Abstract
The development of resistance to linezolid (LZD) in gram-positive bacteria depends on the mutation of a single 23S rRNA gene, followed by homologous recombination and gene conversion of the other alleles. We sought to inhibit this process in Staphylococcus aureus using a range of antibacterial agents, including some that suppress recombination. A model for the rapid selection of LZD resistance was developed which allowed the selection of LZD-resistant mutants with G2576T mutations in all five copies of the 23S rRNA gene following only 5 days of subculture. The emergence of LZD-resistant isolates was delayed by exposing cultures to low concentrations of various classes of antibiotics. All antibiotic classes were effective in delaying the selection of LZD-resistant mutants and, with the exception of fusidic acid (FUS) and rifampin (RIF), prolonged the selection window from 5 to ∼15 days. Inhibitors of DNA processing were no more effective than any other class of antibiotics at suppressing resistance development. However, the unrelated antimicrobials FUS and RIF were particularly effective at preventing the emergence of LZD resistance, prolonging the selection window from 5 to 25 days. The enhanced suppressive effect of FUS and RIF on the development of LZD resistance was lost in a recA-deficient host, suggesting that these drugs affect recA-dependent recombination. Furthermore, FUS and RIF were shown to be effective inhibitors of homologous recombination of a plasmid into the staphylococcal chromosome. We suggest that RIF or FUS in combination with LZD may have a role in preventing the emergence of LZD resistance.
The development of resistance to linezolid (LZD) in clinical isolates of Staphylococcus aureus and enterococci involves the generation of mutations in one of the multiple 23S rRNA gene copies targeted by the drug, followed by homologous recombination between the remaining 23S rRNA gene copies, i.e., a process of gene conversion (15, 18, 30). In addition to LZD resistance, gene conversion has also been implicated in the emergence of resistance to penicillin in pneumococci (4, 23) and, possibly, also in the emergence of resistance to macrolides in staphylococci, which, like LZD resistance, appears to result from alterations to rRNA operons (26).
There has been previous interest in exploring ways in which the development or spread of resistance to an antimicrobial agent might be blocked at the genetic level, e.g., by suppressing the emergence of point mutations with antimutagenic agents, disrupting horizontal DNA transfer (5, 25, 32, 33), or promoting the curing of plasmids by chemical agents (3, 16, 24, 32, 33). However, the possibility of inhibiting gene conversion events to suppress the emergence of resistance has not been explored. We have examined a variety of antimicrobial agents, including those that inhibit DNA processing, for their ability to interfere with the development of LZD resistance in S. aureus. Spiral plating techniques, which require successive serial transfers in the presence of LZD for many weeks, have been required to detect the homologous recombination events that generate stable LZDr mutants of S. aureus (29). To provide a better method for screening for potential suppressors of LZD resistance, we developed an in vitro system using saturated stationary-phase cell cultures which permitted the rapid selection of LZD resistance in S. aureus. We found that low levels of any secondary drug exerted an effect on the development of LZD resistance. Although antibiotics with a known effect on DNA processing showed no advantage over other antimicrobial classes, rifampin (RIF) and fusidic acid (FUS) were particularly effective at suppressing the emergence of LZD resistance. These agents appear to suppress the recombination-based acquisition of LZD resistance in S. aureus to a greater extent than the other classes of antibiotic.
(Parts of the work presented in this paper were communicated at the 44th and the 45th Interscience Conferences on Antimicrobial Agents and Chemotherapy [19, 20].)
MATERIALS AND METHODS
Bacterial strains, antibiotics, plasmids, chemicals, and growth media.
The strains used in this study are described in Table 1. Unless indicated otherwise, the chemicals and antibiotics used in this study were from Sigma-Aldrich (Poole, United Kingdom). Ciprofloxacin (CIP) and moxifloxacin (MXF) were gifts from Bayer Pharmaceuticals (Leverkeusen, Germany), gemifloxacin (GMF) was a gift from LG Life Sciences (Mt. Prospect, IL), rufloxacin (RUF) was a gift from B. Oliva (University of l'Aquila, Aquila, Italy), and LZD was a gift from Pfizer (Kalamazoo, MI). Mueller-Hinton broth (MHB) and Mueller-Hinton agar were from Fisher (Loughborough, United Kingdom).
