Carbapenem resistance due to the acquisition of metallo-β-lactamase (MβL)-encoding genes has increased rapidly among Pseudomonas aeruginosa strains in Asia, Europe, and Latin America (7). However, these resistance genes are rarely detected in carbapenem-resistant P. aeruginosa strains from North America, particularly in the United States, where only three well-documented occurrences of MβL-producing P. aeruginosa strains have been reported (1, 4, 6).
The first MβL from a P. aeruginosa strain to be identified in the United States was VIM-7. This enzyme was originally detected in a hospital strain from Houston, TX, and showed low homology to other VIM variants (6). VIM-7 was subsequently identified in additional isolates from the same institution (1), but it has not been detected in other regions of the world. In 2003, an outbreak of infection with VIM-2-producing P. aeruginosa was identified in a hospital in Chicago, IL (4), and more recently, an IMP-18-producing P. aeruginosa strain from Las Cruces, NM, was reported (2). In the study presented here, we characterize an IMP-15-producing P. aeruginosa strain isolated in a medical center in Kentucky. This strain likely originated in Mexico.
(This work was presented in part as an abstract at the 44th Annual Meeting of the Infectious Diseases Society of America, Toronto, Canada, 2006.)
A total of 50 P. aeruginosa isolates nonsusceptible to meropenem (MICs, ≥8 μg/ml) were recovered from patients hospitalized at University of Kentucky HealthCare (UKHC), Lexington, from 2001 to 2005 and were selected for the evaluation of MβL production and other mechanisms of resistance. These isolates were subjected to susceptibility testing against 10 antimicrobials (ertapenem, imipenem, meropenem, cefepime, ceftazidime, piperacillin-tazobactam, gentamicin, tobramycin, ciprofloxacin, and tetracycline) by using E-test strips according to the instructions of the manufacturer (AB Biodisk, Solna, Sweden). The results showed elevated MICs of imipenem and meropenem (≥8 μg/ml for both) and ceftazidime (≥16 μg/ml) for 37 isolates (indicating nonsusceptibility), and these isolates were further screened for MβL production by using MβL Etest strips (AB Biodisk). Among these tested strains, the MIC of imipenem for only one strain, 1750J (2.0% overall), exhibited a significant reduction when the carbapenem was combined with EDTA (the MIC decreased from >256 to 8 μg/ml), indicating MβL production. This isolate was resistant to ciprofloxacin (MIC, >32 μg/ml), piperacillin-tazobactam (MIC, >256 μg/ml), gentamicin (MIC, >256 μg/ml), and tobramycin (MIC, >256 μg/ml), in addition to carbapenems (MICs of imipenem, ertapenem, and meropenem, >32 μg/ml) and ceftazidime (MIC, >256 μg/ml).
Isolate 1750J was recovered from a previously healthy 22-year-old Hispanic male who was involved in a motor vehicle accident in March 2005 while in Mexico, where he was initially treated. In July 2005, the patient returned to the United States and noted swelling and pain in the area of the surgical wound. After visiting a local physician, he was referred to UKHC. The patient had surgery and was empirically started on vancomycin plus ampicillin-sulbactam. A methicillin-resistant Staphylococcus aureus isolate, an ampicillin-susceptible Enterococcus faecalis isolate, and a multidrug-resistant P. aeruginosa isolate (susceptible only to polymyxin B) were recovered from bone tissue and drainage fluid collected at the time of the operation. Shortly after the surgical procedure, the patient developed a fever and the antimicrobial treatment was changed to polymyxin B, vancomycin, and ampicillin. The patient subsequently underwent two additional wound debridement procedures and finally became afebrile. Bone tissue specimens were collected for culture at each surgery and yielded the same three pathogens. The patient was discharged in late August 2005 to receive polymyxin B and vancomycin by outpatient therapy for a total of 6 to 8 weeks.
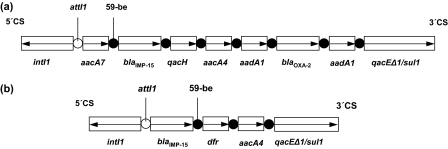
PCR screening of strain 1750J with blaIMP, blaVIM, and blaSPM primers yielded amplicons with blaIMP primers. Both strands of PCR products were sequenced, and nucleotide sequences were analyzed using the Lasergene software package (DNASTAR, Madison, WI) and compared to sequences available on the Internet (http://www.ncbi.nlm.nih.gov/BLAST/). Sequencing revealed that strain 1750J carried blaIMP-15. Primers annealing in the conserved structures of class 1 integrons used in combination with blaIMP primers showed that this MβL gene was located in the second position of a 6-kb integron. The amplification and sequencing of the variable region of this integron showed the presence of six other resistance genes. The first gene cassette in this arrangement was aacA7, followed by blaIMP-15, which was located upstream of qacH, aacA4, and blaOXA-2. The latter gene was flanked on either side by identical copies of aadA1 (Fig. 1a). This class 1 integron also possessed the key elements of the following genetic elements: intI1, located in the 5′ conserved sequence (CS), and qacEΔ1/sul1 in the 3′ CS.
