Abstract
We describe here the further characterization of two DNA aptamers that specifically bind to hepatitis C virus (HCV) RNA polymerase (NS5B) and inhibit its polymerase activity in vitro. Although they were obtained from the same selection procedure and contain an 11-nucleotide consensus sequence, our results indicate that aptamers 27v and 127v use different mechanisms to inhibit HCV polymerase. While aptamer 27v was able to compete with the RNA template for binding to the enzyme and blocked both the initiation and the elongation of RNA synthesis, aptamer 127v competed poorly and exclusively inhibited initiation and postinitiation events. These results illustrate the power of the selective evolution of ligands by exponential enrichment in vitro selection procedure approach to select specific short DNA aptamers able to inhibit HCV NS5B by different mechanisms. We also determined that, in addition to an in vitro inhibitory effect on RNA synthesis, aptamer 27v was able to interfere with the multiplication of HCV JFH1 in Huh7 cells. The efficient cellular entry of these short DNAs and the inhibitory effect observed on human cells infected with HCV indicate that aptamers are useful tools for the study of HCV RNA synthesis, and their use should become a very attractive and alternative approach to therapy for HCV infection.
Hepatitis C virus (HCV) infection causes serious liver diseases, such as chronic hepatitis, which can evolve into cirrhosis and hepatocellular carcinoma (20, 39). The most effective therapy, a combination of pegylated interferon and ribavirin, is efficient in only 50% of patients (32). Therefore, new treatments based on specific and well-tolerated compounds need to be developed.
HCV RNA replication is catalyzed by the viral polymerase NS5B. This RNA-dependent RNA polymerase (RdRp) synthesizes a negative-strand RNA that serves as a template for the synthesis of new positive RNA strands. Viral RNA synthesis can be divided into two major steps: (i) initiation, which corresponds to the formation of a 2- or 3-nucleotide (nt) product and which occurs by a de novo mechanism in vitro as well as in the cellular replicon system that mimics some steps of the in vivo viral cycle (9, 31, 49), and (ii) elongation, which yields a full copy of the template. The transition between these two steps, in which 3-nt to 5-nt products are synthesized, may involve conformational changes of the viral polymerase that are not yet understood (14). The structure of the enzyme, as determined by X-ray crystallography, revealed, like for many other RNA and DNA polymerases, a right-handed-like structure with finger, palm, and thumb domains (1, 7, 28). It has been shown that the NS5B protein may adopt a closed and active conformation. The contact between the two loops extending from the fingers and the thumb links these two domains and closes the back of the enzyme to form a tunnel that constitutes the entry site for ribonucleotides. This suggests that a concerted movement of the thumb and fingertips occurs during the polymerization steps. The NS5B polymerase presenting an open conformation has been described to be inactive (1, 5). Moreover, HCV NS5B possesses a β-hairpin in the thumb domain that protrudes toward the active site. This β-hairpin plays an important role in positioning the 3′ end of the viral RNA genome for the accurate initiation of replication (19). The essential role of NS5B in the HCV life cycle makes this protein an attractive target for the development of specific anti-HCV drugs. Several molecules have been described to inhibit RdRp activity in vitro as well as in the replicon cell culture system (30). These compounds include nucleoside analogues and nonnucleoside inhibitors (for a review, see reference 48).
An in vitro selection procedure, called the selective evolution of ligands by exponential enrichment strategy, allows the isolation of nucleic acid ligands termed aptamers that display high degrees of affinity and specificity for a wide array of targets, ranging from large proteins to small molecules (45). Many selections have been directed against viral proteins, bacterial proteins, and oncoproteins (36). In addition to potential therapeutic applications, aptamers can also be used for diagnostic assays (22) and structure-function analyses (8, 21). In a previous work, we selected DNA aptamers able to bind to HCV NS5B (3). We showed that one of these aptamers, oligodeoxynucleotide (ODN) 27v, is a strong inhibitor of RNA synthesis by NS5B in vitro and interferes with the binding of the RNA template to the enzyme. In this work, we report that a second aptamer, ODN 127v, isolated during the same selection experiment, is able to inhibit RNA synthesis in vitro. This aptamer shares a consensus region with ODN 27v; nevertheless, biochemical and enzymatic analyses revealed that the mechanism of inhibition of RNA synthesis was different for these two aptamers. Moreover, using the recently described HCV replication system in human cells (47), we show the distinct potential of these aptamers to inhibit RNA synthesis and HCV particle (strain JFH1) production in cell culture.
MATERIALS AND METHODS
Proteins and nucleic acids.
The recombinant HCV NS5BΔ21 of HCV isolates H77 (genotype 1a) and JFH1 (genotype 2a) and GB virus B (GBV-B) NS5BΔ19 were expressed in Escherichia coli and purified as described previously (2, 38). The RdRp of poliovirus (3Dpol) was a generous gift from E. Wimmer (State University of New York, Stony Brook).
ODNs were purchased from MWG Biotech (Ebersberg, Germany). Prior to use, the DNA aptamers were purified on a G25 Sephadex spin column (Amersham Biosciences, United Kingdom) and/or precipitated by the use of 0.3 M sodium acetate, 7.5 mM MgCl2, and 1 volume isopropanol. RNA oligonucleotides (ORNs) were synthesized by Dharmacon (Chicago, IL).
RNA template.
The RNA template that was used, named (−)IRES, corresponds to the first 341 nt of the 3′ end of the viral RNA minus strand. It was synthesized by in vitro transcription (MEGAscript kit; Ambion, Austin, TX) of DNA obtained by PCR amplification from pGEM9Zf(−) containing the 341 nt of the 5′ untranslated region (UTR) of HCV H77, as described previously (38). The 23-nt RNA template (21 nt from the 3′ UTR with two C residues added to the 3′ end), 5′-UGGCCUCUCUGCAGAUCAUGUCC-3′, obtained as a 2′-acetoxy ethyl orthoester-protected RNA, was deprotected by acid treatment at 60°C, according to the manufacturer's instructions (Dharmacon).