TABLE 1.
Bacterial strains used in this study
| Strain | LZD MIC (μg/ml) | Parent strain | Genotype | Reference or source |
|---|---|---|---|---|
| RN4220 | 4 | LZDs, recA positive | 8 | |
| EMRSA-15 | 4 | LZDs, recA positive | 13 | |
| KB103 | 4 | RN4220 | LZDs, recA deficient | 1 (Ken Bayles, University of Idaho) |
| KM50 | 32 | RN4220 | LZDr (five copies of G2576T 23S rRNA gene) | This study |
| KM51 | 32 | RN4220 | LZDr (five copies of G2576T 23S rRNA gene) | This study |
| KM52 | 32 | KB103 | LZDr (three copies of G2576T 23S rRNA gene, one copy of T2500A, one copy of G2447T) | This study |
| This study | ||||
| KM53 | 32 | KB103 | LZDr (three copies of G2576T 23S rRNA gene, one copy of G2447T, one unknown mutation) | This study |
| KM183 | 16 | RN4220 | LZDr (three copies of G2576T 23S rRNA gene) | This study |
| KM187 | 32 | RN4220 | LZDr (five copies of G2576T 23S rRNA gene) | This study |
Determination of susceptibility to antimicrobial agents.
MICs were determined by broth microdilution according to BSAC guidelines (2) in MHB. The MIC was defined as the lowest concentration of antibiotic that completely inhibited visible growth after 18 to 24 h of incubation at 37°C. The concentrations of antibiotics causing a 50% reduction in the bacterial growth rate (IC50s) were determined by culture absorbance methods, as described previously (21).
Development of LZDr mutants.
Bacteria (Table 1) were grown in MHB to stationary phase and then exposed to LZD at 4× the MIC for 6 h. Following LZD exposure, an aliquot (5 μl) of the culture was transferred into fresh drug-free medium and the cycle was repeated. Following each cycle, the MIC of LZD was determined for a sample of the culture. When the MIC reached 32 μg/ml, the culture displayed a LZD resistance phenotype equivalent to that for strain KM187 (which has five mutated copies of the 23S rRNA gene carrying the G2576T mutation) (Table 1) and the subculture was terminated. Each subculture experiment was performed four times, and the time to the emergence of resistance was averaged to the nearest day.
Suppression of LZD resistance.
To examine the potential for secondary agents to suppress the emergence of LZD resistance, subculture experiments were performed as described above in the presence of 0.25× the IC50 of one or more secondary agents. IC50s were used in preference to MICs due to their greater reproducibility.
Independent genotyping of all five 23S rRNA gene alleles.
The 23S rRNA gene alleles were amplified by PCR, and the amplicons were sequenced by the method of Tsiodras et al. (31).
Determination of recombination frequencies.
Recombination assays were modified from the method described by Prunier and Leclercq (26). The sodA gene from Staphylococcus saprophyticus, which is 85% identical to the corresponding gene in S. aureus RN4220, was amplified by PCR with the following oligonucleotide primers: 5′-AAGCTTTGGACGTTTATTTTGGTATT and 5′-AAGCTTTTACCTTATGGTTTTGATGC (the engineered restriction sites are underlined). The resulting PCR amplicon was ligated into pCL52.1 (14), propagated in Escherichia coli XL1-Blue, and then introduced into RN4220 by electroporation (27). The frequency with which the S. saprophyticus sodA gene recombined with the chromosomal sodA gene in RN4220 after 24 h of growth at 42°C (the restrictive temperature for plasmid replication) was measured by recovering putative integrants on agar containing tetracycline (TET; 3 μg/ml). Antimicrobial agents were tested for their effects on recombination by adding them to the 42°C culture at a low concentration (0.25× the IC50).
RESULTS AND DISCUSSION
Rapid recovery of LZDr mutants from strain RN4220 and further phenotypic and genotypic analyses.
