochemical studies using peptidoglycan-mimetic compounds.
FIG. 5.
(A)Chemical structures of compound 1, a cephalosporin that mimics the acceptor strand of a Streptomyces sp. strain R61 peptidoglycan. A diaminopimelate surrogate is shown in red. (B) Superpositioned active-site regions of the S. pneumoniae PBP 2X-biapenem complex (light blue) and the Streptomyces dd-peptidase/transpeptidase-compound 1 complex (orange) structures. Biapenem, the side chains of the Trp 374 and Thr526 residues, and compound 1 are shown as thick sticks. Atoms are colored as described for Fig. 2A, except that the carbon atoms in PBP 2X and the dd-peptidase/transpeptidase complex structures are light blue and orange, respectively.
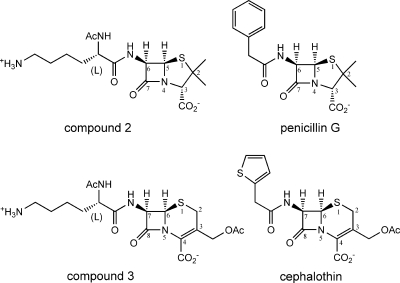
We suspect that the contribution of the C-6 side chain of the carbapenems to the binding of PBPs is less than that of the penicillins and cephalosporins. A previous study indicated that a penicillin with a peptidoglycan-mimetic side chain (Fig. 6) is less effective than penicillin G both in inhibiting soluble PBPs 2X, 1B, and 3 from S. pneumoniae and in antibacterial activity against S. pneumoniae (18, 21). A cephalosporin with a peptidoglycan-mimetic side chain (Fig. 6) is also less effective than cephalothin both in inhibiting a soluble PBP 1B from S. pneumoniae and in antibacterial activity against S. pneumoniae. These results suggest that the chemical structures of the C-6 side chain of penicillin G and the C-7 side chain of cephalothin, but not the peptidoglycan-mimetic side chain, have a striking effect on binding to PBPs from S. pneumoniae. In contrast, the C-6 side chains of carbapenems are smaller than the corresponding side chains of penicillins and cephalosporins, and they form a conserved hydrogen bond to Asn 397 of PBP 2X or Asn 430 of PBP 1A. Thus, additional chemical modifications of the C-6 side chains of carbapenems may enhance their binding affinities for PBPs.
FIG. 6.
Chemical structures of compound 2, compound 3, penicillin G, and cephalothin, with the atomic numbering scheme of the penicillin and cephalosporin skeleton.
Finally, superpositioning of the PBP 2X-carbapenem complex structures with PBP 2X in complex with cefditoren, the cephalosporin antibiotic with a (Z)-2-(4-methylthiazol-5- yl)ethenyl group as its C-3 side chain, revealed that the C-2 side chains of the carbapenems correspond to the C-3 side chain of cefditoren, although a conformational change of the Trp374 side chain occurred only upon cefditoren binding (38). The acquisition of new structural information regarding different interactions between β-lactams and PBPs will be useful for rational designs of new β-lactams.
Acknowledgments
We thank Nobutaka Shimizu and Masahide Kawamoto for help with data collection on the BL41XU beam line at the SPring-8 synchrotron facility and Yoshio Katsuya and Izumi Wada for help with data collection on the BL32B2 beam line at the SPring-8 synchrotron facility. We thank Kazue Nagano for technical assistance with protein expression and purification. We also thank Takao Abe for discussions and comments on the manuscript.
Footnotes
Published ahead of print on 7 April 2008.