Nucleic acid labeling.
Oligonucleotides (10 pmol) were labeled with T4 polynucleotide kinase and 50 μCi [γ-32P]ATP (3,000 Ci·mmol−1; Amersham Biosciences) by incubation in a total volume of 10 μl for 30 min at 37°C, according to the manufacturer's instructions (Promega). Unincorporated [γ-32P]ATP was removed by use of a G25 Sephadex spin column (Amersham Biosciences). RNAs were labeled by in vitro transcription by use of the MEGAscript kit and 15 μCi [α-32P]UTP (Amersham Biosciences). The DNA templates were digested with DNase I for 15 min at 37°C. To measure the amount of radioactivity incorporated into the nucleic acids, 2-μl aliquots were precipitated with 10% trichloroacetic acid (TCA), and the radioactivity was counted in a Wallac scintillation counter. After phenol-chloroform extraction, the RNAs were precipitated with isopropanol and dissolved in water.
In vitro RNA polymerase (RdRp) assay.
The in vitro RNA polymerase (RdRp) assay was performed in a total volume of 20 μl containing 20 mM Tris (pH 7.5); 1 mM dithiothreitol (DTT); 17 U RNasin; 40 mM NaCl; 5 mM MgCl2; 0.5 mM each of the three nucleoside triphosphates (NTPs) ATP, CTP, and GTP; 86 nM of the RNA template (−)IRES; 150 nM of purified NS5B from HCV H77 or JFH1 or from GBV-B; and either 10 μCi [α-32P]UTP and 2 μM UTP or 2 μCi [3H]UTP (46 Ci·mmol−1). The reaction mixture was incubated for 2 h at 30°C, and the radiolabeled products were precipitated by the addition of 10% TCA. The radioactivity incorporated was quantified by counting in a Wallac scintillation counter. For analysis of the 32P-labeled RNA, synthesis was stopped by adding 6.25 mM EDTA, 10 mM Tris-HCl (pH 7.5), and 0.125% sodium dodecyl sulfate (SDS). The products were purified by phenol-chloroform extraction (1:1; vol/vol) and precipitated with 1 volume of isopropanol in the presence of 0.5 M ammonium acetate. The RNAs were dissolved in 95% formamide, 0.5 mM EDTA, 0.025% SDS, 0.025% bromophenol blue, and 0.025% xylene cyanol and then heated for 2 min at 94°C and loaded onto a 6% polyacrylamide denaturing gel containing 7 M urea in TBE (Tris-borate-EDTA) buffer. After electrophoresis, the gel was dried and autoradiographed with Kodak X-Omat-AR-5 film.
For single-round RNA synthesis, HCV NS5B RNA and (−)IRES RNA were preincubated 30 min at 30°C in the same incubation mixture described above, except that ATP and [32P]UTP were omitted. Heparin was then added to a final concentration of 200 μg·ml−1 before addition of ATP and [α-32P]UTP. The reaction mixture was further incubated at 30°C for 2 h. The RNA products were precipitated with 10% TCA, and the radioactivity incorporated into the RNA was quantified as described above.
(i) Poliovirus 3Dpol assay.
Poliovirus 3Dpol (150 nM) was incubated for 2 h at 30°C with 20 μg·ml−1 polyribo-adenosine template and 2 μg·ml−1 primer oligoU in a reaction mixture containing 20 mM Tris (pH 7.5), 1 mM DTT, 17 U RNasin, 30 mM NaCl, 5 mM MgCl2, and 2 μCi [3H]UTP (46 Ci·mmol−1). The newly synthesized products were precipitated by the addition of 10% TCA. The radioactivity incorporated into the RNA was quantified as described above.
(ii) HIV-1 RT assay.
Human immunodeficiency virus (HIV) type 1 (HIV-1) reverse transcriptase (RT) was from clone HIV SF2, an HIV isolate from San Francisco, CA, previously referred to as ARV-2 (40). HIV-1 RT (150 nM) was incubated for 10 min at 37°C with 20 μg·ml−1 polyribo-adenosine and oligo(dT) (5:1) in a reaction mixture containing 50 mM Tris pH 7.9, 1 mM DTT, 40 mM KCl, 5 mM MgCl2, 10 μM dTTP, and 0.5 μCi [3H]dTTP (56 Ci·mmol−1) (12). The reaction was arrested by the addition of 10% TCA, and the amount of radioactivity incorporated into the DNA was determined as described above.
Inhibition of polymerase activity by DNA aptamers.
For multiple-round replication assays, increasing concentrations of aptamer were added to the complete RdRp reaction mixtures. After a 2-h incubation at 30°C, the amount of labeled product was determined as described above. The 50% inhibitory concentration (IC50) was calculated as the aptamer concentration that reduced the polymerase activity by 50%.
For single-round RNA synthesis, HCV NS5B (strain H77) was preincubated for 30 min at 30°C with (−)IRES RNA, CTP, and GTP. Different concentrations of aptamers were then added and incubated for 30 min at 30°C or not incubated before the addition of heparin, ATP, and [32P]UTP. The reaction mixture was further incubated for 2 h at 30°C.
Gel-based initiation and elongation assays.