Successive exposure of stationary-phase S. aureus cultures to LZD facilitated the rapid recovery of resistant mutants (Fig. 1A). Accordingly, within 5 days of exposure of S. aureus strain RN4220 to LZD, cultures contained organisms able to survive in the presence of 16 μg LZD/ml; i.e., there was a fourfold decrease in susceptibility compared to that of the original starting culture. Individual colonies recovered at the 5-day time point were designated strains KM50, KM51, and KM187; and one strain, recovered at 3 days, was designated KM183 (Table 1). These strains were further characterized. The MIC of LZD for each of the strains recovered was 32 μg/ml. None of the strains exhibited cross-resistance to other agents (CIP, MXF, GMF, RUF, novobiocin [NOV], chloramphenicol [CHL], clindamycin [CLI], TET, gentamicin [GNT], FUS, RIF, erythromycin [ERY], cefotaxime [CTX]) (data not shown).
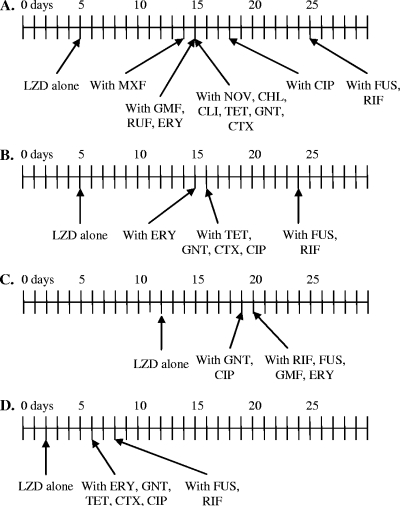
FIG. 1.
Timeline for emergence of mutants resistant to LZD during subculture in the presence of LZD (4× the MIC) and the presence or absence of a second agent or agents (0.25× the IC50). (A) Laboratory strain RN4220; (B) clinical strain EMRSA-15; (C) recA-deficient RN4220 derivative KB103; (D) KM183, an intermediate-level LZDr RN4220 derivative with three G2576T 23S rRNA mutant copies.
PCR amplification and sequencing revealed that strains KM50, KM51, and KM187 possessed a G2576T mutation in all five copies of the 23S rRNA gene. The G2576T mutation has previously been implicated in the resistance of clinical isolates to LZD (11, 18), and the possession of multiple G2576T mutations is believed to occur by recombination-dependent bacterial gene conversion (18). In contrast, KM183 had three copies carrying the G2576T mutation.
Recombination status influences recovery and nature of LZDr mutants.
The prominence of multiple G2576T mutations in the 23S rRNA genes of the RN4220 LZDr derivatives recovered during the successive subculture procedure suggested that recombination-dependent gene conversion was the principal mechanism for the emergence of LZD resistance. Further evidence for this hypothesis was obtained by using recA-deficient strain KB103 (Table 1). In contrast to the situation with RN4220, 12 days of subculture was required to recover LZDr mutants from the recA-deficient mutant, i.e., a further 7 days over that required to recover LZDr mutants from RN4220 (Fig. 1A and C). Two individual LZDr derivatives of strain KB103, designated strains KM52 and KM53 (Table 1), were recovered for further analysis.
The MIC of LZD for strains KM52 and KM53 was 32 μg/ml. KM52 possessed three 23S genes with a G2576T mutation, one 23S gene with a T2500A mutation, and one 23S gene with a G2447T mutation. KM53 also had three 23S genes with the G2576T mutation and one gene with a G2447T mutation; however, the final gene had no mutations in the resistance-defining region. Therefore, in contrast to the LZDr derivatives of strain RN4220, the KB103 derivatives displayed a heterogeneous pattern of resistance genotypes and appeared to depend partially upon mutations other than those involving gene conversion for the expression of LZD resistance.
Delaying the emergence of LZD resistance by incubation in the presence of other antimicrobial agents at sub-MICs.
