Abstract
Members of the genera Enterovirus and Rhinovirus (family Picornaviridae) cause a wide range of human diseases. An established vaccine is available only for poliovirus, and no effective therapy is available for the treatment of infections caused by any pathogenic picornavirus. Peptide-conjugated phosphorodiamidate morpholino oligomers (PPMO) are single-stranded DNA-like antisense agents that readily enter cells. A panel of PPMO was tested for their antiviral activities against various picornaviruses. PPMO targeting conserved internal ribosome entry site (IRES) sequence were highly active against human rhinovirus type 14, coxsackievirus type B2, and poliovirus type 1 (PV1), reducing PV1 titers by up to 6 log10 in cell cultures. Comparative sequence analysis led us to design a PPMO (EnteroX) targeting 22 nucleotides of IRES sequence that are perfectly conserved across greater than 99% of all human enteroviruses and rhinoviruses. EnteroX reduced PV1 replication in cell culture to an extent similar to that of other IRES-specific PPMO. Resistant PV1 arose in cell cultures after 12 passages in the presence of EnteroX and were found to have two mutations within the EnteroX target sequence. Nevertheless, cPVR transgenic mice treated once daily by intraperitoneal (i.p.) injection with EnteroX before and/or after i.p. infection with 3 × 108 PFU (three times the 50% lethal dose) of PV1 had an approximately 80% higher rate of survival than the controls. The viral titer in tissues taken at day 5 postinfection showed that animals in the EnteroX-treated group averaged over 3, 4, and 5 log10 less virus in the small intestine, spinal cord, and brain, respectively, than the amount in the control animals. These results suggest that EnteroX may have broad therapeutic potential against entero- and rhinoviruses.
Rhinovirus and Enterovirus are closely related genera within the family Picornaviridae, and both cause significant global disease burdens (34). Infections with human rhinoviruses (HRVs) are rarely fatal, but they are the most prevalent cause of the common cold and are responsible for a great deal of respiratory tract illness, with considerable associated economic loss (11, 22). The genus Enterovirus includes the polio-, coxsackie-, echo-, and enteroviruses and can cause a wide variety of diseases, ranging from non-life-threatening enteric or respiratory illness or rash (hand-foot-and-mouth disease) to severe and potentially fatal aseptic meningitis, encephalitis, neonatal systemic disease, hemorrhagic conjunctivitis, and poliomyelitis (39). Enteroviral infections result in 30,000 to 50,000 hospitalizations yearly in the United States (38). Although effective vaccines for the three serotypes of poliovirus are available, over 100 other enterovirus serotypes (39) and 100 rhinovirus serotypes (11) have been identified, making vaccines and other therapies which rely on an effective adaptive immune response difficult to develop. There are currently no commercially available therapeutics that have been approved by the FDA for use against any of the entero- or rhinoviruses (10, 28).
The picornavirus genome is an approximately 7.5-kb single-stranded RNA molecule of positive polarity possessing a covalently linked protein (VPg) at the 5′ terminus and a 3′-terminal poly(A) tail (2). The 5′ and 3′ untranslated regions (UTRs) flank the single open reading frame and are known to have various important roles in viral translation, RNA synthesis, and virion assembly (7). Picornavirus replication takes place in the cytoplasm of the host cell, and while viral proteins affect nuclear activity (46), no event of the viral life cycle is known to occur in the nucleus. Viral genomic RNA is translated into a single large polyprotein, which is subsequently cleaved into 10 mature proteins by virus-encoded proteases (2, 44). The 5′ UTRs of all picornaviruses are unusually long, averaging about 700 nucleotides (nt), and contains an internal ribosome entry site (IRES), a region that mediates the initiation of viral RNA translation several hundred nucleotides downstream of the 5′ terminus in a cap-independent manner (16). Four classes of picornaviral IRES configurations have been defined on the basis of RNA secondary structure and biological properties (5, 16). Picornavirus species of the genera Enterovirus and Rhinovirus possess a class I IRES, and those of the genera Cardiovirus and Aphtovirus possess a class II IRES. Although their functional roles are apparently identical, the various classes of IRES share little similarity in sequence or secondary structure (5, 44, 61).
Considering the morbidity caused by entero- and rhinoviruses, as well as the modest results of past drug development efforts and the paucity of FDA-approved drugs, new approaches to the development of therapeutics are needed. The small-molecule compounds in development thus far appear to have limited promise, due variously to high toxicity or low efficacy, stability, or solubility (28). A number of compounds designed to inhibit picornavirus infections through an antisense-mediated mechanism have generated positive preclinical results but are still early in the development process. Antisense phophorothioate DNA has been used against coxsackievirus type B3 (CVB3) in vitro (57) and in vivo (62), antisense RNA has been used against foot-and-mouth disease virus (45), and small interfering RNA has been used against several picornaviruses (3, 20, 21, 26, 42, 47), all with some success. Phosphorodiamidate morpholino oligomers (PMO) are composed of individual subunits consisting of a DNA base connected to a morpholine ring and phophorodiamidate-linked backbone (50). PMO are uncharged at physiological pH and are highly nuclease resistant and water soluble. PMO interfere with gene expression by base pairing to complementary RNA sequence, thereby forming a steric block (48, 49). PMO designed as antisense agents against positive-strand viruses often target viral mRNA sequence involved in one or more of the major early events in translation: preinitiation, ribosomal complex scanning of the 5′ UTR, or initiation at the AUG start site (14, 54, 63). PMO have been conjugated to various cell-penetrating peptides (CPPs) in order to enhance both PMO uptake into cells (14, 33, 63) and antisense efficacy (35). Peptide PMO (PPMO) have demonstrated considerable efficacy against a number of RNA viruses (14, 19, 27, 36, 40, 54), including foot-and-mouth disease virus (53) in cell cultures and against Ebola virus (17), West Nile virus (13), murine coronaviruses (9), and CVB3 (63) in both cell culture and animal models.
For this study, PPMO were designed against HRV type 14 (HRV14) and human poliovirus 1 (PV1), with each individual PPMO targeting a different viral RNA sequence of interest. In cell culture challenges, each PPMO produced a specific level of antiviral activity, ranging from none to high. The most highly active PPMO in each set were those designed to target sequence in the 3′ region of the IRES of the respective target virus. One such PPMO, HRV-EnteroX, designed to target sequence in the IRES that is highly conserved across human entero- and rhinoviruses, was active in cell culture experiments not only against HRV14 but also against clinical isolates of CVB2 and PV1. A nearly identical PPMO, PV-EnteroX, effectively inhibited PV1 replication in infected mice and is a candidate for development as a therapeutic agent with potentially broad utility.
MATERIALS AND METHODS
PPMO synthesis.
PMO is a single-stranded DNA analog with a morpholine ring in place of each riboside moiety and phosphorodiamidate instead of phosphorodiester intersubunit linkages (50). All PMO were synthesized at AVI BioPharma Inc. by previously described methods (50). An arginine-rich CPP, either R9F2C or (RXR)4XB (abbreviated P3 and P7, respectively, in this report; where R is arginine, F is phenylalanine, C is cysteine, X is 6-aminohexanoic acid, and B is beta-alanine), was covalently conjugated to the 5′ end of each PMO to produce PPMO. The synthesis, conjugation, purification, and analysis of all PPMO were identical to procedures described previously (1, 33). Each PPMO in this study was 19 to 24 bases in length. Two sets of PPMO were designed: one set complementary to target sequences in the HRV14 genome and the other set complementary to the PV1 (Mahoney strain) genome. See Table 1, Fig. 1, and the Results for the PPMO sequences and target locations. To control for the non-sequence-specific activity of the two types of PPMO chemistry, a nonsense control PPMO (Scr) sequence with an ∼50% G+C content was randomly generated (Table 1), screened against both primate mRNA and picornaviral sequences by using the BLAST program (http://www.ncbi.nlm/.nih.gov/BLAST/), and prepared in a manner identical to that of the antisense PPMO. Lyophilized PPMO were diluted to 2 mM with filter-sterilized distilled water and were stored at 4°C.
TABLE 1.
PPMO sequences and targets
| PPMOa | Viral target region | PPMO sequence (5′-3′) | Location of targetb |
|---|---|---|---|
| HRV-5′hp | 5′ UTR hairpin | CAAAGGTACATAGTACCAGAG | 49-69 |
| HRV-EnteroX | IRES stem-loop 5 | GAGAAACACGGACACCCAAAGTAG | 551-574 |
| HRV-cre | cre | GTCTGTTTCGTTTTCTCAACG | 2359-2379 |
| HRV-AUG | AUG of HRV14/ΔP1Luc | GGCGTCTTCCATGATCACAGT | NAc,d |
| PV-5′term | 5′ genomic terminus | GGTACAACCCCTGTGCTGTTTTAA | 1-24 |
| PV-L4 | IRES stem-loop 4 | CAACGCAGCCTGGACCACCGTCAC | 343-366 |
| PV-L5 | IRES stem-loop 5 | CCTCGGACTTGCGCGTTACG | 512-531 |
| PV-AUG | Initiator AUG | GCACCCATTATGATACAATTG | 730-750 |
| PV-cre | cre | TACGGTGTTTGCTCTTGAACT | 4461-4481 |
| PV-stop | Open reading frame stop codon | TCGACTGAGGTAGGGTTACTA | 7370-7390 |
| PV-pA | 3′ genomic terminus | TTTTTTTCTCCGAATTAAA | 7429-7440+ |
| PV-EnteroX | 3′ base of IRES stem-loop 5 | GAAACACGGACACCCAAAGTAG | 541-562 |
| Scr | Nonsense (negative control) | AGTCTCGACTTGCTACCTCA | NA |
HRV-PPMO were designed against HRV14 (GenBank accession number K02121). PV-PPMO were designed against PV1 (Mahoney strain; GenBank accession number V01149).