Gel-based initiation and elongation assays were performed essentially as described by Liu et al. (29). These assays make use of a shorter RNA template (23 nt) that allows the better separation of products of various lengths. The assays were carried out in 50 μl of 20 mM Tris (pH 7.4), 50 mM NaCl, 1 mM EDTA, 2 mM MnCl2, 5 mM DTT, 0.4 U/μl RNasin, 1 μM 23-nt RNA template, and 1.13 μM HCV strain H77 NS5B. When the initiation phase was analyzed, 1 mM GTP and 40 μM ATP with 8 μCi [α-32P]ATP (3,000 Ci·mmol−1) were the only NTPs added. The inhibition of the initiation step was examined by quantifying the amount of the 3-nt RNA product synthesized in 30 min at 30°C in the presence of various concentrations of inhibitors. The inhibition of the elongation step was assessed by determining the amount of products whose sizes were equal to and greater than the sizes of the RNA template products in the presence of all four nucleotides with or without inhibitor. At first, a 30-min preincubation with GTP and ATP was performed at 30°C to allow the formation of initiation complexes. Then, after the addition of 40 μM CTP, 40 μM UTP, and 8 μCi [α-32P]ATP with or without aptamer, the reaction mixture was further incubated for 5 min to allow elongation. The reaction was quenched with an equal volume of 50 mM SDS. To 4 μl of the arrested reaction sample, 8 μl of formamide-dye loading buffer was added. After denaturation at 70°C for 5 min, samples were loaded onto a 25% acrylamide gel in TBE buffer containing 7 M urea. The gel was run at 30 W for 3 h and then submitted to electronic autoradiography with an Instant Imager autoradiograph (Packard Instruments).
Electrophoretic mobility shift assay.
The aptamer labeled at the 5′ end with 32P (13 nM, 10,000 cpm) was incubated for 10 min at 30°C in 8 μl of the binding buffer used for the in vitro selection process (20 mM Tris [pH 7.5], 5 mM MgCl2, 1 mM DTT, 40 mM NaCl). Increasing amounts of HCV strain H77 NS5B (31.2 nM to 1 μM in 2 μl) were then added, and incubation was continued for 30 min at 30°C. Two microliters of electrophoresis loading buffer (10 mM Tris [pH 8], 1 mM EDTA, 0.1% bromophenol blue, 0.1% xylene cyanol, 30% glycerol) was added to the samples before they were loaded onto a nondenaturing 10% polyacrylamide gel. The electrophoresis was carried out at 200 V for 6 h at room temperature. The gel was submitted to autoradiography, and the labeled bands corresponding to free aptamers were quantified with the Image J program. The percentage of bound ODN was plotted against the concentration of the protein by using Kaleidagraph (version 3.6) software (Synergy Software). The apparent Kd was defined as the concentration of NS5B that resulted in a 50% shift of the 32P-labeled aptamer.
For competition electrophoretic mobility shift assay experiments, (−)IRES RNA internally labeled with 32P (13 nM) was incubated with HCV strain H77 NS5B (500 nM), and increasing concentrations of aptamer were added to the RdRp reaction mixture in the absence of ribonucleotides.
Thermal denaturation.
The thermal denaturation of the aptamers (a 2.7 μM solution corresponds to an optical density at 260 nm of 1) in 20 mM Tris (pH 7.5), 100 mM NaCl, and 5 mM MgCl2 was monitored on a Cary 1 UV/visible spectrophotometer interfaced with a Peltier effect device that controls the temperature within ±0.1°C. A thin film of mineral oil was loaded onto each sample to counteract solvent evaporation at high temperatures. Buffer without aptamer was used as the reference. The rate of temperature increase or decrease was 0.5°C·min−1 from 5 to 90°C; and the absorbance at 260 nm, 295 nm, and 405 nm was recorded every 0.5°C. The first-derivative plots of the thermal melting profiles were calculated by using Kaleidagraph (version 3.6) software.
Chemical and enzymatic probing.
The 5′-end-labeled (50,000 cpm) aptamers were partially digested at 15, 20, 25, or 30°C with different concentrations of P1 nuclease in the binding buffer used during the in vitro selection process containing 0.5 μg salmon sperm DNA. Aliquots were taken at various times and mixed with an equal volume of loading buffer (95% formamide, 0.5 mM EDTA, 0.025% SDS, 0.1% bromophenol blue, 0.1% xylene cyanol). The cleaved products were analyzed on 20% polyacrylamide-7 M urea denaturing gels. For chemical probing, the labeled ODNs were incubated for 10 min with either 0.42 mM KMnO4 or 0.2% dimethyl sulfate (DMS) in 10 μl of binding buffer. One microliter of allylic alcohol (Sigma) was added to the KMnO4 reaction mixture. After addition of 100 μl of pyrrolidine (1.1 M; Sigma), both reaction mixtures were heated at 90°C for 15 min and dried under vacuum. The pellets were resuspended in 10 μl loading buffer, and the cleaved products were analyzed as described above.
Cell culture.
An HCV-permissive human hepatoma cell line (Huh7) was grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum and gentamicin (50 μg·ml−1) at 37°C in a 5% CO2 atmosphere. These cells (Huh7-QR) were obtained from the Huh7/Rep5.1 cell line cured for the Rep5.1 replicon by treatment with 150 U·ml−1 alpha interferon. After 3 weeks in the presence of this drug and the absence of G418, no replicon RNA could be detected (13).
RNA transfection.
HCV JFH1 genomic RNA was obtained by in vitro transcription from pJFH1-pUC, kindly provided by T. Wakita (47). One microgram of JFH1 RNA was transfected into Huh7 permissive cells with the DMRIE-C reagent according to the manufacturer's instructions (Invitrogen). After six passages, the culture medium (4.6 × 108 viral RNA copies·ml−1) was passed through a 0.45-μm-pore-size nitrocellulose filter and stored at −80°C before being used for the infection assays.
HCV infection and viral RNA quantification.
Twenty-four hours before infection, permissive Huh7 cells were seeded at a density of 5 × 104 cells/well in a 24-well plate. They were infected with 500 μl of the JFH1-producing cell supernatant (5 × 104 infectious particles) in DMEM containing different concentrations of aptamers or no aptamer. After 4 h at 37°C, 500 μl of DMEM-20% fetal calf serum was added, and the infected cells were cultured for 48 h. Total cellular RNAs were extracted with the TRIzol reagent according to the manufacturer's instructions (Invitrogen). Purified RNA was resuspended in 30 μl H2O treated with diethyl pyrocarbonate, and the viral RNA copy number was quantified by real time RT-PCR with an iScript one-step RT-PCR kit (Bio-Rad). The forward primer (5′-AAGTCTTTGGAGCCGTGCAA-3′) and the reverse primer (5′-TGTCTCAACGGGGATGAAAT-3′) were designed to amplify part of the NS3 region of HCV JFH1 RNA. The RT-PCR conditions were 1 cycle of 10 min at 50°C; 1 cycle of 5 min at 95°C; and 45 cycles of 10 s at 95°C, 30 s at 64°C, and 30 s at 72°C.