Recently, Tsakris et al. (30) observed that the exposure of S. aureus to sublethal levels of NOV, an antimicrobial agent that interferes with the supercoiling of DNA (17), suppresses the emergence of LZD resistance. This observation suggests that agents with the ability to interfere with the processing of DNA could inhibit the development of LZD resistance in S. aureus and might also have a wider application in preventing other gene conversion processes that confer antimicrobial resistance. However, the studies reported (30) were limited to the examination of NOV alone, and experiments with further DNA processing inhibitors and other classes of antimicrobial agents were not performed. Consequently, we examined the effects of secondary antimicrobial agents of various classes on the development of LZD resistance in S. aureus strains RN4220, strain KB103, and a clinical isolate, EMRSA-15.
With the exception of FUS and RIF, all of the agents, including those that inhibit DNA processing, suppressed the emergence of resistance for similar periods of time (the emergence of LZDr variants at 14 to 18 days for RN4220 and the emergence of LZDr EMRSA-15 at 15 to 16 days); FUS and RIF both prolonged the length of subculture required for the emergence of LZD resistance to 25 days in the RN4220 background and 24 days in the EMRSA-15 background (Fig. 1A and B). This pattern was also observed for the development of high-level LZD resistance in LZD intermediate-resistant strain KM183 (Fig. 1D). The use of a combination of FUS and RIF did not further delay the emergence of LZD resistance (data not shown). RIF and FUS were also able to extend the period for recovery of LZDr mutants in recA-deficient strain KB103, extending this from 12 days in the absence of a secondary agent to 20 days in the presence of RIF or FUS (Fig. 1C). However, in the recA-deficient strain, GMF and ERY were also equally effective in suppressing the emergence of LZD resistance (Fig. 1C). Inhibitors of DNA processing, including various fluoroquinolones and NOV, were no more effective than other classes of antimicrobial agents in suppressing resistance in our model. Indeed, RIF and FUS, which do not directly interfere with DNA processing, were the most effective agents in suppressing the development of resistance to LZD. This was not simply due to the suppression of growth by the secondary agents because the concentrations used were insufficient to affect the growth rates of the organisms (data not shown). The apparent differences in the efficacies of the secondary agents in suppressing the emergence of LZDr could reflect the selection of variants resistant to the second agent during subculture. However, the coselection of resistance to the second agents did not occur during the subculture period (data not shown).
We believe that our rapid procedure for the selection of LZDr mutants in recombination-proficient hosts reflects gene conversion and is representative of the events that occur under clinical selection pressure. Accordingly, LZDr mutants recovered in vitro with levels of resistance similar to those of the clinical isolates possessed, like their clinical counterparts, G2576T mutations in all rRNA operons. Furthermore, dependency on recombination for the generation of multiple G2576T mutations in wild-type strains was demonstrated by the heterogeneity of the LZD resistance mutations arising in recA-deficient host KB103. These single-site mutations in KB103 are probably the result of independent point mutations in 23S rRNA (15). The G2576T mutations in KB103 might have arisen from successive independent point mutations or may be the consequence of recA-independent recombination activities, which appear to occur in S. aureus (7).
Role of recombination in suppression of resistance.
An assay to determine recombination frequencies based on the system developed by Prunier and Leclercq (26), which involves integration of the sodA gene into the staphylococcal chromosome, was used to examine the level of recombination in strain RN4220 and the influence of exposure to low concentrations of antimicrobial agents (Table 2). TET, GNT, CIP, and NOV, which prevented the emergence of LZD resistance for 14 to 18 days in RN4220, had a moderate effect on recombination, whereas FUS and RIF, which suppressed the emergence of LZD resistance for 25 days, reduced the level of recombination to approximately 30% of that seen in the absence of an antimicrobial. The ability of FUS and RIF to suppress recombination was maintained in the presence of LZD (Table 2).
TABLE 2.