REFERENCES
- 1.Anderson, D. L. 2006. Doripenem. Drugs Today (Barc.) 42:399-404. [DOI] [PubMed] [Google Scholar]
- 2.Asahi, Y., Y. Takeuchi, and K. Ubukata. 1999. Diversity of substitutions within or adjacent to conserved amino acid motifs of penicillin-binding protein 2X in cephalosporin-resistant Streptococcus pneumoniae isolates. Antimicrob. Agents Chemother. 43:1252-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barcus, V. A., K. Ghanekar, M. Yeo, T. J. Coffey, and C. G. Dowson. 1995. Genetics of high level penicillin resistance in clinical isolates of Streptococcus pneumoniae. FEMS Microbiol. Lett. 126:299-303. [DOI] [PubMed] [Google Scholar]
- 4.Bonfiglio, G., G. Maccarone, M. L. Mezzatesta, A. Privitera, V. Carciotto, M. Santagati, S. Stefani, and G. Nicoletti. 1997. In vitro activity of biapenem against recent gram-negative and gram-positive clinical isolates. Chemotherapy 43:393. [DOI] [PubMed] [Google Scholar]
- 5.Contreras-Martel, C., V. Job, A. M. Di Guilmi, T. Vernet, O. Dideberg, and A. Dessen. 2006. Crystal structure of penicillin-binding protein 1a (PBP1a) reveals a mutational hotspot implicated in β-lactam resistance in Streptococcus pneumoniae. J. Mol. Biol. 355:684-696. [DOI] [PubMed] [Google Scholar]
- 6.Dalhoff, A., N. Janjic, and R. Echols. 2006. Redefining penems. Biochem. Pharmacol. 71:1085-1095. [DOI] [PubMed] [Google Scholar]
- 7.Davies, T. A., W. Shang, K. Bush, and R. K. Flamm. 2008. Activity of doripenem and comparator β-lactams against US clinical isolates of Streptococcus pneumoniae with defined mutations in the penicillin-binding domains of pbp1a, pbp2b and pbp2x. J. Antimicrob. Chemother. 61:751-753. [DOI] [PubMed] [Google Scholar]
- 8.Dowson, C. G., T. J. Coffey, and B. G. Spratt. 1994. Origin and molecular epidemiology of penicillin-binding-protein-mediated resistance to β-lactam antibiotics. Trends Microbiol. 2:361-366. [DOI] [PubMed] [Google Scholar]
- 9.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Bustos, J., and A. Tomasz. 1990. A biological price of antibiotic resistance: major changes in the peptidoglycan structure of penicillin-resistant pneumococci. Proc. Natl. Acad. Sci. USA 87:5415-5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goa, K. L., and S. Noble. 2003. Panipenem/betamipron. Drugs 63:913-925. [DOI] [PubMed] [Google Scholar]
- 12.Goffin, C., and J. M. Ghuysen. 1998. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol. Mol. Biol. Rev. 62:1079-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordon, E., N. Mouz, E. Duée, and O. Dideberg. 2000. The crystal structure of the penicillin-binding protein 2x from Streptococcus pneumoniae and its acyl-enzyme form: implication in drug resistance. J. Mol. Biol. 299:477-485. [DOI] [PubMed] [Google Scholar]
- 14.Hikida, M., K. Itahashi, A. Igarashi, T. Shiba, and M. Kitamura. 1999. In vitro antibacterial activity of LJC 11,036, an active metabolite of L-084, a new oral carbapenem antibiotic with potent antipneumococcal activity. Antimicrob. Agents Chemother. 43:2010-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imuta, M., H. Itani, H. Ona, Y. Hamada, S. Uyeo, and T. Yoshida. 1991. Carbapenem and penem antibiotics. VI. Synthesis and antibacterial activity of 2-heteroaromatic-thiomethyl and 2-carbamoyloxymethyl 1-methylcarbapenems. Chem. Pharm. Bull. (Tokyo) 39:663-671. [DOI] [PubMed] [Google Scholar]
- 16.Isoda, T., H. Ushirogochi, K. Satoh, T. Takasaki, I. Yamamura, C. Sato, A. Mihira, T. Abe, S. Tamai, S. Yamamoto, T. Kumagai, and Y. Nagao. 2006. Syntheses and pharmacokinetic studies of prodrug esters for the development of oral carbapenem, L-084. J. Antibiot. (Tokyo) 59:241-247. [DOI] [PubMed] [Google Scholar]
- 17.Job, V., A. M. Di Guilmi, L. Martin, T. Vernet, O. Dideberg, and A. Dessen. 2003. Structural studies of the transpeptidase domain of PBP1a from Streptococcus pneumoniae. Acta Crystallogr. D Biol. Crystallogr. 59:1067-1069. [DOI] [PubMed] [Google Scholar]
- 18.Josephine, H. R., P. Charlier, C. Davies, R. A. Nicholas, and R. F. Pratt. 2006. Reactivity of penicillin-binding proteins with peptidoglycan-mimetic β-lactams: what's wrong with these enzymes? Biochemistry 45:15873-15883. [DOI] [PubMed] [Google Scholar]
