Abstract
A chromosomally encoded class D β-lactamase, OXA-114, was characterized from Achromobacter xylosoxidans strain CIP69598. β-Lactamase OXA-114 shared 56% amino acid identity with the naturally occurring class D β-lactamase of Burkholderia cenocepacia and 42% identity with the acquired oxacillinases OXA-9 and OXA-18. OXA-114 has a narrow-spectrum hydrolysis profile, although it includes imipenem, at a very low level. PCR and sequencing revealed that blaOXA-114-like genes were identified in all A. xylosoxidans strains tested (n = 5), indicating that this β-lactamase is naturally occurring in that species. Induction experiments with imipenem and cefoxitin did not show inducibility of blaOXA-114 expression.
Achromobacter xylosoxidans (previously Alcaligenes xylosoxidans) is a non-glucose-fermenting gram-negative species that is increasingly recognized as a clinically significant nosocomial pathogen. With its tendency to contaminate fluid, many outbreaks have been reported, mostly in immunocompromised hosts, causing serious infections, including bacteremia (4, 9). The organism has variable susceptibility to β-lactams (8). It is mostly resistant to narrow-spectrum penicillins and to several cephalosporins, including cefotaxime, whereas susceptibility to ureidopenicillins and carbapenems varies (8). However, acquisition of metallo-β-lactamases, such as IMP-1, VIM-1, or VIM-2, has been reported for A. xylosoxidans, therefore leading to high-level resistance to carbapenems (12, 17-20).
The intrinsic β-lactamases produced by A. xylosoxidans have been characterized only biochemically. Lévesque et al. reported three types of cephalosporinases, with isoelectric points (pIs) of 7.4, 9.3, and 8.1 and molecular masses of 32.3 kDa, 22.800 kDa, and 36.200 kDa, respectively (13). Fujii et al. characterized a penicillinase with an unusually high pI of 9.8 and a molecular mass of 18 kDa (5). Philippon et al. reported a penicillinase with a pI of 5.7 and an oxacillinase with a pI of 7.7 (3, 15). None of the β-lactamase genes have been identified so far. The present study was designed to identify and to characterize, at the genetic and biochemical levels, the naturally occurring β-lactamase(s) of A. xylosoxidans.
MATERIALS AND METHODS
Bacterial strains and plasmids.
A. xylosoxidans reference strain CIP69598 (Pasteur Institute strain collection) and four clinical isolates recovered at the Bicêtre hospital (MER, TB1, LOL, and DUC) were included in the study. Identification was performed with the API 32-GN system (bioMerieux, Marcy l'Etoile, France). Escherichia coli TOP10 (Invitrogen, Cergy Pontoise, France) was used as the host for cloning experiments. Kanamycin-resistant pBK-CMV plasmid (Stratagene, Amsterdam, The Netherlands) was used as the cloning vector. Bacterial cultures were routinely grown in Trypticase soy (TS) broth at 37°C.
Antimicrobial agents and susceptibility testing.
MICs of various β-lactams were determined by an agar dilution technique, as previously described (16). Results of susceptibility testing were interpreted according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) (1).
Cloning experiments and analysis of recombinant plasmids.
Genomic DNA from A. xylosoxidans CIP69598 was extracted and digested partially with restriction enzyme Sau3AI (New England Biolabs, Ipswich, MA). The resultant DNA fragments were ligated with BamHI-digested pBK-CMV as described previously (16). E. coli strain TOP10 was transformed with these recombinant plasmids by electroporation (Gene Pulser II; Bio-Rad Laboratories, Ivry-sur-Seine, France). Transformants were selected on TS agar containing ticarcillin (50 mg/liter) and kanamycin (30 mg/liter). The recombinant plasmid was purified with a Qiagen plasmid mini kit (Qiagen, Courtaboeuf, France) and sequenced on both strands by use of an ABI 3100 sequencer (Applied Biosystems, Foster City, CA). The nucleotide and deduced protein sequences were analyzed with software available over the Internet (www.ncbi.nlm.nih.gov).
Genotyping comparison of the five A. xylosoxidans isolates was performed using the random amplified polymorphic DNA detection technique as described previously (21).
PCR experiments and genotyping.
The blaOXA genes of the studied strains were amplified by use of external primers OXA-114A (5′-ACGCCTGAACCCTTTTATCC-3′) and OXA-114B (5′-ATCGACAGGCCGCGCAGT-3′) to produce a 1,025-bp fragment. Both strands of the PCR products were then sequenced.