Relative recombination frequencies following exposure of S. aureus RN4220 to various antimicrobial agents alone and in combination
| Antimicrobial agenta | Recombination frequencyb |
|---|---|
| No drug | 1.000 |
| LZD | 0.713 ± 0.060 |
| FUS | 0.284 ± 0.041 |
| RIF | 0.333 ± 0.042 |
| TET | 0.757 ± 0.037 |
| GNT | 0.829 ± 0.029 |
| CIP | 0.901 ± 0.089 |
| NOV | 0.886 ± 0.076 |
| LZD + FUS | 0.279 ± 0.053 |
| LZD + RIF | 0.187 ± 0.069 |
| LZD + TET | 0.812 ± 0.050 |
| LZD + GNT | 0.770 ± 0.047 |
| LZD + CIP | 0.856 ± 0.103 |
| LZD + NOV | 0.821 ± 0.032 |
With the exception of LZD, the drugs were added at 0.25× the IC50; LZD was added at 0.25× the MIC.
Defined as the number of putative integrants recovered on agar containing 3 μg TET/ml as a proportion of the number of the integrants recovered from the drug-free (no drug) control.
The data presented here suggest that FUS and RIF are particularly effective inhibitors of recombination, possibly mediated by effects on RecA activity. Effects on RecA are consistent with the observation that the preferential activity of FUS and RIF in preventing the emergence of LZD resistance in wild-type strains was lost in recA-deficient host strain KB103, since GNT, ERY, and CIP were all as effective as FUS or RIF under these conditions (Fig. 1C). We attempted to measure recA transcription following exposure to FUS, RIF, TET, and LZD using reverse transcription-PCR. The levels of transcription were not significantly different under any of these conditions (data not shown). The complexity of the regulatory system controlling RecA activity (6) suggests that it will be difficult to identify how FUS and RIF affect RecA-mediated processes. However, it is unlikely that antibiotics with different primary modes of action and no structural similarities would have the same effect on RecA-mediated recombination.
Conclusions.
The data presented here further strengthen the conclusion that LZD resistance in S. aureus depends on recombination for rapid development. However, to our knowledge, the evidence presented is the first to show that low concentrations of FUS and RIF are particularly effective in suppressing bacterial recombination. These observations could have clinical utility in preventing the emergence of LZD resistance driven by recombination. Although to date the development of resistance to LZD in S. aureus has been observed only rarely, the potential for resistance clearly exists, particularly with long-term oral therapy when the focus of infection remains (18). FUS, RIF, and LZD all exhibit good tissue penetration (22). Thus, if combination therapy with LZD and either RIF or FUS was employed, bacteria would likely be exposed to relatively high tissue concentrations of secondary agents. This should minimize the chance of mutant selection that is seen when either RIF or FUS is used as monotherapy (9, 10). Furthermore, our results suggest that the ability of RIF and FUS to suppress recombination is maintained in the presence of LZD, which would be a necessary requirement for the proposed combination drug strategies. Our findings should be extended to studies with other bacteria, notably, coagulase-negative staphylococci and enterococci, where LZD resistance has been seen more commonly (12) (18, 28).
Acknowledgments
This work was supported by a research grant from the United Kingdom Department of Health to I. Chopra, M. H. Wilcox, and E. Ingham.
Footnotes
Published ahead of print on 31 March 2008.