The position of the target sequence in the viral genome.
NA, not applicable.
Chimeric sequence: 3′ end of HRV 5′UTR and 5′ end of Luc coding sequence.
FIG. 1.
PPMO target sites in viral RNA and sequence conservation in the stem-loop 5 region of human entero- and rhinoviruses. (A) Schematic of a composite entero- and rhinovirus genome. The VPg protein is indicated by a black dot at the 5′ end of the genome; and the names of other features of the viral genome are in italicized font, including the cloverleaf region (′CL') and the other five stem-loop regions of the 5′ UTR (denoted with roman numerals), the initiator AUG, and the poly(A) tail. The bracketed region before the AUG represents the ∼100-nt difference in length between the HRV and PV1 5′ UTRs. PPMO target sites are in nonitalicized font and are indicated by thin lines and arrows. (B) Histogram showing sequence conservation in the stem-loop 5 region of human entero- and rhinoviruses. A total of 196 full-length genomic sequences were aligned and analyzed (see Materials and Methods). An arbitrary scale is depicted at the left of the histogram, where 1.0 denotes perfect agreement at a nucleotide position across all entries. The relative target sites of the two versions of EnteroX and the PV-L5 PPMO are shown above the histogram.
Cells, viruses, and plasmids.
HeLa cells (ATCC CCL 2.2) were grown in tissue culture flasks in Dulbecco's modified Eagle medium (DMEM)-nutrient mixture F-12 (Ham F-12 medium) (1:1; DMEM-F-12; CellGro) supplemented with 2 mM l-glutamine, 1 mM sodium pyruvate, 100 U/ml of penicillin and streptomycin, and 10% newborn calf serum (GibcoBRL). SK-N-MC cells (ATCC HTB-10) and Vero cells (ATCC CCL-81) were grown in tissue culture flasks in Eagle minimum essential medium supplemented with 2 mM l-glutamine and Earle's balanced salt solution adjusted to contain 1.5 g/liter sodium bicarbonate, 0.1 mM nonessential amino acids, and 1.0 mM sodium pyruvate; 100 U/ml of penicillin and streptomycin; and 10% newborn calf serum. Full-length HRV14 was obtained from an infectious clone as described previously (31). Clinical isolates of CVB2 and PV1 (provided by T. Chonmaitree, Department of Pediatrics, UTMB, Galveston, TX) were identified by serology and were passaged once in Vero and once in HeLa cells prior to their experimental use. The CVB2 and PV1 clinical isolates were not subject to sequence analysis. Plasmid pRib(+)XpA contains the cDNA of PV1 (Mahoney strain) preceded by a cis-acting hammerhead ribozyme sequence and the T7 RNA polymerase promoter (23) and can generate infectious PV1 RNA. pRib(+)XpA-derived PV1 was used throughout this study, except where indicated. Plasmid pRib(+)RLuc contains the cDNA of a PV1 replicon in which the P1 region encoding the capsid proteins is substituted with the gene encoding firefly luciferase (Luc) preceded by the hammerhead ribozyme sequence and the T7 polymerase promoter sequence (23). The ribozyme sequence has been shown to promote the enhanced replication of the viral sequence after transfection (23). Both of the plasmids described above are designed to generate replicable synthetic RNAs. Plasmid pRib(+)T7Luc contains the coding sequence for firefly Luc preceded by the hammerhead ribozyme sequence and the T7 polymerase promoter sequence (23). Plasmid HRV14/ΔP1Luc contains a cDNA of HRV14 in which the P1 region encoding capsid proteins is substituted with firefly Luc coding sequence (31).
Picornavirus sequence alignment and analysis.
A total of 196 full-length genomic sequences for human entero- and rhinoviruses obtained through the NCBI website (http://www.ncbi.nlm.nih.gov/genomes/VIRUSES/12058.html) were aligned (6) and analyzed with MUSCLE multiple-sequence alignment software (http://phylogenomics.berkeley.edu/cgi-bin/muscle/input_muscle.py).
Cell viability assays.
HeLa cells grown in either 24- or 96-well plates to ∼80% confluence were treated with water (mock treatment) or 1 to 100 μM PPMO in serum-free growth medium (a 1:1 mixture of DMEM-F-12) for 24 h or with 10 μM PPMO in DMEM-F-12 for 10 to 69 h. Cell viability was then measured by using the CellTiter 96 AQueous one solution cell proliferation [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS)] assay (Promega) according to the manufacturer's instructions. Cells were incubated with MTS solution for 4 h, and the absorbance was measured at 492 nm with an enzyme-linked immunosorbent assay reader. The percent survival of the PPMO-treated cells was based on a comparison to the survival of mock-treated cells (which was set equal to 100%).
PPMO treatment of virus-infected cell cultures and plaque assays.
The experiments with HRV14 and clinical isolates of CVB2 and PV1 were performed with nearly confluent HeLa cells and employed PPMO applied only as a postinfection (p.i.) treatment. Cells were inoculated with HRV14 at a multiplicity of infection (MOI) of 0.03, with CVB2 at an MOI of 0.05, or with PV1 at an MOI of 0.05. After a 1-h infection period, the inoculum was removed and the cells were replenished with virus growth medium (with 10% calf serum) containing water (as a mock treatment) or PPMO and were incubated at 37°C (except for HRV14, which was incubated at 34°C) in 5% CO2. The viral titers in the culture supernatants were determined at 24 h p.i. (PV1 and CVB2) or 48 h p.i. (HRV14), as described below.
The experiments with cDNA-derived PV1 were performed with nearly confluent HeLa cells and employed preinfection treatment with PPMO, unless otherwise noted. The cells were treated with serum-free growth medium containing water (mock treatment) or PPMO for 6 h (unless indicated otherwise) and were then rinsed with phosphate-buffered saline (PBS) and infected with PV1 at the indicated MOI in serum-free growth medium; after 1 h, the viral inoculum was removed and fresh growth medium with serum was added to the cells. At various time points p.i., the supernatants and cell lysates were collected and stored at −80°C. Viral titer determinations were performed by the plaque assay by seeding HeLa cells into six-well plates (1 × 106 cells/well) and incubating the plates for 24 h at 37°C (34°C for HRV14). Cells at ∼90% confluence were washed with PBS and then overlaid with 250 μl of serial 10-fold dilutions of the supernatant from either clarified cell cultures or tissue lysates. The cells were incubated for 1 h, the supernatants were removed, and the cells were overlaid with 2 ml of sterilized 1% agar and DMEM-F-12 or tragacanth and DMEM. The cells were then incubated at 37°C (34°C for HRV14) for 36 h (PV1 and CVB2) or 72 h (HRV14), fixed with 2% formaldehyde for 30 min, and then stained with 1% crystal violet. The plaques were then counted and the viral PFU/ml was calculated.
In vitro transcription of viral reporter RNAs.
Noncapped PV1-Luc [from Rib(+)RLuc] and HRV14-Luc RNA (from HRV14/ΔP1Luc) transcripts were obtained by using a T7 Megascript transcription kit (Ambion), while capped Luc RNA transcripts [from pRib(+)T7Luc] were obtained by using the mMessage mMachine T7 kit (Ambion) after plasmid linearization. The integrity of the resulting RNAs was confirmed by gel electrophoresis, the RNAs were quantified by spectrophotometry, and the RNA concentration was adjusted to 20 μg/μl.
Evaluation of viral translation by Luc assay.
To investigate the effects of PPMO on the translation of PV1-Luc RNA inside cells, HeLa cells were treated for 4 h with water (mock treatment) or 10 μM PPMO and were then washed three times with PBS and suspended in PBS at 5 × 106 cells/ml. Next, 800 μl (4 × 106) cells was added to a 0.4-cm cuvette with 20 μg of viral reporter RNA and electroporated at 300 V, a 500-μF capacitance, and a 24-Ω resistance with an Electro Cell Manipulator 600 (BTX). Transfected cells were added to 2.2 ml of medium containing 10 μM PPMO or water either with or without 2 mM guanidine hydrochloride (GuaHCl). As a control group, samples of transfected cells were treated with 10 μg/ml puromycin. Next, the transfected cells were dispensed into 24-well plates (at 0.5 ml/well), incubated at 37° in 5% CO2, and at the indicated times harvested for measurement of Luc activity as described previously (23). Briefly, cells were scraped from the wells, clarified, and resuspended in 100 μl of cell culture lysis buffer (Promega); and 10 μl was then analyzed with a Luc assay reagent (Promega) in a luminometer.