IEF analysis and hydrolytic and induction studies.
Isoelectric focusing (IEF) analysis was performed with a pH 3.5 to 9.5 Ampholine polyacrylamide gel (GE Healthcare), using crude extracts of the A. xylosoxidans isolates and of E. coli TOP10 harboring p6S3 (16). The inducibility of β-lactamase production in A. xylosoxidans isolates CIP69598 and LOL was tested in TS broth at 37°C, using imipenem (1 μg/ml) and cefoxitin (50 μg/ml) as inducers, as described previously (11), and hydrolysis was measured with 100 μM of benzylpenicillin or imipenem as the substrate. The β-lactamase activity was defined as follows: 1 unit of enzyme was the amount that hydrolyzed 1 μmol of substrate per min. The total protein content was measured with bovine albumin as the standard (DC protein assay kit; Bio-Rad).
β-Lactamase purification.
The β-lactamase was purified as previously described, with some modifications (11). E. coli TOP10(p6S3) was cultured overnight in 4 liters of TS broth containing 30 μg/ml of kanamycin and 50 μg/ml of ampicillin. After centrifugation, the bacterial pellet was resuspended in 30 ml of 100 mM phosphate buffer (pH 7.0). Crude protein extracts were then obtained by sonication followed by centrifugation. After spermine treatment for precipitation of DNA, the supernatant was subjected to several purification steps, including ion-exchange chromatography with a Q-Sepharose column, first with 20 mM Tris-HCl buffer (pH 8.8), followed by chromatography on an S-Sepharose column equilibrated with 20 mM bis-Tris buffer (pH 6.5). Elution of the β-lactamase was performed with a linear K2SO4 gradient (0 to 500 mM). Peaks of β-lactamase activity were pooled and dialyzed against 50 mM phosphate buffer (pH 7.0). The protein content was measured by the Bio-Rad DC protein assay, and the specific activities of the crude extract and the purified β-lactamase from E. coli TOP10(p6S3) were compared. The protein purification rate and the relative molecular mass of β-lactamase OXA-114a were estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Kinetic studies.
Purified β-lactamase was used for determination of kinetic parameters (kcat and Km) at 30°C in a reaction buffer made of 100 mM Tris-H2SO4-300 mM K2SO4 (pH 7.0), in which NaHCO3 was added to a final concentration of 10 mM in order to avoid biphasic kinetics (14). The initial rates of hydrolysis of β-lactams were determined with a UV spectrophotometer as previously described (10). Due to a too-low Km value for piperacillin, a Ki value was obtained by competition, using benzylpenicillin as a reporter substrate. The 50% inhibitory concentration was determined as the clavulanate, tazobactam, or sulbactam concentration that reduced the hydrolysis rate of 100 μM of cephalothin by 50% under conditions in which the enzyme was preincubated with various concentrations of inhibitor for 3 min at 30°C before addition of the substrate.
Nucleotide sequence accession number.
The nucleotide sequences for OXA-114a reported in this paper have been submitted to the EMBL/GenBank nucleotide sequence database under accession number EU188842.
RESULTS AND DISCUSSION
β-Lactam resistance phenotypes of study strains.
The five studied A. xylosoxidans strains were resistant to cefotaxime, cefoxitin, cefepime, and aztreonam, as usually observed in that species. They were susceptible to piperacillin, moxalactam, and imipenem according to the CLSI criteria (Table 1). Susceptibility to ticarcillin and ceftazidime was variable, with resistance to those β-lactam molecules being observed only for strains LOL and MER. Although all isolates were susceptible to imipenem, various MICs were obtained (Table 1). Since such a phenomenon might have resulted from variable expression of a carbapenemase, imipenem hydrolysis was measured and showed weak interstrain variability, with 3, 5, and 12 mU per mg of protein for isolates CIP, MER, and LOL, respectively, without a correlation with the MICs of imipenem (Table 1). IEF analysis revealed an identical pI value of 8.6 obtained with cultures of each isolate. Two other bands were also detected for those isolates, with pI values of approximately 5.5 and 9.5.
TABLE 1.