Infection of cells with secreted HCV and determination of infectivity.
Huh7 permissive cells were seeded at a density of 5 × 104 cells/well in Lab-tek chamber slides (eight wells). They were infected with 5 × 104 JFH1 infectious particles in 250 μl DMEM with or without aptamers, as described above. The cell monolayers were rinsed to eliminate the input virus and were then incubated for 48 h at 37°C in DMEM containing 10% fetal calf serum. The cell supernatants were harvested from the individual wells, diluted with an equal volume of DMEM containing 10% fetal calf serum, and used to reinfect Lab-tek chamber slides seeded with 25,000 Huh7 permissive cells. After 48 h of culture, the infected Huh7 cells were fixed with 3% paraformaldehyde and then permeabilized with 0.1% Triton X-100 in phosphate-buffered saline (PBS; 140 mM NaCl, 3 mM KCl, 8 mM Na2HPO4, 1.5 mM KH2PO4 [pH 7.4]). After three washes with PBS, the cells were incubated for 3 h at 4°C with rat monoclonal anti-E2 antibody 3/11, kindly provided by Czeslaw Wychowski (CNRS-UPR2511, Lille, France), in PBS containing 10% fetal calf serum (16). After three washes with PBS, the cells were incubated with Alexa Fluor 594-conjugated secondary antibodies. Coverslips were mounted on the slides with the Vectashield mounting reagent (Vector, Burlingame, CA). The foci of fluorescent cells were counted with a Zeiss microscope with a ×40 lens.
Cell proliferation assay.
The cytotoxicities of the aptamers were assessed by the CellTiTer 96 Aqueous One solution cell proliferation assay (Promega) according to the manufacturer's instructions. Briefly, Huh7-QR cells were seeded at a density of 2.5 × 104 cells/well in a 96-well plate. Twenty-four hours later, the cells were washed with DMEM without serum and incubated for 4 h in DMEM containing aptamer 27v, 127v, or IV-04 (an HIV Tar-specific aptamer [6]) at 5 μM or 1 μM. Then, 50 μl of DMEM-20% FCS was added to each well and incubation was continued for 24 h at 37°C. The culture medium was then replaced by DMEM (100 μl) containing 20 μl of CellTiTer 96 reagent. After a 1-h incubation at 37°C, the absorption was measured at 492 nm and compared to that of wells seeded with serial dilutions of cells grown without the ODNs (3,125 to 5 × 104 cells per well).
Confocal microscopy.
Huh7-QR permissive cells were plated on coverslips at 105 cells/well in 24-well plates and were grown for 24 h. The culture medium was discarded, 5′ Cy3-labeled ODNs (27v, 127v, and IV-04; 1 μM in 490 μl DMEM) were added to the culture, and the mixture was incubated for 15 min at room temperature, followed by the addition of 5 × 104 JFH1 virus and further incubation for 4 h at 37°C. The culture medium was adjusted to 1 ml with DMEM containing 20% fetal calf serum, and the cells were cultured for 24 h at 37°C. The cells were then washed three times with PBS and incubated in a solution of biotinylated concanavalin A (Sigma) at a final concentration of 50 μg·ml−1. After a wash with PBS, the cells were fixed with 500 μl of 3.7% formaldehyde for 5 min. The cells were washed again and then incubated with 300 μl of 1 μg·ml−1 4′-6-diaminido-2-phenylindole (Sigma) for 10 min. After three washes with PBS, the cells were incubated for 1 h with 300 μl of 5 μg·ml−1 streptavidin conjugated to Alexa Fluor 488 (Molecular Probes) and washed again with PBS, as described above. Coverslips were sealed onto the glass slides by using a drop of fluorescence mounting medium (Vectashield, hard set; Vector). The cells were observed with a Leica TCS SP5 confocal microscope (Plateforme d'Imagerie Cellulaire, IFR66, Université de Bordeaux 2) and a ×63 HCX PL APO CS lens.
RESULTS
Inhibition of HCV NS5B activity by DNA aptamers.
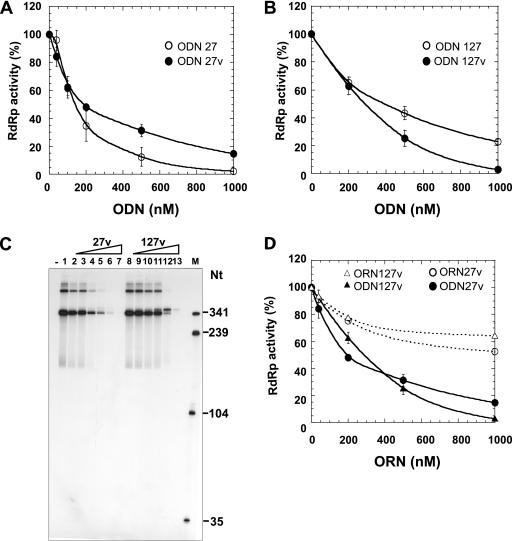
We have previously described the in vitro selection of DNA aptamers (81 nt) constituted by a 35-nt random sequence flanked by two constant sequences intended for PCR amplification (3). Among the 58 sequenced aptamers with a high affinity for HCV NS5B, 30 could be classified into four groups on the basis of their sequence homologies, and two of them (aptamers 27 and 89) were shown to be strong inhibitors of NS5B activity in vitro. The use of deleted forms of these aptamers allowed us to find that the inhibitory potential of ODN 27 was associated with its 35-nt-long random region. Indeed, this shortened aptamer, called ODN 27v, was able to inhibit the HCV H77 RdRp with an IC50 of 196 ± 16 nM, very similar to the value obtained for the original full-length 81-nt aptamer ODN 27 (IC50, 145 ± 5 nM) (Fig. 1A) (3).