To investigate the effects of PPMO on the translation of HRV-Luc RNA in a cell-free assay, in vitro-transcribed HRV-Luc RNA was used to program rabbit reticulocyte lysate (Promega) in the presence of various concentrations of HRV14- or HRV14/Luc-specific PPMO. The results were charted as the relative Luc activity of the indicated PPMO at the indicated molar excess to RNA compared to that in mock-treated control reactions. Each reaction mixture contained the same amount of RNA. Ten-microliter reaction mixtures were incubated at 30°C for 1 h before 1 μl was removed for mixture with 100 μl Luc assay reagent and quantification in a luminometer.
Generation, isolation, and sequencing of PPMO-resistant virus and construction of recombinant PV1.
PV1 (MOI, 0.1) was sequentially passaged in HeLa cells under conditions similar to those used in the virus inhibition experiments described above (in “PPMO treatment of virus-infected cell cultures and plaque assays”) by using 10 μM PPMO (unless noted otherwise) until a complete cytopathic effect (CPE) was observed (24 to 96-plus h). The cells and virus were then frozen and thawed three times; the virus was then clarified, the titer determined, and the virus used to initiate the next round of infection. Infections were repeated independently six times with each PPMO treatment and three times for the mock-treated controls. Subsequently, HeLa cells (2 × 106) were infected with either passage 3 (P3) or P12 PPMO-escape-mutant virus at an MOI of 10 for 1 h at 37°C in 5% CO2. The cells were rinsed with PBS to remove the remaining virus, and DMEM-F-12 medium was added. At 4 h p.i., the medium was removed, the cells were rinsed twice with ice-cold PBS, and total cytoplasmic RNA was collected in cytoplasmic lysis buffer (PBS with 0.5% Triton X-100). The RNA was purified with phenol-chloroform-isoamyl alcohol and quantified by spectrophotometery, and cDNA was synthesized with a ThermoScript reverse transcription-PCR system (Invitrogen) and poly(dT) primers. The PCR-amplified cDNA was used for direct sequencing (Elim Biopharmaceuticals, Inc.), and multiple samples were analyzed by using multiple-sequence alignment and Chromas software. To create PV1 cDNA plasmids containing mutations observed in the viral PPMO-escape-mutant populations, the various mutations were recombined into pRib(+)XpA by site-directed mutagenesis with the QuikChange mutagenesis XLII kit (Stratagene), and appropriate primers were then transformed in SURE electroporation-competent cells (Stratagene). The mutant virus plasmid nomenclature is based on the following: virus type, the PPMO treatment that generated the mutant, and the nucleotide identity and the location of nucleotide mutational substitutions (with position 1 being at the 5′ end of the PPMO target site in the virus) (e.g., PV1/L4/A21 is a PV1 mutant generated with the PV-L4 PPMO and has an A substitution at nt 21 of the target).
One-step-growth curve analysis of escape mutants.
To determine single-cycle growth characteristics, wild-type (wt) and mutagenized plasmids were used to synthesize viral RNA by in vitro transcription reactions (as described above). Cells were transfected (as described above), immediately transferred to 2 ml of medium containing 10 μM PV-EnteroX, and incubated at 37° in 5% CO2 until complete CPE was observed. The cell lysates and supernatants were then clarified as described above, and the viral titers were determined. HeLa cells (2 × 106) subject to treatment with 10 μM PPMO or left untreated were infected with the P0 of wt or mutant PV1 at an MOI of 10. Following infection, progeny virus were harvested at various time points by the freeze-thaw method, and the titers on fresh monolayers were determined by plaque assay, as described above.
PPMO treatment and viral infection of mice.
To determine survival rates, ∼10-week-old poliovirus receptor transgenic (cPVR) mice (12) were treated for 48 and 24 h before infection, in addition to 1 h p.i., and every 24 h afterwards for 5 days (eight treatments in total) or were treated after infection only (six treatments) with either PV-EnteroX PPMO, Scr P7-PMO, or PBS. Eighteen mice per group were given i.p. injections with 500 μl sterile PBS or 200 μg (∼10 mg/kg of body weight) of PPMO in 500 μl sterile PBS by using 5-ml syringes. It was previously shown that multiple doses of ∼10 mg/kg PPMO were well tolerated and were effectively antiviral when they were administered to mice either intravenously (63) or i.p. (13). The mice were infected with 3 × 108 PFU of wt virus (approximately three times the 50% lethal dose [12]) and were monitored daily for the onset of paralysis and euthanized when nearly total paralysis was reached. A group of six noninfected cPVR mice received the same regimen of PV-EnteroX PPMO as the infected animals (one dose per day for 8 consecutive days), and three noninfected cPVR mice received no PPMO. The appearance and behavior of all mice were monitored daily. All mice were housed at the UCSF Mission Bay Animal Care Facility and were cared for in accordance with UCSF IACUC guidelines.
For the tissue tropism experiments, 18 mice per group were treated and infected as described above. On days 2, 3, and 5 p.i., six mice from each group were euthanized and their tissues were removed, washed, and placed on dry ice. The organs were immediately homogenized, and the homogenates were clarified and frozen at −80°C. The titers of virus on HeLa cells in the tissue homogenates were subsequently determined by standard plaque assay. Viral RNA from the muscle of PV-EnteroX-infected mice was isolated by TRIzol extraction; and the cDNA was then synthesized, sequenced, and analyzed as described above.
Statistical analysis.
Data analysis was carried out with Prism 4 software (GraphPad Software, Inc.). The results are expressed as means ± standard deviations.
RESULTS
PPMO design.
A set of four PPMO (prepared with P3 peptide) was designed to target specific RNA sequences in HRV14 or HRV14/ΔP1Luc, while a second set of eight PPMO (prepared with P7 peptide) was designed against PV1 genomic sequence. All PPMO were designed to base pair with viral positive-strand genomic RNA sequences thought to be important for different events in viral translation, RNA synthesis, or encapsidation (Table 1 and Fig. 1A). Three of the four HRV14 PPMO and four of the eight PV1 PPMO were designed to target sequences in the respective viral 5′ UTRs. In entero- and rhinoviruses, the 5′-most 90 nt or so of the genome form a region of highly ordered structure referred to as the “cloverleaf,” or domain I. It has been shown that this region of RNA interacts with viral proteins, forming a ribonucleoprotein complex, and is integral to the choreography of viral RNA synthesis (reviewed in reference 41). PV-5′term targets the 5′-terminal 24 nt of the PV1 genome, and HRV-5′hp targets 21 of the 27 nt that constitute the HRV cloverleaf stem-loop d, a region especially critical in ribonucleoprotein formation (41).
The class I IRES region comprises positions from approximately nt 130 to nt 600 of the 5′ UTR and folds into five domains (domains II to VI), based on RNA secondary structure (reviewed in reference 16). PPMO PV-L4 targets a sequence in domain IV of PV1, while PV-L5 and PV-EnteroX target the domain V region of PV1 and HRV-EnteroX targets the domain V region of HRV (Fig. 1A and B). Both domain IV and domain V are essential for the recruitment and positioning of the various components required for viral translation, such as the 40S ribosomal subunit, initiator tRNA, and a number of translation-initiation-associated proteins (16). The IRES-targeting PPMO in this study were designed to interfere with the interaction of various translation-initiation factors with the viral IRES and thereby disrupt the process of translation. Specifically, PV-L4 was designed to bind just 3′ of a poly(C) tract known to interact with the cellular protein PCBP2 (55, 56). PV-L5 was designed to bind within domain V, including the 14-nt stem-loop (nt 511 to 524) structurally adjacent to a sequence previously shown to be involved both in the recruitment of the canonical translation initiation factor eIF4G (37) and in the reduction of neurovirulence of the Sabin vaccine strains of poliovirus (through single-nucleotide mutations in this region) (30). PV-EnteroX and HRV14-EnteroX target a sequence that is highly conserved across both human entero- and rhinoviruses (Fig. 1B) at the 3′ base region of stem-loop 5 in the IRES. The PV-AUG PPMO was designed to target the sequence flanking and including the translation initiator AUG codon of PV1 genomic RNA. Although multiple AUG codons are present in the 5′ UTR of picornaviruses, each viral species employs one predominant AUG for translation initiation, and its identity in HRV14 (51) and PV1 (15) has been established. PPMO HRV-AUG is complementary to the 9 nt at the 3′ terminus of the HRV14 5′ UTR and the first 12 nt of the Luc-coding sequence present in HRV14/ΔP1Luc.
The genomic RNA of entero- and rhinoviruses contains an RNA stem-loop structure of 60 to 80 nt, the cis-acting replication element (cre), within the protein-coding region, which is required for RNA synthesis. HRV cre and PV1 cre differ in their sequences, structures, and locations within their respective genomes, yet they have been shown to have similar functions, namely, urydylylation of the peptide primer VPg (41). PPMO HRV-cre and PV-cre were designed to base pair with sequences located within the cre genes of HRV and PV1, respectively.