MICs of β-lactams for A. xylosoxidans strain CIP69598, E. coli TOP10 harboring recombinant plasmid p6S3, expressing the β-lactamase OXA-114a, and E. coli TOP10 (reference strain)
| β-Lactam(s)a | MIC (μg/ml) for organism
|
||||||
|---|---|---|---|---|---|---|---|
| A. xylosoxidans CIP69598 | A. xylosoxidans MER | A. xylosoxidans LOL | A. xylosoxidans DUC | A. xylosoxidans TB1 | E. coli TOP10(p6S3) | E. coli TOP10 | |
| Amoxicillin | 16 | 2 | >64 | 1 | 8 | 16 | 2 |
| Amoxicillin plus CLA | 4 | 1 | 8 | 1 | 8 | 16 | 2 |
| Ticarcillin | 4 | 1 | >64 | 1 | 1 | 4 | 2 |
| Ticarcillin plus CLA | 4 | 1 | >64 | 1 | 1 | 4 | 2 |
| Piperacillin | 1 | 1 | 4 | 1 | 2 | 4 | 1 |
| Piperacillin plus TZB | 0.5 | 0.5 | 4 | 0.5 | 1 | 4 | 1 |
| Cephalothin | >64 | 32 | >64 | 32 | 32 | 8 | 2 |
| Cefuroxime | >64 | >64 | >64 | >64 | >64 | 4 | 2 |
| Ceftazidime | 4 | 16 | 4 | 8 | 4 | 0.06 | 0.06 |
| Cefotaxime | >64 | >64 | >64 | >64 | >64 | 0.25 | 0.12 |
| Cefepime | 64 | >64 | >64 | >64 | >64 | 0.12 | 0.06 |
| Cefoxitin | >64 | >64 | >64 | >64 | >64 | 4 | 4 |
| Moxalactam | 2 | 2 | 1 | 2 | 2 | 1 | 1 |
| Aztreonam | >64 | >64 | >64 | >64 | >64 | 0.12 | 0.12 |
| Imipenem | 4 | 1 | 4 | 1 | 1 | 0.06 | 0.06 |
CLA, clavulanic acid at a fixed concentration of 4 μg/ml; TZB, tazobactam at a fixed concentration of 4 μg/ml.
Cloning and sequencing of the β-lactamase gene.
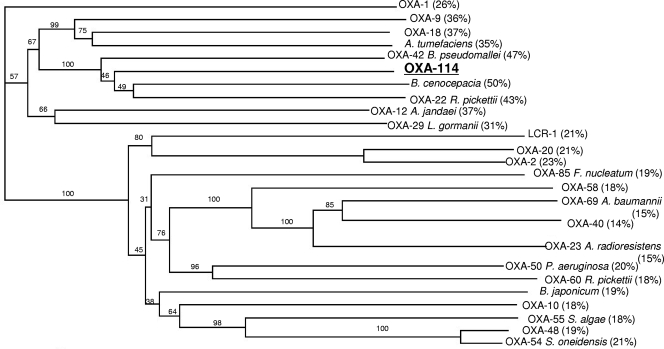
Repeated cloning experiments yielded only a single E. coli clone after selection on ticarcillin- and kanamycin-containing plates. Recombinant strain E. coli TOP10(p6S3) possessed a 1,176-bp plasmid insert and was resistant to amoxicillin and piperacillin, a resistance pattern which was not inhibited by clavulanic acid and tazobactam, respectively (Table 1). Sequencing of plasmid p6S3 revealed an open reading frame consisting of 828 bp, corresponding to a 275-amino-acid protein. Sequence analysis indicated that this open reading frame corresponded to a novel class D β-lactamase. The closest identity was observed with putative naturally occurring β-lactamases of Burkholderia cenocepacia and Delftia acidovorans, with up to 50% amino acid sequence identity (GenBank accession no. ABK10118.1 and EAV77675.1, respectively) and 37% identity with the acquired oxacillinases OXA-9 and OXA-18 (Fig. 1) (16, 22). This novel β-lactamase from A. xylosoxidans was designated OXA-114a (since other variants were subsequently identified) (see below). OXA-114a possessed the conserved motifs of serine β-lactamases, such as S-T-F-K at positions 70 to 73 and the K-T-G motif at positions 216 to 218, according to the class D β-lactamase numbering scheme (2). The Y-G-N and Q-X-X-X-L motifs, characteristic of an oxacillinase, were identified at positions 144 to 146 and 176 to 180, respectively. The pI value of the mature OXA-114a protein was 8.6, and its molecular mass was ca. 27 kDa.