FIG. 1.
Effects of ODNs and ORNs on RdRp activity of NS5B. RdRp assays were performed as described in Materials and Methods. Strain H77 NS5B at 150 nM was incubated in a 20-μl RdRp reaction mixture with 86 nM (−)IRES RNA and increasing amounts of 81-nt ODNs (ODN 27 or 127) or 35-nt ODNs (ODN 27v or 127v). (A) ODN 27 (open circles) and ODN 27v (closed circles); (B) ODN 127 (open circles) and ODN 127v (closed circles). The amount of RNA synthesis was determined by measuring the TCA-precipitable radioactivity. The results, which correspond to the mean values of three independent experiments, are expressed as relative activity, in which 100% activity corresponds to the enzymatic activity in the absence of ODN aptamers. (C) The RdRp assay was performed in the presence of [α-32P]UTP without an ODN (lane 1) or with 100 nM, 200 nM, 400 nM, 800 nM, 2 μM, and 4 μM ODN 27v (lanes 2 to 7, respectively) or ODN 127v (lanes 8 to 13, respectively). The 32P-labeled reaction products were denatured and loaded onto a 6% denaturing polyacrylamide gel. Lane −, incubation without enzyme; lane M, RNA size marker. (D) Increasing concentrations of DNA aptamers 27v (closed circles) and 127v (closed triangles) or oligonucleotides corresponding to the RNA version of aptamers 27v (open circles) and 127v (open triangles) were added to the polymerase reaction mixture. Error bars represent the standard deviations.
In order to identify additional aptamers able to inhibit HCV NS5B, we sequenced additional clones isolated from the same selection procedure and tested their effects on RNA synthesis in vitro. Hence, we found that one aptamer, ODN 127, inhibited HCV NS5B with an IC50 of 401 ± 46 nM (Fig. 1B). Aptamer 127 displayed an 11-nt consensus sequence (GCNNATTGTCC) which is also present in aptamers 27 and 89. An ODN corresponding to its 35-nt random region (ODN 127v) was synthesized and tested in vitro. Its IC50 was 322 ± 48 nM, with the total inhibition of RNA synthesis occurring at 1 μM (Fig. 1B). The inhibition of RNA synthesis was further examined by analyzing the 32P-labeled RNA products on denaturing polyacrylamide gels. Two products whose sizes were equal to or greater than the size of the template RNA were observed. As described previously (3), the amount of these products decreases with increasing concentrations of 27v (Fig. 1C, lanes 2 to 7). Addition of serial dilutions of 127v also lowered the amounts of these products, but at higher concentrations (0.8 to 4 μM), it induced the synthesis of a new product slightly longer than the template (Fig. 1C, lanes 8 to 13). Next, we checked if the DNA nature of the aptamers was important in the inhibition of the HCV NS5B activity. Thus, two ORNs whose nucleotide sequences were identical to those of ODNs 27v and 127v were synthesized, and their effects on RNA synthesis were determined. The results, presented in Fig. 1D, showed that ORNs corresponding to 27v and 127v showed weak inhibition of RNA synthesis by NS5B. We have focused our study on ODNs 27v and 127v, since they proved to be inhibitors as good as the selected aptamers but contained only the variable region.
Specificities of ODNs 27v and 127v for HCV NS5B.
We previously showed that ODN 27v did not inhibit the polymerase activity of poliovirus 3Dpol (3). To further analyze the specificity of HCV NS5B inhibition by these aptamers, we tested the effects of ODNs 27v and 127v on the activities of several viral polymerases, in addition to HCV H77 NS5B: HIV-1 RT and GBV-B NS5B for ODNs 27v and 127v and poliovirus 3Dpol for ODN 127v. As shown in Fig. 2A and B, HIV-1 RT was not affected or was only slightly affected by high concentrations of aptamers 27v and 127v. Similarly, ODN 127v did not inhibit 3Dpol RdRp.
FIG. 2.
Effects of ODNs 27v and 127v on the activities of different viral polymerases. Polymerase assays were performed as described in Materials and Methods. (A and B) Effects of ODN 27v (A) and ODN 127v (B) on the activities of the HIV-1 RT (open squares), HCV H77-NS5B (closed circles), poliovirus 3Dpol (closed triangles), and GBV-B NS5B (open triangles); (C) effects of ODN 27v (circles) and ODN 127v (triangles) on the activities of genotype 1a (closed symbols) and genotype 2a (open symbols) NS5B. The results correspond to the mean values of four independent experiments. They are expressed as relative activity, with 100% activity corresponding to the enzymatic activity in the absence of ODN. Error bars represent the standard deviations.
As HIV and poliovirus belong to virus families quite different from the family Flaviviridae, we analyzed the effects of the aptamers on the polymerase of GBV-B, a member of the family Flaviviridae closely related to HCV (35). ODNs 27v and 127v were able to inhibit the RNA synthesis directed by the GBV-B NS5B (Fig. 2A and B). However, this inhibition was only partial compared to the effect on HCV NS5B, as 33% and 47% of the enzyme activity was still observed with 1 μM ODN 27v and ODN 127v, respectively.
Aptamers 27 and 127 were selected by using the NS5B of an HCV genotype 1a strain (strain H77) as the target. We analyzed the effects of these aptamers on the RNA synthesis catalyzed by NS5B from an HCV genotype 2a strain (strain JFH1, used in the cell infection studies). As seen in Fig. 2C, the latter enzyme was as sensitive to the effect of aptamer 27v as the NS5B from the genotype 1a strain (compare the black and open circles in Fig. 2C). On the contrary, aptamer 127v inhibition of the polymerase activity of the JFH1 NS5B was weaker (compare the black and open triangles in Fig. 2C).