Two PPMO were designed to disrupt critical structures in the 3′-terminal region of the viral genome. PPMO PV-stop was designed to target the PV translation stop codon region and thereby interfere with efficient translation termination. The poly(A) tail at the 3′ end of picornaviral RNA is thought to participate in the initiation of negative-strand RNA synthesis (24, 41) and in virus RNA stability (43). PPMO PV-pA is designed to duplex with the 3′-terminal 12 nt of the PV genome and the first 7 A residues of the poly(A) tail.
PPMO cause little cytotoxicity at antiviral concentrations in cell cultures.
To evaluate the effect of PPMO on cell viability, noninfected HeLa cells were treated with the various P7 PMO used in this study. Figure 2A shows the results obtained with Scr, PV-L4, PV-L5, and PV-L4 and PV-L5 in combination over a concentration range of 5 to 25 μM. The results shown in Fig. 2A are representative of those obtained with the other P7 PMO as well (data not shown). After 24 h of treatment at a concentration of 10 μM, none of the P7 PMO caused more than a 10% reduction of cell viability, as measured by the MTS assay (Fig. 2A and data not shown). A similar experiment was performed with P3 PMO. HeLa cells cultured under the same conditions employed in the antiviral experiments with P3-PMO described below had less than a 10% impact on cell viability after 24 h of treatment with 1 to 10 μM of any of the P3 PMO (data not shown).
FIG. 2.
Evaluation of PPMO cytotoxicity in cell cultures. (A) HeLa cell viability was measured by the MTS assay after 24 h of treatment with the indicated concentrations of the indicated PPMO. (B) HeLa cells were incubated with 10 μM of the indicated PPMO, and the cell viability then measured at the indicated times after the initiation of treatment. The percentage of viable cells was obtained by comparison to the number of mock (water)-treated cells, which was set equal to 100%. Each treatment in both of the experiments is the average for 12 wells.
To determine if longer periods of treatment with P7 PMO had any effect on cell viability, we used the MTS assay with cells after 10 to 69 h of treatment with 10 μM PPMO. With the exception of PV-L4, none of the PPMO reduced the MTS assay values by more than 10% at any time point; PV-L4 caused an approximately 15 to 20% reduction in the viability of cells exposed to it for 48 h or more (Fig. 2B).
PPMO inhibition of multiple enteroviruses.
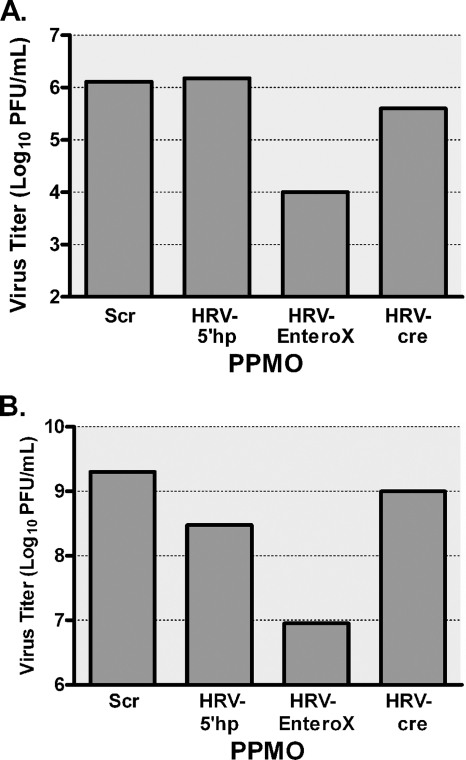
Initially, we determined the effectiveness of the HRV-specific PPMO against HRV14 and against the clinical isolates of PV1 and CVB2. HeLa cells were treated with PPMO at 2.5 or 5 μM after infection with the respective viruses. Virus inhibition was measured by a standard plaque assay at 24 h p.i. (PV1 and CVB2) or 48 h p.i. (HRV14). Of the PPMO tested, only HRV-EnteroX significantly inhibited the viral titers (Fig. 3A and B and data not shown), producing an approximately 2-log10 reduction when used at 5 μM. It was not surprising that HRV-5′hp and HRV-cre had little efficacy against CVB2 or PV1, since these viruses share little homology with HRV at the target sites of these PPMO. The viral titers obtained with Scr PPMO were equivalent to those of infected and mock-treated cells (data not shown).
FIG. 3.
EnteroX PPMO inhibits HRV14 and CVB2 in cell cultures. HeLa cells were treated with 5 μM of the indicated PPMO after a 1-h period of infection with HRV14 at an MOI of 0.03 (A) or a clinical isolate of CVB2 at an MOI of 0.05 (B). Viral titers were determined from samples taken at 24 h p.i. (CVB2) or 48 h p.i. (HRV14) by plaque assay. In both of the experiments, mock (water)-treated cells produced titers that were almost identical to those produced by Scr PPMO-treated cells. The average of two experiments is shown.
The antiviral efficacy of each of the PV-PPMO against cDNA-derived PV1 (Mahoney strain) was determined. HeLa cells were pretreated for 6 h with PPMO at 2, 5, and 10 μM before infection with PV1. Virus inhibition was measured by standard plaque assay at 10 h p.i. Under these conditions, PPMO targeting the IRES region were highly effective, resulting in 2- to 3-log10 reductions at 10 μM (Fig. 4A and data not shown). The other PPMO produced few antiviral effects.
FIG. 4.
PPMO targeting the IRES sequence inhibit PV1, and PPMO can be used in combination to more effectively limit PV1 infections of cell cultures. HeLa cells were incubated with 10 μM of the indicated PPMO before and after infection with PV1 (MOI, 0.1), and the viral titers at 10 h p.i. (A) or the indicated time points p.i. (B) were determined by plaque assay. In each of the experiments, the average of three experiments is shown. (C) HeLa cells were treated with the indicated concentrations of PV-EnteroX before and after infection with PV1 (MOI, 0.1). Viral titers were determined by the plaque assay at 21 h p.i. (▪). As well, the viability of noninfected HeLa cells was measured by the MTS assay 21 h after the initiation of treatment with the indicated concentrations of PV-EnteroX (▴). The average for triplicate samples is shown.
Previous reports indicated that treatment with two PPMO was more effective than treatment with a single PPMO at inhibiting influenza A virus (19) or porcine reproductive and respiratory syndrome virus (40). We therefore examined whether treatment with two PPMO could be more effective than treatment with a single PPMO at inhibiting PV1. Indeed, use of a combination of two PPMO (PV-L4 and PV-L5) resulted in a greater reduction in the viral titer (by over 1 log10 at some time points) than that obtained with an amount of either constituent PPMO that was equimolar to that used in the combination treatment (Fig. 4B and data not shown). Certain other combinations of PPMO (PV-L4-EnteroX and PV-L5-EnteroX) yielded similar results (data not shown).
The selectivity index (SI), which is measured as a ratio of the concentration of a compound that is lethal to 50% of the cultured cells divided by the concentration of a compound that is lethal to 50% of the virus in cell culture, or the 50% cytotoxic concentration (CC50)/50% inhibitory concentration (IC50), is a standard method for scoring the relationship between the efficacy and toxicity of a potential therapeutic agent. Because the target of PV-EnteroX is almost perfectly conserved across human entero- and rhinoviruses and the targets of PV-L4 and PV-L5 are not, we determined the SI of PV-EnteroX. To find the IC50 of PV-EnteroX, HeLa cells were infected with PV1 and treated p.i. with 2 to 100 μM of PV-EnteroX, and the viral titers were than determined (by plaque assay) from the supernatant taken 21 h later (Fig. 4C). The IC50 was determined to be 2 μM. Similar results under similar experimental conditions were obtained with SK-N-MC cells (a human neuroepithelioma cell line) (data not shown). To determine the CC50 of PV-EnteroX for HeLa cells in cell culture, noninfected HeLa cells were treated with various concentrations of PV-EnteroX for 21 h before the MTS assay (Fig. 4C). The CC50 of PV-EnteroX under these conditions was 52 μM. Similar results were obtained with SK-N-MC cells (data not shown). The SI of PV-EnteroX was therefore determined to be 26 (52 μM/2 μM) under these conditions.
IRES-targeting PPMO specifically inhibit translation.