FIG. 1.
Schematic representation of the class 1 integrons containing blaIMP-15: (a) 6-kb integron In95 found in P. aeruginosa isolate 1750J from Lexington, KY, and also in a P. aeruginosa isolate from Guadalajara, Mexico (GenBank accession no. EF184216); (b) blaIMP-15-carrying integron found in a P. aeruginosa isolate in Thailand (GenBank accession no. AY553333). The horizontal arrows indicate the gene cassettes and their respective translation orientations. 59-be, 59-base element.
The gene encoding IMP-15 was initially observed in a P. aeruginosa strain from Thailand (GenBank accession no. AY553333). This MβL gene was carried in the first position of a class 1 integron also harboring a dihydrofolate reductase gene (dfr) and an aminoglycoside acetyltranferase gene (aac) (Fig. 1b), but these findings have not been reported in the literature. More recently, a P. aeruginosa isolate from Guadalajara, Mexico, was found to carry blaIMP-15 (5). This gene was embedded in an integron named In95, with a structure identical to that of the integron in isolate 1750J from Kentucky described here. Epidemiologic typing (by pulsed-field gel electrophoresis) comparing the isolate 1750J with the IMP-15-producing Mexican strain showed that these two isolates were identical (data not shown). This finding suggests that the infection in the patient treated at UKHC was probably acquired in Mexico when the patient was initially injured and hospitalized.
The UKHC facilities initiated a formal antimicrobial control program in 1998. Key interventions have included the restriction of the use of meropenem (as formulary carbapenem only) to situations in which the infection is caused by a documented extended-spectrum β-lactamase-producing organism or pathogens resistant to other reasonable therapeutic alternatives. Since the implementation of this program, the rate of meropenem resistance among P. aeruginosa strains has remained well below the national average of 25% (3), with only 11% of the P. aeruginosa isolates being nonsusceptible to meropenem in 2006.
Carbapenem resistance due to MβL production remains very unusual in the United States, but MβL screening methods (7) should be performed on multidrug-resistant, carbapenem-resistant strains, especially for patients returning from geographic areas where MβL-producing organisms are endemic. The use of these simple tests would facilitate the implementation of appropriate infection control measures and prevent the dissemination of MβL genes, important resistance mechanisms carried on highly mobile genetic elements that facilitate the spread of resistance to numerous antibiotics.
Acknowledgments
We thank the following individuals for assistance in testing and/or manuscript preparation: R. P. Rapp, S. B. Overman, and R. N. Jones.
Footnotes
Published ahead of print on 24 March 2008.
REFERENCES
- 1.Aboufaycal, H., H. S. Sader, K. Rolston, L. M. Deshpande, M. Toleman, G. Bodey, I. Raad, and R. N. Jones. 2007. blaVIM-2 and blaVIM-7 carbapenemase-producing Pseudomonas aeruginosa isolates detected in a tertiary care medical center in the United States: report from the MYSTIC Program. J. Clin. Microbiol. 45:614-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanson, N. D., A. Hossain, L. Buck, E. S. Moland, and K. S. Thomson. 2006. First occurrence of a Pseudomonas aeruginosa isolate in the United States producing an IMP metallo-β-lactamase, IMP-18. Antimicrob. Agents Chemother. 50:2272-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones, R. N., C. Mendes, P. J. Turner, and R. Masterton. 2005. An overview of the Meropenem Yearly Susceptibility Test Information Collection (MYSTIC) Program: 1997-2004. Diagn. Microbiol. Infect. Dis. 53:247-56. [DOI] [PubMed] [Google Scholar]
- 4.Lolans, K., A. M. Queenan, K. Bush, A. Sahud, and J. P. Quinn. 2005. First nosocomial outbreak of Pseudomonas aeruginosa producing an integron-borne metallo-β-lactamase (VIM-2) in the United States. Antimicrob. Agents Chemother. 49:3538-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sader, H. S., L. M. Deshpande, R. Morfin-Otero, U. Garza-Ramos, J. Silva-Sanchez, and R. N. Jones. 2007. Dissemination of IMP-type metallo-β-lactamases among Pseudomonas aeruginosa and emergence of VIM-2 in Klebsiella oxytoca in Mexico: report from the SENTRY Antimicrobial Surveillance Program, abstr. C2-2061, p. 150. Abstr. 47th Intersci. Conf. Antimicrob. Agents Chemother., Chicago, IL.
- 6.Toleman, M. A., K. Rolston, R. N. Jones, and T. R. Walsh. 2004. blaVIM-7, an evolutionarily distinct metallo-β-lactamase gene in a Pseudomonas aeruginosa isolate from the United States. Antimicrob. Agents Chemother. 48:329-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walsh, T. R., M. A. Toleman, L. Poirel, and P. Nordmann. 2005. Metallo-β-lactamases: the quiet before the storm? Clin. Microbiol. Rev. 18:306-25. [DOI] [PMC free article] [PubMed] [Google Scholar]