Binding properties of ODNs 27v and 127v.
In order to determine the binding affinities of ODNs 27v and 127v for the HCV strain H77 NS5B, we performed gel shift experiments using a fixed concentration of [32P]ODN and increasing amounts of enzyme. As shown in Fig. 3A and B, when the NS5B from the HCV genotype 1a strain was present at a high concentration (1 μM), 90% of ODNs 27v and 127v was bound. The dissociation constants (Kds) of the complex determined from the curves (Fig. 3) were 132.3 ± 20 nM for ODN 27v and 320.6 ± 21 nM for ODN 127v. A change in the slope of the ODN 127v binding curve was observed at between 100 and 200 nM NS5B, suggesting the presence of different molecular species in the ODN 127v preparation (see below). It should be noted that in the gel shift experiments described in a previous report (3), only 50% of ODN 27v was bound to the NS5B from a genotype 1a strain at concentrations as high as 1 μM. The introduction of a further purification step of the aptamers by using either precipitation by isopropanol or exclusion chromatography (G25 Sephadex spin column) explains the increased binding. The last step was essential to obtaining reproducible results with ODN 127v.
FIG. 3.
Determination of parameters of binding of aptamers 27v and 127v to HCV NS5B. The 5′-end-labeled ODN 27v (A) and ODN 127v (B) were incubated in 10 μl of selection buffer with increasing amounts of H77-NS5B (31.2 nM to 1 μM). After incubation, the NS5B-ODN complexes were separated from the free ODNs by electrophoresis on a nondenaturing polyacrylamide gel (left panels). The percentage of bound ODNs (right panels) was determined as described in Materials and Methods. The results correspond to the mean values of four independent experiments. Error bars represent the standard deviations.
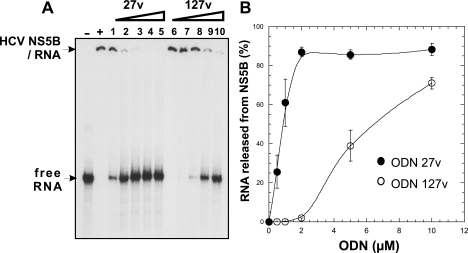
To determine if the binding of ODN 127v to the enzyme affected the formation of a RNA template-NS5B complex, competition experiments were carried out. Fixed concentrations of [32P]RNA template (13 nM) and HCV strain H77 NS5B (500 nM) were incubated with increasing concentrations of ODN 127v or ODN 27v as a control. The reaction products were analyzed by electrophoresis on nondenaturing polyacrylamide gels (Fig. 4A). In the absence of ODN, all of the RNA template was bound to NS5B (Fig. 4A, lane +). Only a small amount of RNA was released from NS5B when 1 μM ODN 127v was used (Fig. 4A, lane 7). The dissociation of the NS5B-RNA complex was readily visible with 5 μM ODN 127v and reached 70% at 10 μM (Fig. 4A, lane 10, and Fig. 4B). In agreement with the findings described in a previous report (3), 1 μM ODN 27v displaced about 60% of the (−)IRES RNA from the strain H77 NS5B. These results indicated that the binding of 127v to NS5B allowed the dissociation of the template RNA, but with an eightfold lower efficiency than that achieved with ODN 27v (Fig. 4A, lanes 1 to 5, and Fig. 4B).
FIG. 4.
Gel shift competition assay with HCV NS5B. (A) H77-NS5B (500 nM) incubated in 10 μl RdRp reaction mixture with 10,000 cpm 32P-labeled (−)IRES RNA (13 nM) and increasing amounts of ODN 27v (0.5 μM, 1 μM, 2 μM, 5 μM, and 10 μM [lanes 1 to 5, respectively]) or ODN 127v (0.5 μM, 1 μM, 2 μM, 5 μM, and 10 μM [lanes 6 to 10, respectively]) After incubation, the complexes were separated from free RNA by electrophoresis on a nondenaturing polyacrylamide gel. Lane −, no NS5B; lane +, no ODN plus NS5B. (B) The labeled bands corresponding to free RNA were quantified, and the percentage was plotted against the concentration of the DNA aptamers (closed circles, ODN 27v; open circles, ODN 127v). The data correspond to the mean values of three independent experiments. Error bars represent the standard deviations.
Mechanism of RNA synthesis inhibition by aptamers.
The fact that ODN 27v efficiently displaced the template RNA from the enzyme suggests that it inhibits initiation, the first step of RNA synthesis. To test this hypothesis, RdRp assays were performed by using single-cycle synthesis and heparin. Heparin can trap the free enzyme and therefore prevent the reinitiation of RNA synthesis, allowing direct study of the effect of ODN on the enzyme already engaged in initiation complexes. For this purpose, the initiation complex, comprising NS5B, the (−)IRES template, and the two initiating nucleotides CTP and GTP, was preformed for 30 min. Heparin was then added, followed by the addition of ATP and [32P]UTP, which allowed the elongation reaction to proceed. Several conditions were tested by adding the aptamer before or after the formation of the initiation complex. When the aptamer was added before the formation of the initiation complex, ODN 27v was able to fully inhibit RNA synthesis, as expected (Fig. 5A). When this aptamer was added after the formation of the initiation complex, immediately before the start of the elongation reaction (Fig. 5A), the inhibitory effect on RNA synthesis clearly decreased. Almost 60% of the RdRp activity persisted, whereas 5% of the RdRp activity persisted when ODN 27v was added before the formation of the initiation complex. When ODN 27v was incubated with the preformed initiation complex for an additional 30 min before the beginning of the elongation reaction (Fig. 5A), the decrease in inhibition caused by this ODN was less marked, since at 5 μM the enzyme activity was 40% of that of the control reaction performed without ODN. This difference was likely due to the fact that during the 30-min incubation preceding the beginning of elongation, ODN 27v might have partially dissociated the RNA-NS5B complex and/or induced structural changes in the enzyme, leading to stronger inhibition.