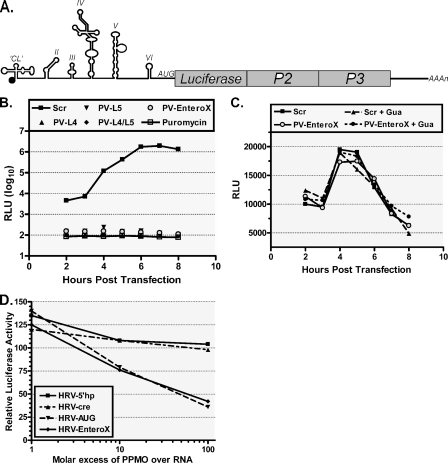
To investigate the antiviral mechanism of action of the effective PPMO, HeLa cells were treated with various PPMO and then transfected with PV1-Luc RNA [transcribed from pRib(+)RLuc in vitro] (Fig. 5A), which utilizes IRES-mediated translation, or with Luc RNA [from pRib(+)T7Luc], which utilizes cap-dependent translation. Transfected cells were then placed in medium containing either PPMO, PPMO and GuaHCl, or puromycin, and translational activity was determined at 1-h intervals after transfection by measuring Luc production. GuaHCl is an inhibitor of PV1 replication (52), while puromycin is an inhibitor of translation (60), and both were used as control reagents in this experiment. As measured by the level of Luc production, treatment with 10 μM PV-EnteroX (as well as other PV1-IRES-directed PPMO) inhibited the translation of PV1-Luc RNA as effectively as treatment with 10 μg/ml puromycin (Fig. 5B and data not shown). In contrast, neither PV-EnteroX nor any of the other PV-PPMO inhibited the translation of Luc RNA (Fig. 5C), indicating that the PPMO inhibition of translation was sequence specific and not global in nature. We also examined the ability of PPMO to inhibit the IRES-mediated translation of HRV-Luc RNA produced from HRV14/ΔP1Luc in rabbit reticulocyte lysates. Only PPMO targeting regions of sequence thought to be directly involved in the process of translation (HRV-EnteroX and HRV-AUG) were able to inhibit translation effectively, whereas PPMO designed to target sequences thought to participate in the regulation of RNA synthesis (HRV-5′hp and HRV-cre) were not effective (Fig. 5D). Interestingly, the PV-AUG PPMO, designed against the AUG region of PV1, had little activity against PV1 RNA (Fig. 4A), whereas the HRV-AUG PPMO, directed against the AUG region of HRV14/ΔP1Luc, was quite effective (Fig. 5D). We note that HRV14/ΔP1Luc consists of a chimeric HRV-Luc sequence in the initiator AUG region (see “PPMO design” above and Table 1).
FIG. 5.
PPMO targeting the IRES and AUG region of viral reporter replicon RNA restrict translation in a potent and specific manner. (A) Schematic of the PV1-Luc replicon pRib(+)RLuc. (B) In-cell translation assay. Cells treated with indicated PPMO at 10 μM were transfected with PV1-Luc RNA [produced from pRib(+)RLuc] and then treated with GuaHCl (Gua), an inhibitor of viral replication. The cells were then assayed for their Luc levels at the indicated times. The translation of PV1-Luc RNA was severely limited by PPMO that specifically target the IRES, as measured by a decrease in the total reflective light units (RLU). The level of suppression produced by antisense PPMO was similar to that observed with puromycin, a known inhibitor of translation. (C) PPMO failed to inhibit the translation of capped Luc RNA [produced from pRib(+)T7Luc] under the same conditions described above for panel B, indicating that PPMO inhibition of translation is sequence specific. (D) Cell-free Luc assay. HRV14-Luc RNA transcripts (produced from HRV14/ΔP1Luc; see Materials and Methods) and the indicated PPMO were mixed in rabbit reticulocyte lysate translation reaction mixtures. The Luc activity is charted in comparison to that from the mock-treated control reactions. PPMO targeting the IRES and AUG region effectively suppressed the translation of HRV-Luc RNA, while PPMO targeting HRV elements thought to have a regulatory role in HRV RNA synthesis did not suppress translation. The average of two experiments is shown in each graph.
PV1 escapes the antiviral effects of PPMO by target-specific mutations.
RNA viruses such as PV1 are well known for their ability to escape the effects of antiviral compounds through mutations that change the drug target site. We explored whether PV1 could become resistant to treatment with some of the PPMO described in this study. PV1 was repeatedly passaged in HeLa cells in the presence of PV-L4, PV-L5, PV-L4 and PV-L5 in combination, or PV-EnteroX PPMO. PV1 variants resistant to PPMO PV-L4 or PV-L5 were isolated after three passages (Fig. 6A and Table 2). PV1 variants resistant to the two-PPMO treatment (i.e., PV-L4 and PV-L5) or to PV-EnteroX were isolated after 12 passages. With the PV-L4 and PV-L5 combination treatment, during some passages, unusually long periods after infection (96-plus h) were required to produce CPE. Also, PPMO at 5 μM, a concentration below that which had been highly effective in the viral inhibition experiments described above, was required in the first two passages to obtain escape mutants from the PV-L4 and PV-L5 combination treatment.
FIG. 6.
PV1 can escape the antiviral effects of PPMO in cell cultures through target-specific mutations. (A) PV1 (MOI, 0.1) was sequentially passaged in HeLa cells in the presence of 5 to 10 μM PPMO until complete CPE was observed. Virus was then isolated and used to initiate the next round of infection. After 12 passages, PV-EnteroX-resistant PV1 was isolated and sequenced. Two nucleotide mutations were found in the PV-EnteroX target region, and these were then cloned either singly or doubly into pRib(+)XpA. Mutant clones (see Materials and Methods for the nomenclature) were used to generate mutant viral RNA, which was transfected into HeLa cells treated with 10 μM PV-EnteroX (EX) and allowed to grow until CPE was observed. These P0 populations were then used at an MOI of 10 to infect HeLa cells that either had not (B) or had (C) been treated with 10 μM PV-EnteroX. Virus was taken at the indicated times (hours) p.i., and the titers determined by plaque assay. The viral titers shown are the averages of three experiments.
TABLE 2.
Comparison of PPMO-resistant PV1 recovered from PPMO-treated HeLa cells
| Virus source/PPMO | No. of passagesa | Location(s) of mutant nucleotidesb | Emergent nucleotide sequenceb |
|---|---|---|---|
| PV1/PV-L4 | P0 | GUGACGGUGGUCCAGGCUGCGUUG (343-366) | |
| PV1/PV-L4 | P3 | 363 | --------------------A--- |
| PV1/PV-L4 | P12 | 345, 363 | --A-----------------A--- |
| PV1/PV-L5 | P0 | CGUAACGCGCAAGUCCGAGG (512-531) | |
| PV1/PV-L5 | P3 | 529 | -----------------U-- |
| PV1/PV-L5 | P12 | 517, 529 | -----U-----------U-- |
| PV1/PV-EnteroX | P0 | CUACUUUGGGUGUCCGUGUUUC (541-562) | |
| PV1/PV-EnteroX | P3 | ---------------------- | |
| PV1/PV-EnteroX | P12 | 544, 558 | ---U-------------A---- |
| No PPMO | P12 | ---------------------- |
PV1 was passaged and selected in the presence of the indicated PPMO, as described in Materials and Methods and Results. The passages were conducted in the presence of the indicated PPMO.
Numbering is based on the PV1 (Mahoney) genomic sequence. Dashes denote the same nucleotide identity as the original sequence.
To investigate the determinant(s) of resistance to PPMO, viral RNA was isolated from resistant populations and the entire PV1 genome of each population was sequenced. This analysis showed specific PV1 mutations in the PPMO target regions. Interestingly, after PV1 was passaged in six independent series with each of the four PPMO treatments, only a single mutation was found after 3 passages and two mutations were found after 12 passages. PV1 treated with the PPMO PV-L4 generated two unique escape mutations, both of which were G-to-A nucleotide changes at residues near the 5′ and 3′ ends of the PPMO target site. Mutant virus PV1/L4/A3 was present after 3 passages, and PV1/L4/A21 was present after 12 passages. PV1 treated concurrently with both PV-L4 and PV-L5 also contained those same two mutations after P12. PV1 treated with PPMO PV-L5 alone also generated only two mutations, one in the 5′ end of the PPMO target (PV1/L5/U6), which was present after 3 passages, and one in the 3′ end of the PPMO target (PV1/L5/U18), which was present after 12 passages. In the PV-EnteroX-treated populations, mutations were also found toward the 5′ and 3′ ends of the PPMO target site. No mutations were found in the target site of virus treated with PV-EnteroX until P12. Of the six different PV-EnteroX-generated mutations found, three were G-to-A mutations that generated C-A mismatches, two were C-to-U changes that led to G-U mismatches, and one was an A-to-U mutation that caused a T-U mismatch (Table 2).
To establish whether any or all of these mutations were indeed the determinants of escape from PPMO inhibition, we produced recombinant viral plasmids containing either single or double PPMO escape mutations. Mutant plasmids were confirmed by sequencing and were used to generate viral RNA that was transfected into HeLa cells pretreated with the appropriate PPMO. Each P0 population of mutant virus was then evaluated in growth kinetics experiments. In the absence of PPMO, all of the mutant viruses grew to or near the wt virus levels (Fig. 6B and data not shown). In the presence of PPMO, all mutants produced much higher titers than the wt virus did (Fig. 6C) and reached titers similar to the wt virus titer in the absence of PPMO (compare Fig. 6C with Fig. 6B), indicating that the mutations that they contained were responsible for escape from the action of the PPMO tested.
PV-EnteroX inhibits PV1 production in vivo and protects mice from a lethal infectious dose of virus.