FIG. 1.
Dendrogram obtained for representative class D β-lactamases by neighbor-joining analysis. The alignment used for tree calculation was performed with ClustalX. Branch lengths are drawn to scale and are proportional to the number of amino acid changes. The distance along the vertical axis has no significance. The amino acid identity of each β-lactamase compared to OXA-114a is indicated in parentheses. Acquired β-lactamases in gram-negative organisms are OXA-1, OXA-9, OXA-18, OXA-20, OXA-2, OXA-58, OXA-40, OXA-23, OXA-10, and OXA-48, whereas the others are naturally occurring ones.
Detection of OXA-114-like β-lactamase genes in A. xylosoxidans isolates.
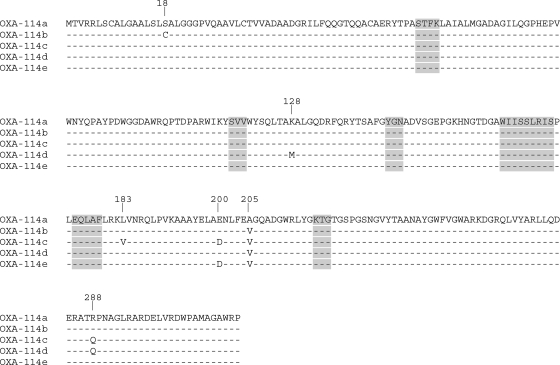
Four additional A. xylosoxidans isolates were screened for the presence of blaOXA-114a. Preliminary random amplified polymorphic DNA genotyping showed that the five strains were not clonally related (data not shown). PCR amplification performed with primers specific for blaOXA-114a yielded amplicons from all the studied strains. Sequencing identified four other OXA-114-like sequences (OXA-114b to OXA-114e) sharing high amino acid identity, with a maximum four-amino-acid difference between each other. None of the variable residues were located in conserved sequences typical of oxacillinases (Fig. 2).
FIG. 2.
Amino acid sequence comparison of OXA-114-like determinants. OXA-114a is from A. xylosoxidans isolate CIP69598, OXA-114b is from isolate MER, OXA-114c is from isolate TB1, OXA-114d is from isolate LOL, and OXA-114e is from isolate DUC. Dashes show identical amino acid residues, and the critical motifs for class D β-lactamases are shaded in gray. Numbering is done according to class D nomenclature (2).
Kinetic parameters of OXA-114a.
After purification, the purity of the OXA-114a extract was estimated to be >90% by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, with a yield of 250 μg per liter of culture. The purification coefficient was estimated to be 35-fold. Kinetic measurements of purified OXA-114a indicated an overall weak catalytic efficiency toward β-lactams. However, efficient hydrolysis of piperacillin and, to a lesser extent, ticarcillin was noticed (Table 2). Oxacillin hydrolysis was not detected with OXA-114a, as previously observed for other oxacillinases, such as the naturally occurring OXA-50 from Pseudomonas aeruginosa (6). No hydrolysis was detected for expanded-spectrum cephalosporins, such as ceftazidime, cefoxitin, and cefepime. Thus, the resistance observed for the last two substrates in all the clinical isolates was likely not related to the β-lactamase OXA-114. Interestingly, a detectable hydrolysis of imipenem was observed. However, kcat values could not be obtained for imipenem because of a very high Km value (>2,000 μM), indicating a very poor affinity for that substrate. Hydrolysis of imipenem by OXA-114a was in accordance with the hydrolysis of that substrate determined with culture extracts obtained from the clinical isolates. This result suggested that this β-lactamase could be expressed in those isolates, even at a low level. As observed for most oxacillinases, β-lactamase OXA-114a was not inhibited by class A β-lactamase inhibitors.
TABLE 2.
Kinetic parameters of purified β-lactamase OXA-114aa
| Substrate | kcat (s−1) | Km (μM)c | kcat/Km (mM−1·s−1) |
|---|---|---|---|
| Benzylpenicillin | 0.1 | 10 | 10 |
| Ticarcillin | 0.1 | 40 | 2.5 |
| Piperacillin | 0.5 | 1b | 100 |
| Cephalothin | 0.5 | 15 | 30 |
| Ceftazidime | <0.01 | ND | |
| Cefotaxime | <0.01 | ND | |
| Cefoxitin | <0.01 | ND | |
| Imipenem | >0.1 | >2,000 | 0.05 |
| Oxacillin | <0.01 | ND |
Data are the means for three independent experiments. Standard deviations were within 10% of the means.