FIG. 5.
RdRp activity in a single-cycle round of synthesis. (A and B) Inhibition curves for ODN 27v (A) and ODN 127v (B) under single-cycle conditions. H77-NS5B (150 nM), (−)IRES template (86 nM), and CTP and GTP (0.5 mM each) were preincubated for 30 min at 30°C. Heparin was added, and elongation was started by the addition of 0.5 mM ATP plus [α-32P]UTP. Various concentrations of ODN were added at the same time as RNA and NS5B (closed circles) or as ATP and [α-32P]UTP at the beginning of elongation (closed triangles). In a third experiment (open circles), ODN was added after the formation of the RNA-NS5B complex and the mixture was incubated for 30 min at 30°C before the addition of heparin and ATP plus [α-32P]UTP. The data correspond to the means of four independent experiments. Error bars represent the standard deviations.
This drop in the inhibition of RNA polymerase activity suggested that the ODN failed to act on preformed NS5B-viral RNA-GTP-CTP complexes. However, had ODN 27v targeted only the initiation of RNA synthesis, no inhibition should have been observed. Thus, the residual inhibition observed is most probably due to an effect on elongation. We can thus conclude that ODN 27v acts on both initiation and elongation.
Similar experiments were performed with ODN 127v. As expected, when ODN 127v was added before the formation of the initiation complex, it fully inhibited RNA synthesis (Fig. 5B). However, when this aptamer was added after the formation of the initiation complex and immediately before the start of the elongation reaction (Fig. 5B), no inhibition was observed. When ODN 127v was incubated with the preformed initiation complex for an additional 30 min before the beginning of the elongation reaction (Fig. 5B), RNA synthesis was inhibited. Altogether, these results suggest that ODN 127v inhibits initiation but has no effect on the elongation step. As inhibition was also observed when the preformed initiation complex was incubated with ODN 127v (Fig. 5B), it is likely that it also prevents the transition step.
To confirm these data, we studied the effects of ODNs 27v and 127v on initiation and elongation by the gel-based assay described before (29). In this assay, initiation and elongation products can be analyzed separately. Inhibition of the initiation step was monitored by the synthesis of a 3-nt RNA product synthesized from the 3′ end ( —UCC-3′) of a 23-nt RNA template (Fig. 6A) by using [32P]ATP and GTP in the presence of various concentrations of aptamer for 30 min. At first we used Mn2+ as the divalent cation, as described by Liu et al. (29). When the concentration of ODN 127v was increased, the amount of the 3-nt product as well as that of a more slowly migrating product diminished (Fig. 6B, lanes 1 to 4) compared to the amounts in a control reaction performed without the aptamer (Fig. 6B, lanes 5 and 10). When the same experiment was performed with various concentrations of ODN 27v, the decrease in the 3-nt product was less pronounced even at high concentration, whereas the more slowly migrating band completely disappeared (Fig. 6B, lanes 6 to 9). As the 3′ sequence of aptamer 27v is TCC-3′, we hypothesized that part of the 3-nt product could be the result of an initiation that occurs on the ODN 27v used as the template. To test this hypothesis, we repeated the same experiment without template RNA. As shown in Fig. 6C, a labeled band migrating as a 3-nt product was clearly visible when NS5B was incubated with ODN 27v at 1 μM to 25 μM, whereas no signal above the background was visible when NS5B was incubated alone (Fig. 6C, lanes 5 and 10) or with ODN 127v (Fig. 6C, lanes 1 to 4). As the 3′ sequence of ODN 127v is TGCCG-3′, the absence of detectable initiation could result from the lack of CTP during the incubation period. To further confirm that NS5B cannot initiate RNA synthesis at the 3′ end of ODN 127v, RNA synthesis was examined by the synthesis of short RNA products by using [32P]ATP, CTP, and GTP in the presence of various concentrations of ODN 27v or 127v for 30 min without the 23-nt RNA template. Labeled RNA products were detected only in the presence of ODN 27v (Fig. 6D, lanes 6 to 9), whereas only the background signal was found with ODN 127v (Fig. 6D, lanes 1 to 4).
FIG. 6.
Analysis of ODN 27v and 127v inhibition in gel-based initiation and elongation assays. (A) Sequence of the 23-nt RNA template used in gel-based initiation and elongation assay. (B) Inhibition of initiation step; 1.13 μM H77-NS5B was incubated for 30 min with 1 μM 23-nt RNA template, [32P]ATP, and GTP without aptamer (lanes 5 and 10) or with 0.2 μM, 1 μM, 5 μM, or 25 μM ODN 127v (lanes 1 to 4, respectively) or ODN 27v (lanes 6 to 9, respectively). (C and D) Analysis of the initiation products synthesized without RNA template with [32P]ATP and GTP (C) or with [32P]ATP, CTP, and GTP (D). The lanes are as described above for panel B. (E) Inhibition of the elongation step; 1.13 μM H77-NS5B was incubated for 30 min with 1 μM 23-nt RNA template, ATP, and GTP without aptamer. Elongation was started by addition of UTP, CTP, [32P]ATP, and aptamers. The lanes corresponding to the elongation reaction with aptamer dilutions or without aptamers are as described above for panel B.