Since some PPMO tested were highly effective at inhibiting PV1 infections in cell cultures, we explored whether PV1 infection could also be inhibited in vivo in a mouse model. cPVR transgenic mice (18 per group) were treated with sterile PBS or 200 μg (∼10 mg/kg) of either the Scr or the PV-EnteroX PPMO at 48 and 24 h before i.p. infection with wt PV1. After infection, the mice were further treated with PBS or 200 μg of PPMO at 1 h p.i. and daily on days 1 to 5 p.i. Six mice from each treatment group were killed on days 2, 3, and 5 p.i., and tissue samples were collected from typically infected organs for viral titer determination by plaque assays. Interestingly, while the viral titers in some tissues, such as the large intestine, spleen, and muscle, were similar between the PV-EnteroX-treated group and the control groups (which were treated with PBS or Scr PPMO), other tissues, such as small intestine, spinal cord, and brain, from the PV-EnteroX-treated mice showed greatly decreased viral titers compared to those in the control groups (Fig. 7A to C and data not shown). The experiment described above was repeated under the same conditions for the evaluation of animal survivorship. Mice treated with PV-EnteroX were strongly protected from the paralytic symptoms of infection (data not shown) and from subsequent death (Fig. 7D). Sixteen of 18 mice In the PBS-treated group died; in contrast, only 1 of 18 mice in the PV-EnteroX-treated group died. Noninfected/nontreated and noninfected/PV-EnteroX-treated mice displayed normal behavior and appearance throughout the duration of the survivorship experiment. In a subsequent experiment, employing the same conditions described above except without the two preinfection treatments, PV-EnteroX PPMO protected five of six mice from death, whereas five of six mice in both the PBS- and Scr-treated groups died by day 8 p.i. (data not shown).
FIG. 7.
PV-EnteroX PPMO treatment limits viral titers in infected tissues and protects PV1-infected mice from death. Viral titers from the small intestines (A), spinal cords (B), and brain tissues (C) of cPVR mice receiving i.p. treatment with PBS or 200 μg (∼10 mg/kg) of the indicated PPMO at 48 and 24 h before, 1 h after, and then once a day on days 1 to 5 after intraperitoneal infection with 1 × 108 PFU of wt poliovirus (n = 6 per group per time point) are shown. (D) Survival plot of cPVR mice after treatment and infection, as described above (n = 18 per group).
DISCUSSION
Although vaccines effective against poliovirus were introduced in the mid-1950s and early 1960s, poliomyelitis is still endemic in some areas of the world (4) and outbreaks of vaccine-derived poliovirus are a continuing problem (32). Additionally, the incidence of other medically significant enterovirus infections is increasing (38), and rhinovirus-associated illness remains an inscrutable feature of the public health landscape worldwide. A therapeutic agent that could contribute to the moderation of disease caused by any of these viruses would be a welcome development.
PPMO inhibit viral replication in cell cultures at noncytotoxic doses.
In this study, PPMO targeting IRES sequences were highly effective against infectious virus in cell cultures at noncytotoxic doses, producing titer reductions of up to 6 log10 compared to the titers for the controls (Fig. 4A and B). IRES-directed PPMO specifically reduced viral replication whether they were applied to cultured cells before or soon after the virus inoculation period. Importantly, the EnteroX PPMO inhibited not only DNA-derived PV1 and HRV14 but also clinical isolates of PV1 and CVB2. These results are in agreement with those of Yuan et al., who productively targeted CVB3 with a PPMO of nearly identical sequence to that of the EnteroX PPMO (63). HRV-EnteroX and PV-EnteroX both target sequence in the stem-loop 5 region of the IRES and are identical, except that HRV-EnteroX has two additional bases at the 5′ end compared to the sequence of PV-EnteroX (Table 1). The two additional bases of HRV-EnteroX are not as well conserved across strains of entero- and rhinoviruses as the other 22 bases (which comprise PV-EnteroX) (Fig. 1B). PV-EnteroX matches the targeted genomic sequence of over 99% of the entero- and rhinoviruses used in our alignment.
All of the PPMO used in this study were shown to have low levels of or no cytotoxicity at the doses that produced potent antiviral activity in two types of cells in culture, Vero cells and SK-N-MC cells, a cell line of human neurogenic origin (Fig. 2A and B, Fig. 4C, and data not shown). An SI of 26 was determined for PV-EnteroX after a 21-h-p.i. treatment period in cell culture (Fig. 4C). However, it is likely that a shorter treatment time would yield a substantially higher SI (19). Future study will likely include evaluation of PV-EnteroX PPMO made with alternate CPPs, in the hopes of improving the SI. We also found that the use of a combination of PPMO can be more effective than the use of a single PPMO (Fig. 4B). Even so, while multiple PPMO had antiviral efficacy superior to that of a single PPMO in cell cultures in some instances, we found that PV-EnteroX by itself was highly effective at inhibiting PV1 infection in mice (Fig. 7). The dosing regimen of P7 PMO that we employed was apparently subtoxic in our study with cPVR mice, as it had been in a previous study with C3H/HeN mice (13).
PPMO mechanism of action is through interference with viral translation.
Our results with reporter RNAs suggest that the mechanism of action by which the IRES-targeting PPMO worked was by sequence-specific interference in the process of translation (Fig. 5). Interestingly, two highly and nearly equally effective PPMO, PV-L4 and PV-L5 (Fig. 4B), target regions with quite different secondary structures. While the target of PV-L5 consists primarily of the open region of stem-loop 5, the target of PV-L4 is extensively paired with the opposite side of the stem-loop 4 structure. Whether PV-L4 is able to invade the RNA secondary structure in this area and/or binds before stem formation occurs in nascent genomes is presently not known. Interestingly, PV-L4 targets the stem sequence just 3′ of the known PCB2 binding site (55, 56). Whether the PV-L5 and PV-EnteroX target sites are protein binding sites is not presently known. We note, however, that mutations in the PV-L5 target region have previously been shown to affect viral growth in neuronal cells (29).
PV1 mutates in response to PPMO treatment in cell cultures.
The high mutation rate of RNA viruses raises the concern of potential viral escape from the action of any antiviral compound being considered as a prospective therapeutic agent. As we were never able to reduce the number of virions to less than ∼103 in cell culture after a single passage with PPMO treatment at a noncytotoxic dose and since we know that the mutation rate for poliovirus is approximately two mutations per genome per replication cycle, we hypothesized that if escape mutants could arise in the population, they would likely be apparent after few passages. Interestingly, while escape mutants generated by treatment with PV-L4 or PV-L5 alone arose after P3, the PV-L4 and PV-L5 combination or PV-EnteroX treatment did not generate escape mutants until after P12. This delay suggests that treatment with a combination of PPMO may be less likely than that with a single PPMO to produce resistant virus and that the EnteroX target region in PV1 is critical in nature (as also suggested by its high degree of sequence conservation). We speculate that the escape mutations observed emerged de novo during our experiments, as a result of PPMO treatment, rather than from propagation of preexisting mutations in input virus. The input PV1 was produced from cDNA by transcription with T7 RNA polymerase, which has an average error frequency of about 1 in 2 × 104 (25), and with only one exception (mutant PV1/L5/U18), none of the observed mutations appeared in our search of the GenBank database (data not shown). Mutations in the PPMO target sites of PPMO-resistant PV1 were shown to be the determinants of resistance (Fig. 6C).
Surprisingly, none of the PPMO escape mutations appeared to have a strongly attenuating affect on PV1 growth in cell culture, although the titers of the PV-EnteroX escape mutants never quite equaled the titer of the wt virus (Fig. 6B). An interesting characteristic of all of the PPMO escape mutants found after 12 passages was that they each contained two mutations, with one mutation in the 5′ end region and the other in the 3′ end region of the PPMO target site. Mutations in the terminal regions, along with the observation that the majority of PV1 mutations were either G to A or C to U, suggest that specific architectural and structural properties of a PPMO target duplex affect the efficacy of PPMO. While both the propensity for PV1 to generate escape mutants in the presence of PPMO in cell cultures and the ability of recombinant virus with but a single mutation in the target region to resist the action of PV-EnteroX (Fig. 6C) are noteworthy, the likelihood that PV1 resistant to PV-EnteroX will arise in a therapeutic setting in vivo is not clear at this point. We observed that although PV1 was able to escape the effects of PV-EnteroX after multiple passages in cell culture, treatment with the same PPMO protected mice from a usually lethal dose of PV1. Sequenced virus recovered from the muscle tissue of PV-EnteroX-treated mice that survived PV1 infection showed no mutations in the region of the EnteroX target site (data not shown). As well, the high level of protection provided to PV1-infected animals by PV-EnteroX treatment further indicates that compensatory mutations did not occur or that the mutations induced yielded virus variants with poor fitness in vivo.
EnteroX PPMO treatment of mice resulted in reduced PV1 titers in tissues of the central nervous system and protection from a lethal outcome.
Poliovirus is naturally transmitted only between humans, although nonhuman primates and cPVR mice are susceptible to parenteral infection. The infection resulting from i.p. inoculation of cPVR mice has been shown to closely mimic the infection seen in humans and other primates (12, 58, 59). In humans, poliovirus first infects and replicates in the gut mucosa and from there drains into cervical and mesenteric lymph nodes and the blood, causing a transient viremia (8). Usually, poliovirus causes minor nonspecific symptoms such as fever and malaise; however, in ∼1 to 2% of infected individuals, the virus invades the central nervous system, and the subsequent death of motor neurons within the spinal cord can lead to muscle paralysis and sometimes death.