The Ki value was determined, with benzylpenicillin as the substrate.
ND, no detectable hydrolysis (<0.01 s−1) for a maximum amount of 5 μg of purified enzyme and up to 500 μM of substrate.
Induction experiments.
Using imipenem and cefoxitin as inducers, no significant increase in hydrolysis was noticed for both benzylpenicillin and imipenem as substrates, likely indicating that expression of OXA-114 was not inducible. This result fits with the lack of induction of many naturally occurring oxacillinases, as observed for OXA-50 in P. aeruginosa (6) or OXA-51/69 in Acinetobacter baumannii (11), whereas inducibility of oxacillinase OXA-60 from another gram-negative aerobe, Ralstonia pickettii, was observed (7).
Conclusions.
This study contributed to unraveling at least one of the natural mechanisms of resistance to β-lactams in A. xylosoxidans. This work identified the naturally occurring β-lactamase of A. xylosoxidans as being neither a penicillinase nor a cephalosporinase, as previously suggested (5, 13), but rather a narrow-spectrum class D β-lactamase. It is likely that the contribution of OXA-114-like enzymes to the final β-lactam resistance profile of most A. xylosoxidans strains may be secondary, as previously demonstrated for naturally occurring oxacillinases of other gram-negative aerobic species, such as OXA-50-like enzymes from P. aeruginosa or OXA-51/69 from A. baumannii. However, since it is known that expression of that latter β-lactamase gene could be enhanced by the presence of insertion sequence elements providing promoter sequences (ISAba1 in A. baumannii), it might be hypothesized that similar events could also be expected for blaOXA-114-like genes in A. xylosoxidans, leading to a higher level of OXA-114-mediated β-lactam resistance. The present study strengthens the observation that numerous gram-negative aerobes harbor naturally occurring class D β-lactamase genes in their chromosomes and constitute possible sources of resistance genes.
Acknowledgments
We thank H. Mammeri, A. Carrër, and G. Cuzon for technical help.
This work was funded by a grant from the Ministère de l'Education Nationale et de la Recherche (UPRES-EA3539), Université Paris XI, Paris, France, and mostly by a grant from the European Community (LSHM-CT-2005-018705). Y.D. was funded in part by NIH training grant T32AI007333.
Footnotes
Published ahead of print on 24 March 2008.
REFERENCES
- 1.Clinical and Laboratory Standards Institute. 2008. Performance standards for antimicrobial susceptibility testing; 18th informational supplement M100-S18. Clinical and Laboratory Standards Institute, Wayne, PA.
- 2.Couture, F., J. Lachapelle, and R. C. Lévesque. 1992. Phylogeny of LCR-1 and OXA-5 with class A and class D β-lactamases. Mol. Microbiol. 6:1693-1705. [DOI] [PubMed] [Google Scholar]
- 3.Decré, D., G. Arlet, E. Bergogne-Bérézin, and A. Philippon. 1995. Identification of a carbenicillin-hydrolyzing beta-lactamase in Alcaligenes denitrificans subsp. xylosoxydans. Antimicrob. Agents Chemother. 39:771-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duggan, J. M., S. J. Goldstein, C. E. Chenoweth, C. A. Kauffman, and S. F. Bradley. 1996. Achromobacter xylosoxidans bacteremia: report of four cases and review of the literature. Clin. Infect. Dis. 23:569-576. [DOI] [PubMed] [Google Scholar]
- 5.Fujii, T., K. Sato, M. Inoue, and S. Mitsuhashi. 1985. Purification and properties of a β-lactamase from Alcaligenes denitrificans subsp. xylosoxydans. J. Antimicrob. Chemother. 16:297-304. [DOI] [PubMed] [Google Scholar]