REFERENCES
- 1.Bayles, K. W., E. W. Brunskill, J. J. Iandolo, L. L. Hruska, S. Huang, P. A. Pattee, B. K. Smiley, and R. E. Yasbin. 1994. A genetic and molecular characterization of the recA gene from Staphylococcus aureus. Gene 147:13-20. [DOI] [PubMed] [Google Scholar]
- 2.BSAC. 1991. A guide to sensitivity testing; report of the working party on antibiotic sensitivity testing of the British Society for Antimicrobial Chemotherapy. J. Antimicrob. Chemother. 27(Suppl. D):1-50. [PubMed] [Google Scholar]
- 3.Burman, L. G. 1977. R-plasmid transfer and its response to nalidixic acid. J. Bacteriol. 131:76-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chambers, H. F. 1999. Penicillin-binding protein-mediated resistance in pneumococci and staphylococci. J. Infect. Dis. 179:S353-S359. [DOI] [PubMed] [Google Scholar]
- 5.Chopra, I., A. J. O'Neill, and K. Miller. 2003. The role of mutators in the emergence of antibiotic-resistant bacteria. Drug Resist. Updat. 6:137-145. [DOI] [PubMed] [Google Scholar]
- 6.Cox, M. M. 2007. Regulation of bacterial RecA protein function. Crit. Rev. Biochem. Mol. Biol. 42:41-63. [DOI] [PubMed] [Google Scholar]
- 7.Dutra, B. E., V. A. Sutera, Jr., and S. T. Lovett. 2007. RecA-independent recombination is efficient but limited by exonucleases. Proc. Natl. Acad. Sci. USA 104:216-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fairweather, N., S. Kennedy, T. J. Foster, M. Kehoe, and G. Dougan. 1983. Expression of a cloned Staphylococcus aureus alpha-hemolysin determinant in Bacillus subtilis and Staphylococcus aureus. Infect. Immun. 41:1112-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falagas, M. E., I. A. Bliziotis, and K. N. Fragoulis. 2007. Oral rifampin for eradication of Staphylococcus aureus carriage from healthy and sick populations: a systematic review of the evidence from comparative trials. Am. J. Infect. Control 35:106. [DOI] [PubMed] [Google Scholar]
- 10.Howden, B., and M. L. Grayson. 2006. Dumb and dumber—the potential waste of a useful antistaphylococcal agent: emerging fusidic acid resistance in Staphylococcus aureus. Clin. Infect. Dis. 42:394-400. [DOI] [PubMed] [Google Scholar]
- 11.Howe, R. A., A. Noel, K. E. Bowker, V. I. Enne, T. R. Walsh, and A. P. MacGowan. 2002. Abstr. 42nd Intersci. Conf. Antimicrob. Agents Chemotherapy, abstr. CI-1608.
- 12.Jones, R. N., T. R. Fritsche, H. S. Sader, and J. E. Ross. 2007. LEADER surveillance program results for 2006: an activity and spectrum analysis of linezolid using clinical isolates from the United States (50 medical centers). Diagn. Microbiol. Infect. Dis. 59:309. [DOI] [PubMed] [Google Scholar]
- 13.Kumari, D. N., V. Keer, P. M. Hawkey, P. Parnell, N. Joseph, J. F. Richardson, and B. Cookson. 1997. Comparison and application of ribosome spacer DNA amplicon polymorphisms and pulsed-field gel electrophoresis for differentiation of methicillin-resistant Staphylococcus aureus strains. J. Clin. Microbiol. 35:881-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin, W. S., T. Cunneen, and C. Y. Lee. 1994. Sequence analysis and molecular characterization of genes required for the biosynthesis of type 1 capsular polysaccharide in Staphylococcus aureus. J. Bacteriol. 176:7005-7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lobritz, M., R. Hutton-Thomas, S. Marshall, and L. B. Rice. 2003. Recombination proficiency influences frequency and locus of mutational resistance to linezolid in Enterococcus faecalis. Antimicrob. Agents Chemother. 47:3318-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lujan, S. A., L. M. Guogas, H. Ragonese, S. W. Matson, and M. R. Redinbo. 2007. Disrupting antibiotic resistance propagation by inhibiting the conjugative DNA relaxase. Proc. Natl. Acad. Sci. USA 104:12282-12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maxwell, A. 1993. The interaction between coumarin drugs and DNA gyrase. Mol. Microbiol. 9:681-686. [DOI] [PubMed] [Google Scholar]
- 18.Meka, V. G., H. S. Gold, A. Cooke, L. Venkataraman, G. M. Eliopoulos, R. C. Moellering, Jr., and S. G. Jenkins. 2004. Reversion to susceptibility in a linezolid-resistant clinical isolate of Staphylococcus aureus. J. Antimicrob. Chemother. 54:818-820. [DOI] [PubMed] [Google Scholar]
- 19.Miller, K., A. J. O'Neill, M. H. Wilcox, E. Ingham, and I. Chopra. 2005. Development of linezolid resistance in a recA mutant of Staphylococcus aureus, abstr. C1-1413. Abstr. 45th Intersci. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, DC.