PPMO have been documented to localize in the liver (9) and in the gut and muscle (18) when they are delivered i.p. and in the heart, kidney, and pancreas when delivered intravenously (63). It is unclear whether PPMO can enter the cells of the nervous system in vivo. Even so, PV-EnteroX effectively limited the PV1 titer in both the spinal cord and the brain (Fig. 7B and C), perhaps due to the concomitant decrease in the amount of virus in the small intestine (Fig. 7A), an organ known to be a primary site of poliovirus replication early in the course of an infection. Importantly, mice that received only p.i. treatments with PPMO had a rate of survivorship similar to that of mice that received both preinfection and p.i. treatments, and noninfected/PPMO-treated mice appeared to suffer no ill effects. Further studies need to be carried out to characterize the antiviral efficacies of various p.i. treatment regimens.
The results of this study show that the PV-EnteroX PPMO is highly effective at inhibiting PV1 infections in mice and that this PPMO appears to have therapeutic potential against a broad spectrum of human enteroviruses and rhinoviruses.
Acknowledgments
We thank the Chemistry Group at AVI Biopharma for the production of all PPMO used in this study, Robert Blouch and Yiming Bao for expert technical assistance, and Stanley Lemon for critical discussion.
This work was supported by an NIH fellowship to J.K.S.
Footnotes
Published ahead of print on 17 March 2008.
REFERENCES
- 1.Abes, S., H. M. Moulton, P. Clair, P. Prevot, D. S. Youngblood, R. P. Wu, P. L. Iversen, and B. Lebleu. 2006. Vectorization of morpholino oligomers by the (R-Ahx-R)4 peptide allows efficient splicing correction in the absence of endosomolytic agents. J. Control. Release 116:304-313. [DOI] [PubMed] [Google Scholar]
- 2.Agol, V. I. 2002. Picornavirus genome: an overview, p. 127-148. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, DC.
- 3.Ahn, J., E. S. Jun, H. S. Lee, S. Y. Yoon, D. Kim, C. H. Joo, Y. K. Kim, and H. Lee. 2005. A small interfering RNA targeting coxsackievirus B3 protects permissive HeLa cells from viral challenge. J. Virol. 79:8620-8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aylward, R. B. 2006. Eradicating polio: today's challenges and tomorrow's legacy. Ann. Trop. Med. Parasitol. 100:401-413. [DOI] [PubMed] [Google Scholar]
- 5.Bakhshesh, M., E. Groppelli, M. M. Willcocks, E. Royall, G. J. Belsham, and L. O. Roberts. 2008. The picornavirus avian encephalomyelitis virus possesses a hepatitis C virus-like internal ribosome entry site element. J. Virol. 82:1993-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bao, Y., S. Federhen, D. Leipe, V. Pham, S. Resenchuk, M. Rozanov, R. Tatusov, and T. Tatusova. 2004. National center for biotechnology information viral genomes project. J. Virol. 78:7291-7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bedard, K. M., and B. L. Semler. 2004. Regulation of picornavirus gene expression. Microbes Infect. 6:702-713. [DOI] [PubMed] [Google Scholar]
- 8.Bodian, D., and D. H. Horstmann. 1965. Polioviruses, p. 430-473. In F. L. Horsfall and I. Tamm (ed.), Viral and rickettsial infections of man. Lippincott, Philadelphia, PA.
- 9.Burrer, R., B. W. Neuman, J. P. Ting, D. A. Stein, H. M. Moulton, P. L. Iversen, P. Kuhn, and M. J. Buchmeier. 2007. Antiviral effects of antisense morpholino oligomers in murine coronavirus infection models. J. Virol. 81:5637-5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charles, C. H., M. Yelmene, and G. X. Luo. 2004. Recent advances in rhinovirus therapeutics. Curr. Drug Targets Infect. Disord. 4:331-337. [DOI] [PubMed] [Google Scholar]
- 11.Couch, R. B. 2001. Rhinoviruses, p. 777-798. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, vol. 2, 4th ed. Lippincott, Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 12.Crotty, S., L. Hix, L. J. Sigal, and R. Andino. 2002. Poliovirus pathogenesis in a new poliovirus receptor transgenic mouse model: age-dependent paralysis and a mucosal route of infection. J. Gen. Virol. 83:1707-1720. [DOI] [PubMed] [Google Scholar]
- 13.Deas, T. S., C. J. Bennett, S. A. Jones, M. Tilgner, P. Ren, M. J. Behr, D. A. Stein, P. L. Iversen, L. D. Kramer, K. A. Bernard, and P. Y. Shi. 2007. In vitro resistance selection and in vivo efficacy of morpholino oligomers against West Nile virus. Antimicrob. Agents Chemother. 51:2470-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deas, T. S., I. Binduga-Gajewska, M. Tilgner, P. Ren, D. A. Stein, H. M. Moulton, P. L. Iversen, E. B. Kauffman, L. D. Kramer, and P. Y. Shi. 2005. Inhibition of flavivirus infections by antisense oligomers specifically suppressing viral translation and RNA replication. J. Virol. 79:4599-4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dorner, A. J., L. F. Dorner, G. R. Larsen, E. Wimmer, and C. W. Anderson. 1982. Identification of the initiation site of poliovirus polyprotein synthesis. J. Virol. 42:1017-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ehrenfeld, E., and N. L. Teterina. 2002. Initiation of translation of picornavirus RNAs: structure and function of the internal ribosome entry site, p. 159-169. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, DC.
- 17.Enterlein, S., K. L. Warfield, D. L. Swenson, D. A. Stein, J. L. Smith, C. S. Gamble, A. D. Kroeker, P. L. Iversen, S. Bavari, and E. Muhlberger. 2006. VP35 knockdown inhibits Ebola virus amplification and protects against lethal infection in mice. Antimicrob. Agents Chemother. 50:984-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fletcher, S., K. Honeyman, A. M. Fall, P. L. Harding, R. D. Johnsen, J. P. Steinhaus, H. M. Moulton, P. L. Iversen, and S. D. Wilton. 2007. Morpholino oligomer-mediated exon skipping averts the onset of dystrophic pathology in the mdx mouse. Mol. Ther. 15:1587-1592. [DOI] [PubMed] [Google Scholar]
- 19.Ge, Q., M. Pastey, D. Kobasa, P. Puthavathana, C. Lupfer, R. K. Bestwick, P. L. Iversen, J. Chen, and D. A. Stein. 2006. Inhibition of multiple subtypes of influenza A virus in cell cultures with morpholino oligomers. Antimicrob. Agents Chemother. 50:3724-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gitlin, L., S. Karelsky, and R. Andino. 2002. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature 418:430-434. [DOI] [PubMed] [Google Scholar]
- 21.Gitlin, L., J. K. Stone, and R. Andino. 2005. Poliovirus escape from RNA interference: short interfering RNA-target recognition and implications for therapeutic approaches. J. Virol. 79:1027-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greenberg, S. B. 2003. Respiratory consequences of rhinovirus infection. Arch. Intern. Med. 163:278-284. [DOI] [PubMed] [Google Scholar]
- 23.Herold, J., and R. Andino. 2000. Poliovirus requires a precise 5′ end for efficient positive-strand RNA synthesis. J. Virol. 74:6394-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herold, J., and R. Andino. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang, J., L. G. Brieba, and R. Sousa. 2000. Misincorporation by wild-type and mutant T7 RNA polymerases: identification of interactions that reduce misincorporation rates by stabilizing the catalytically incompetent open conformation. Biochemistry 39:11571-11580. [DOI] [PubMed] [Google Scholar]
- 26.Kahana, R., L. Kuznetzova, A. Rogel, M. Shemesh, D. Hai, H. Yadin, and Y. Stram. 2004. Inhibition of foot-and-mouth disease virus replication by small interfering RNA. J. Gen. Virol. 85:3213-3217. [DOI] [PubMed] [Google Scholar]
- 27.Kinney, R. M., C. Y. Huang, B. C. Rose, A. D. Kroeker, T. W. Dreher, P. L. Iversen, and D. A. Stein. 2005. Inhibition of dengue virus serotypes 1 to 4 in Vero cell cultures with morpholino oligomers. J. Virol. 79:5116-5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Magden, J., L. Kaariainen, and T. Ahola. 2005. Inhibitors of virus replication: recent developments and prospects. Appl. Microbiol. Biotechnol. 66:612-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malnou, C. E., T. A. Poyry, R. J. Jackson, and K. M. Kean. 2002. Poliovirus internal ribosome entry segment structure alterations that specifically affect function in neuronal cells: molecular genetic analysis. J. Virol. 76:10617-10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malnou, C. E., A. Werner, A. M. Borman, E. Westhof, and K. M. Kean. 2004. Effects of vaccine strain mutations in domain V of the internal ribosome entry segment compared in the wild type poliovirus type 1 context. J. Biol. Chem. 279:10261-10269. [DOI] [PubMed] [Google Scholar]
- 31.McKnight, K. L., and S. M. Lemon. 1996. Capsid coding sequence is required for efficient replication of human rhinovirus 14 RNA. J. Virol. 70:1941-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Minor, P. D. 2004. Polio eradication, cessation of vaccination and re-emergence of disease. Nat. Rev. Microbiol. 2:473-482. [DOI] [PubMed] [Google Scholar]
- 33.Moulton, H. M., M. H. Nelson, S. A. Hatlevig, M. T. Reddy, and P. L. Iversen. 2004. Cellular uptake of antisense morpholino oligomers conjugated to arginine-rich peptides. Bioconjug. Chem. 15:290-299. [DOI] [PubMed] [Google Scholar]
- 34.Murray, P. R., K. S. Rosenthal, G. S. Kobayashi, and M. A Pfaller. 2005. Picornaviruses, p. 450-461. In M. Brown (ed.), Medical microbiology, 5th ed. Mosby Elsevier, St. Louis, MO.