- 6.Girlich, D., T. Naas, and P. Nordmann. 2004. Biochemical characterization of the naturally occurring oxacillinase OXA-50 of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 48:2043-2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Girlich, D., T. Naas, and P. Nordmann. 2004. OXA-60, a chromosomal, inducible, and imipenem-hydrolyzing class D β-lactamase from Ralstonia pickettii. Antimicrob. Agents Chemother. 48:4217-4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glupczynski, Y., W. Hansen, J. Freney, and E. Yourassowsky. 1988. In vitro susceptibility of Alcaligenes denitrificans subsp. xylosoxidans to 24 antimicrobial agents. Antimicrob. Agents Chemother. 32:276-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gomez-Cerezo, J., I. Suarez, J. J. Rios, P. Pena, M. J. Garcia de Miguel, M. de Jose, O. Monteagudo, P. Linares, A. Barbado-Cano, and J. J. Vazquez. 2003. Achromobacter xylosoxidans bacteremia: a 10-year analysis of 54 cases. Eur. J. Clin. Microbiol. Infect. Dis. 22:360-363. [DOI] [PubMed] [Google Scholar]
- 10.Héritier, C., L. Poirel, D. Aubert, and P. Nordmann. 2003. Genetic and functional analysis of the chromosome-encoded carbapenem-hydrolyzing oxacillinase OXA-40 of Acinetobacter baumannii. Antimicrob. Agents Chemother. 47:268-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Héritier, C., L. Poirel, P. E. Fournier, J. M. Claverie, D. Raoult, and P. Nordmann. 2005. Characterization of the naturally occurring oxacillinase of Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:4174-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iyobe, S., H. Kusadokoro, A. Takahashi, S. Yomoda, T. Okubo, A. Nakamura, and K. O'Hara. 2002. Detection of a variant metallo-β-lactamase, IMP-10, from two unrelated strains of Pseudomonas aeruginosa and an Alcaligenes xylosoxidans strain. Antimicrob. Agents Chemother. 46:2014-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lévesque, R., R. Letarte, and J. C. Pechère. 1983. Comparative study of the β-lactamase activity found in Achromobacter. Can. J. Microbiol. 29:819-826. [DOI] [PubMed] [Google Scholar]
- 14.Maveyraud, L., D. Golemi, L. P. Kotra, S. Tranier, S. Vakulenko, S. Mobashery, and J. P. Samama. 2000. Insights into class D β-lactamases are revealed by the crystal structure of the OXA-10 enzyme from Pseudomonas aeruginosa. Structure 8:1289-1298. [DOI] [PubMed] [Google Scholar]
- 15.Philippon, A., K. Mensah, G. Fournier, and J. Freney. 1990. Two resistance phenotypes to β-lactams of Alcaligenes denitrificans subsp. xylosoxydans in relation to β-lactamase types. J. Antimicrob. Chemother. 25:698-700. [DOI] [PubMed] [Google Scholar]
- 16.Philippon, L. N., T. Naas, A. T. Bouthors, V. Barakett, and P. Nordmann. 1997. OXA-18, a class D clavulanic acid-inhibited extended-spectrum β-lactamase from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 41:2188-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riccio, M. L., L. Pallecchi, R. Fontana, and G. M. Rossolini. 2001. In70 of plasmid pAX22, a blaVIM-1-containing integron carrying a new aminoglycoside phosphotransferase gene cassette. Antimicrob. Agents Chemother. 45:1249-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Senda, K., Y. Arakawa, S. Ichiyama, K. Nakashima, H. Ito, S. Ohsuka, K. Shimokata, N. Kato, and M. Ohta. 1996. PCR detection of metallo-β-lactamase gene blaIMP in gram-negative rods resistant to broad-spectrum β-lactams. J. Clin. Microbiol. 34:2909-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shibata, N., Y. Doi, K. Yamane, T. Yagi, H. Kurokawa, K. Shibayama, H. Kato, K. Kai, and Y. Arakawa. 2003. PCR typing of genetic determinants for metallo-β-lactamases and integrases carried by gram-negative bacteria isolated in Japan, with focus on the class 3 integron. J. Clin. Microbiol. 41:5407-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sofianou, D., A. Markogiannakis, E. Metzidie, S. Pournaras, and A. Tsakris. 2005. VIM-2 metallo-β-lactamase in Achromobacter xylosoxidans in Europe. Eur. J. Clin. Microbiol. Infect. Dis. 24:854-855. [DOI] [PubMed] [Google Scholar]
- 21.Telenius, H., N. P. Carter, C. E. Bebb, M. Nordenskjöld, B. A. Ponder, and A. Tunnacliffe. 1992. Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics 13:718-725. [DOI] [PubMed] [Google Scholar]
- 22.Tolmasky, M. E. 1990. Sequencing and expression of aadA, bla, and tnpR from the multiresistance transposon Tn1331. Plasmid 24:218-226. [DOI] [PubMed] [Google Scholar]