- 19.Keating, G. M., and C. M. Perry. 2005. Ertapenem: a review of its use in the treatment of bacterial infections. Drugs 65:2151-2178. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi, R., M. Konomi, K. Hasegawa, M. Morozumi, K. Sunakawa, and K. Ubukata. 2005. In vitro activity of tebipenem, a new oral carbapenem antibiotic, against penicillin-nonsusceptible Streptococcus pneumoniae. Antimicrob. Agents Chemother. 49:889-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar, I., H. R. Josephine, and R. F. Pratt. 2007. Reactions of peptidoglycan-mimetic β-lactams with penicillin-binding proteins in vivo and in membranes. ACS Chem. Biol. 2:620-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar, I., and R. F. Pratt. 2005. Transpeptidation reactions of a specific substrate catalyzed by the streptomyces R61 DD-peptidase: characterization of a chromogenic substrate and acyl acceptor design. Biochemistry 44:9971-9979. [DOI] [PubMed] [Google Scholar]
- 23.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 24.Lee, W., M. A. McDonough, L. P. Kotra, Z. H. Li, N. R. Silvaggi, Y. Takeda, J. A. Kelly, and S. Mobashery. 2001. A 1.2-Å snapshot of the final step of bacterial cell wall biosynthesis. Proc. Natl. Acad. Sci. USA 98:1427-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lowe, M. N., and H. M. Lamb. 2000. Meropenem: an updated review of its use in the management of intra-abdominal infections. Drugs 60:619-646. [DOI] [PubMed] [Google Scholar]
- 26.Macheboeuf, P., C. Contreras-Martel, V. Job, O. Dideberg, and A. Dessen. 2006. Penicillin binding proteins: key players in bacterial cell cycle and drug resistance processes. FEMS Microbiol. Rev. 30:673-691. [DOI] [PubMed] [Google Scholar]
- 27.Macheboeuf, P., A. M. Di Guilmi, V. Job, T. Vernet, O. Dideberg, and A. Dessen. 2005. Active site restructuring regulates ligand recognition in class A penicillin-binding proteins. Proc. Natl. Acad. Sci. USA 102:577-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDonough, M. A., J. W. Anderson, N. R. Silvaggi, R. F. Pratt, J. R. Knox, and J. A. Kelly. 2002. Structures of two kinetic intermediates reveal species specificity of penicillin-binding proteins. J. Mol. Biol. 322:111-122. [DOI] [PubMed] [Google Scholar]
- 29.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240-255. [DOI] [PubMed] [Google Scholar]
- 30.Nagao, Y., Y. Nagase, T. Kumagai, H. Matsunaga, T. Abe, O. Shimada, T. Hayashi, and Y. Inoue. 1992. β-Lactams. 3. Asymmetric total synthesis of new non-natural 1β-methylcarbapenems exhibiting strong antimicrobial activities and stability against human renal dehydropeptidase-I. J. Org. Chem. 57:4243-4249. [Google Scholar]
- 31.Nishio, M., M. Hirota, and Y. Umezawa. 1998. The CH/π interaction: evidence, nature, and consequences. WILEY-VCH, New York, NY.
- 32.Pares, S., N. Mouz, Y. Pétillot, R. Hakenbeck, and O. Dideberg. 1996. X-ray structure of Streptococcus pneumoniae PBP2x, a primary penicillin target enzyme. Nat. Struct. Biol. 3:284-289. [DOI] [PubMed] [Google Scholar]
- 33.Perry, C. M., and T. Ibbotson. 2002. Biapenem. Drugs 62:2221-2234. [DOI] [PubMed] [Google Scholar]
- 34.Rodloff, A. C., E. J. Goldstein, and A. Torres. 2006. Two decades of imipenem therapy. J. Antimicrob. Chemother. 58:916-929. [DOI] [PubMed] [Google Scholar]
- 35.Sanbongi, Y., T. Ida, M. Ishikawa, Y. Osaki, H. Kataoka, T. Suzuki, K. Kondo, F. Ohsawa, and M. Yonezawa. 2004. Complete sequences of six penicillin-binding protein genes from 40 Streptococcus pneumoniae clinical isolates collected in Japan. Antimicrob. Agents Chemother. 48:2244-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ubukata, K., M. Hikida, M. Yoshida, K. Nishiki, Y. Furukawa, K. Tashiro, M. Konno, and S. Mitsuhashi. 1990. In vitro activity of LJC10,627, a new carbapenem antibiotic with high stability to dehydropeptidase I. Antimicrob. Agents Chemother. 34:994-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vagin, A., and A. Teplyakov. 1997. MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 30:1022-1025. [Google Scholar]
- 38.Yamada, M., T. Watanabe, T. Miyara, N. Baba, J. Saito, Y. Takeuchi, and F. Ohsawa. 2007. Crystal structure of cefditoren complexed with Streptococcus pneumoniae penicillin-binding protein 2X: structural basis for its high antimicrobial activity. Antimicrob. Agents Chemother. 51:3902-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao, G., T. I. Meier, J. Hoskins, and K. A. McAllister. 2000. Identification and characterization of the penicillin-binding protein 2a of Streptococcus pneumoniae and its possible role in resistance to β-lactam antibiotics. Antimicrob. Agents Chemother. 44:1745-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]