- 35.Nelson, M. H., D. A. Stein, A. D. Kroeker, S. A. Hatlevig, P. L. Iversen, and H. M. Moulton. 2005. Arginine-rich peptide conjugation to morpholino oligomers: effects on antisense activity and specificity. Bioconjug. Chem. 16:959-966. [DOI] [PubMed] [Google Scholar]
- 36.Neuman, B. W., D. A. Stein, A. D. Kroeker, M. J. Churchill, A. M. Kim, P. Kuhn, P. Dawson, H. M. Moulton, R. K. Bestwick, P. L. Iversen, and M. J. Buchmeier. 2005. Inhibition, escape, and attenuated growth of severe acute respiratory syndrome coronavirus treated with antisense morpholino oligomers. J. Virol. 79:9665-9676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ochs, K., A. Zeller, L. Saleh, G. Bassili, Y. Song, A. Sonntag, and M. Niepmann. 2003. Impaired binding of standard initiation factors mediates poliovirus translation attenuation. J. Virol. 77:115-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palacios, G., and M. S. Oberste. 2005. Enteroviruses as agents of emerging infectious diseases. J. Neurovirol. 11:424-433. [DOI] [PubMed] [Google Scholar]
- 39.Pallansch, M. A., and R. P. Roos. 2001. Enteroviruses, polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses, p. 723-776. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, vol. 2, 4th ed. Lippincott, Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 40.Patel, D., T. Opriessnig, D. A. Stein, P. G. Halbur, X. J. Meng, P. L. Iversen, and Y. J. Zhang. 2008. Peptide-conjugated morpholino oligomers inhibit porcine reproductive and respiratory syndrome virus replication. Antivir. Res. 77:95-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paul, A. V. 2002. Possible unifying mechanism of picornavirus genome replication, p. 227-246. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, DC.
- 42.Phipps, K. M., A. Martinez, J. Lu, B. A. Heinz, and G. Zhao. 2004. Small interfering RNA molecules as potential anti-human rhinovirus agents: in vitro potency, specificity, and mechanism. Antivir. Res. 61:49-55. [DOI] [PubMed] [Google Scholar]
- 43.Polacek, C., and A. M. Lindberg. 2001. Genetic characterization of the coxsackievirus B2 3′ untranslated region. J. Gen. Virol. 82:1339-1348. [DOI] [PubMed] [Google Scholar]
- 44.Racaniello, V. R. 2001. Picornaviridae: the viruses and their replication, p. 685-722. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, vol. 2, 4th ed. Lippincott, Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 45.Rosas, M. F., E. Martinez-Salas, and F. Sobrino. 2003. Stable expression of antisense RNAs targeted to the 5′ non-coding region confers heterotypic inhibition to foot-and-mouth disease virus infection. J. Gen. Virol. 84:393-402. [DOI] [PubMed] [Google Scholar]
- 46.Sharma, R., S. Raychaudhuri, and A. Dasgupta. 2004. Nuclear entry of poliovirus protease-polymerase precursor 3CD: implications for host cell transcription shut-off. Virology 320:195-205. [DOI] [PubMed] [Google Scholar]
- 47.Sim, A. C., A. Luhur, T. M. Tan, V. T. Chow, and C. L. Poh. 2005. RNA interference against enterovirus 71 infection. Virology 341:72-79. [DOI] [PubMed] [Google Scholar]
- 48.Stein, D., E. Foster, S. B. Huang, D. Weller, and J. Summerton. 1997. A specificity comparison of four antisense types: morpholino, 2′-O-methyl RNA, DNA, and phosphorothioate DNA. Antisense Nucleic Acid Drug Dev. 7:151-157. [DOI] [PubMed] [Google Scholar]
- 49.Summerton, J. 1999. Morpholino antisense oligomers: the case for an RNase H-independent structural type. Biochim. Biophys. Acta 1489:141-158. [DOI] [PubMed] [Google Scholar]
- 50.Summerton, J., and D. Weller. 1997. Morpholino antisense oligomers: design, preparation, and properties. Antisense Nucleic Acid Drug Dev. 7:187-195. [DOI] [PubMed] [Google Scholar]
- 51.Todd, S., J. S. Towner, and B. L. Semler. 1997. Translation and replication properties of the human rhinovirus genome in vivo and in vitro. Virology 229:90-97. [DOI] [PubMed] [Google Scholar]
- 52.Tolskaya, E. A., L. I. Romanova, M. S. Kolesnikova, A. P. Gmyl, A. E. Gorbalenya, and V. I. Agol. 1994. Genetic studies on the poliovirus 2C protein, an NTPase. A plausible mechanism of guanidine effect on the 2C function and evidence for the importance of 2C oligomerization. J. Mol. Biol. 236:1310-1323. [DOI] [PubMed] [Google Scholar]
- 53.Vagnozzi, A., D. A. Stein, P. L. Iversen, and E. Rieder. 2007. Inhibition of foot-and-mouth disease virus infections in cell cultures with antisense morpholino oligomers. J. Virol. 81:11669-11680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van den Born, E., D. A. Stein, P. L. Iversen, and E. J. Snijder. 2005. Antiviral activity of morpholino oligomers designed to block various aspects of equine arteritis virus amplification in cell culture. J. Gen. Virol. 86:3081-3090. [DOI] [PubMed] [Google Scholar]
- 55.Walter, B. L., J. H. Nguyen, E. Ehrenfeld, and B. L. Semler. 1999. Differential utilization of poly(rC) binding protein 2 in translation directed by picornavirus IRES elements. RNA 5:1570-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walter, B. L., T. B. Parsley, E. Ehrenfeld, and B. L. Semler. 2002. Distinct poly(rC) binding protein KH domain determinants for poliovirus translation initiation and viral RNA replication. J. Virol. 76:12008-12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, A., P. K. Cheung, H. Zhang, C. M. Carthy, L. Bohunek, J. E. Wilson, B. M. McManus, and D. Yang. 2001. Specific inhibition of coxsackievirus B3 translation and replication by phosphorothioate antisense oligodeoxynucleotides. Antimicrob. Agents Chemother. 45:1043-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wenner, H. A., and P. Kamitsuka. 1957. Primary sites of virus multiplication following intramuscular inoculation of poliomyelitis virus in cynomolgus monkeys. Virology 3:429-443. [DOI] [PubMed] [Google Scholar]
- 59.Wenner, H. A., P. Kamitsuka, M. Lenahan, and I. Archetti. 1960. The pathogenesis of poliomyelitis. Sites of multiplication of poliovirus in cynomolgus monkeys after alimentary infection. Arch. Gesamte Virusforsch. 9:537-558. [DOI] [PubMed] [Google Scholar]
- 60.Westerberg, U. B., G. Bolcsfoldi, and E. Eliasson. 1976. Control of transfer RNA synthesis in the presence of inhibitors of protein synthesis. Biochim. Biophys. Acta 447:203-213. [DOI] [PubMed] [Google Scholar]
- 61.Witwer, C., S. Rauscher, I. L. Hofacker, and P. F. Stadler. 2001. Conserved RNA secondary structures in Picornaviridae genomes. Nucleic Acids Res. 29:5079-5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yuan, J., P. K. Cheung, H. Zhang, D. Chau, B. Yanagawa, C. Cheung, H. Luo, Y. Wang, A. Suarez, B. M. McManus, and D. Yang. 2004. A phosphorothioate antisense oligodeoxynucleotide specifically inhibits coxsackievirus B3 replication in cardiomyocytes and mouse hearts. Lab. Investig. 84:703-714. [DOI] [PubMed] [Google Scholar]
- 63.Yuan, J., D. A. Stein, T. Lim, D. Qiu, S. Coughlin, Z. Liu, Y. Wang, R. Blouch, H. M. Moulton, P. L. Iversen, and D. Yang. 2006. Inhibition of coxsackievirus B3 in cell cultures and in mice by peptide-conjugated morpholino oligomers targeting the internal ribosome entry site. J. Virol. 80:11510-11519. [DOI] [PMC free article] [PubMed] [Google Scholar]