- 20.Miller, K., A. J. O'Neill, M. H. Wilcox, E. Ingham, and I. Chopra. 2004. Suppression of linezolid resistance in Staphylococcus aureus using drug combinations, abstr. CI-1498. Abstr. 44th Intersci. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, DC.
- 21.Muhammad, I., X. C. Li, M. R. Jacob, B. L. Tekwani, D. C. Dunbar, and D. Ferreira. 2003. Antimicrobial and antiparasitic (+)-trans-hexahydrodibenzopyrans and analogues from Machaerium multiflorum. J. Nat. Prod. 66:804-809. [DOI] [PubMed] [Google Scholar]
- 22.Nathwani, D., G. D. Barlow, K. Ajdukiewicz, K. Gray, J. Morrison, B. Clift, A. J. France, and P. Davey. 2003. Cost-minimization analysis and audit of antibiotic management of bone and joint infections with ambulatory teicoplanin, in-patient care or outpatient oral linezolid therapy. J. Antimicrob. Chemother. 51:391-396. [DOI] [PubMed] [Google Scholar]
- 23.Normark, B. H., and S. Normark. 2002. Evolution and spread of antibiotic resistance. J. Intern. Med. 252:91-106. [DOI] [PubMed] [Google Scholar]
- 24.Palomares, J., R. Prados, and E. Perea. 1987. Effect of subinhibitory concentrations of ampicillin on the R plasmid transfer in Escherichia coli. Chemioterapia 6:256-260. [PubMed] [Google Scholar]
- 25.Pillai, S. P., C. A. Pillai, D. M. Shankel, and L. A. Mitscher. 2001. The ability of certain antimutagenic agents to prevent development of antibiotic resistance. Mutat. Res. 496:61-73. [DOI] [PubMed] [Google Scholar]
- 26.Prunier, A.-L., and R. Leclercq. 2005. Role of mutS and mutL genes in hypermutability and recombination in Staphylococcus aureus. J. Bacteriol. 187:3455-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schenk, S., and R. Laddaga. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 73:133-138. [DOI] [PubMed] [Google Scholar]
- 28.Schulte, B., A. Heininger, I. B. Autenrieth, and C. Wolz. 2007. Emergence of increasing linezolid-resistance in enterococci in a post-outbreak situation with vancomycin-resistant Enterococcus faecium. Epidemiol. Infect. [Epub ahead of print.] doi: 10.1017/50950268807009508. [DOI] [PMC free article] [PubMed]
- 29.Shinabarger, D. L. 1999. Mechanism of action of the oxazolidinone antibacterial agents. Expert Opin. Investig. Drugs 8:1195-1202. [DOI] [PubMed] [Google Scholar]
- 30.Tsakris, A., S. K. Pillai, H. S. Gold, C. Thauvin-Eliopoulos, L. Venkataraman, C. Wennersten, R. C. Moellering, Jr., and G. M. Eliopoulos. 2007. Persistence of rRNA operon mutated copies and rapid re-emergence of linezolid resistance in Staphylococcus aureus. J. Antimicrob. Chemother. 60:649-651. [DOI] [PubMed] [Google Scholar]
- 31.Tsiodras, S., H. S. Gold, G. Sakoulas, G. M. Eliopoulos, C. Wennersten, L. Venkataraman, R. C. Moellering, and M. J. Ferraro. 2001. Linezolid resistance in a clinical isolate of Staphylococcus aureus. Lancet 358:207-208. [DOI] [PubMed] [Google Scholar]
- 32.Viljanen, P., and J. Boratynski. 1991. The susceptibility of conjugative resistance transfer in gram-negative bacteria to physicochemical and biochemical agents. FEMS Microbiol. Rev. 8:43-54. [DOI] [PubMed] [Google Scholar]
- 33.Wooley, R., H. Dickerson, K. Simmons, E. Shotts, and J. Brown. 1986. Effect of EDTA-Tris on an Escherichia coli isolate containing R plasmids. Vet. Microbiol. 12:65-75. [DOI] [PubMed] [Google Scholar]