We then assessed the effects of ODNs 27v and 127v on elongation. For that, NS5B was preincubated with the 23-nt RNA, ATP, and GTP for 30 min before the addition of aptamers and the additional NTPs (CTP, UTP, and radiolabeled ATP). The elongation was monitored for the synthesis of a 23-nt product as well as higher-molecular-weight species resulting from template switching (29). The results of such an experiment are shown in Fig. 6E. In agreement with the results obtained by a single round of synthesis, they demonstrated that ODN 127v did not inhibit elongation. It should be noted that at the highest concentration tested (25 μM; Fig. 6E, lane 4), in addition to products whose sizes were equal to and greater than the size of the template product, an RNA slightly shorter than the template was also observed. On the contrary, ODN 27v strongly inhibited the elongation of RNA products initiated on the 23-nt RNA template (Fig. 6E, lanes 6 to 9). Moreover, at high concentrations (5 and 25 μM), aptamer 27v was used as a template by NS5B polymerase and copied into RNA (the bands indicated by an asterisk in lanes 8 and 9 of Fig. 6E). HCV NS5B has previously been shown to be able to synthesize RNA on a single-stranded DNA template, even though the efficiency of RNA synthesis was lower than that on an RNA template (10, 24). The experiments described above were performed in the presence of Mn2+, which is known to enhance the activities of polymerases but to decrease their fidelity compared to the fidelity achieved when Mg2+ is used. In addition, Mg2+ is the divalent cation most probably used in the cell. The effects of ODNs 27v and 127v on initiation and elongation were therefore reinvestigated in a gel-based assay, but in the presence of 5 mM MgCl2 instead of 2 mM MnCl2. As observed with Mn2+, ODN 127v inhibited only initiation, whereas ODN 27v prevented both initiation and elongation from the 23-nt RNA template. However, in the presence of Mg2+, the level of RNA synthesis from ODN 27v was low (data not shown). These results are in agreement with those presented in Fig. 1C, where no RNA product corresponding to the size of ODN 27v was visible.
Secondary structures of the aptamers.
To gain insight into the structural features of the aptamers, we combined UV melting experiments with chemical and enzymatic probing. Both melting curves obtained at 260 nm (Fig. 7A and B) displayed a single, fully reversible transition, indicating that both aptamers adopted a folded structure. First derivatives of the melting curves provided melting temperatures (Tms) of 38 and 35°C for ODNs 27v and 127v, respectively. These values are in agreement with the Tms predicted for the most stable structures of the aptamers by use of the mfold program (41) with parameter values set to the conditions used in the selection process (temperature, 30°C; [Na+], 0.1 M; [Mg2+], 0.005 M). Absorbance values were simultaneously recorded at 260 and 295 nm. No transition was detected at the latter wavelength, indicating the absence of G-quartet folding (33) or some other peculiar noncanonical assemblies, such as i motifs or triplex structures (34). Although the nucleotide sequences of ODNs 27v and 127v do not allow prediction of the formation of intramolecular G quartets, intermolecular G-quartet arrangements, such as dimers of aptamers (37), had to be considered since ODN 27v contains three G clusters in its sequence. Probing experiments were performed with DMS, since position N-7 of the G nucleotides participating in Hoogsteen or reverse Hoogsteen pairings, involved in the formation of G quartets or triplexes, are known to become nonreactive to methylation by DMS. The results from these experiments showed that all the G residues present in the sequences of both aptamers were identically reactive to the alkylating agent DMS (see Fig. S1 in the supplemental material). This led us to conclude that G residues involved in G-quartet structures or other triplexes were absent. Probing experiments by the use of either chemical modification of T residues with KMnO4 (T residues participating in a double helix are underreactive to KMnO4 in comparison to the reactivities of T residues in single-stranded regions) or enzymatic mapping with the single-strand-specific nuclease P1 did not provide us with patterns of protection compatible with any of the individual structures proposed by the mfold program (data not shown).
FIG. 7.
UV melting curves for the aptamers. The thermal denaturation of ODNs 27v (A) and ODN 127v (B) was monitored at 260 nm, as described in Materials and Methods. Denaturation and renaturation curves are shown by closed and open circles, respectively, and their corresponding first derivatives are indicated by continuous and dotted lines. OD, optical density. (C and D) Secondary structures of ODN 27v (C) and ODN 127v (D) predicted by mfold software.
This prompted us to consider that each aptamer may adopt two mutually exclusive alternative folding patterns with similar stabilities. The proposed structures of ODNs 27v and 127v are represented in Fig. 7C and D, respectively. They correspond to the two best structures calculated by the mfold program for each aptamer, as their predicted Tms matched the values determined experimentally. These two alternative conformations would be in equilibrium with each other, and at least one of the two forms could be recognized by the enzyme and drive equilibrium toward the conformation that binds to the protein. It can be noted that the consensus sequence common to both aptamers is located in a 3′ stem-loop in one predicted structure of ODN 27v and in a 5′ stem-loop in both predicted structures of ODN 127v.
Determination of aptamer regions involved in binding and/or inhibition.
In order to identify the minimal domain required for binding and/or inhibition, truncated versions of ODNs were generated. Table 1 summarizes the binding and inhibitory properties of ODN 27v mutants. We first tested 3′ truncated ODNs, called ODNs 27v_del1 and 27v_del2. A deletion of 10 nt (ODN 27v_del2) did not affect the affinity for NS5B, whereas a deletion of 19 nt abolished the binding of the ODN, suggesting that the minimal region for NS5B binding is localized between nucleotides 16 and 25. Even though deletion of the 3′ 10 nt in ODN 27v_del2 did not affect binding to the enzyme, the inhibitory potential was reduced by a factor of 3 (IC50, 629 ± 100 nM), suggesting that some elements involved in NS5B inhibition are localized in this aptamer domain.
TABLE 1.
Binding and inhibitory properties of truncated ODN 27va
| ODN | Sequence (5′-3′) | Kd (nM) | IC50 (μM) |
|---|---|---|---|
| 27v | ACGTACACTAGTGGTCCGGGCGGGGCTCATTGTCC | 132.3 ± 20 | 0.19 ± 0.1 |
| 27v_del1 | ACGTACACTAGTGGTCC------------------ | —b | — |
| 27v_del2 | ACGTACACTAGTGGTCCGGGCGGGG---------- | 140.4 ± 13 | 0.63 ± 0.1 |
| 27v_del3 | --------------------CGGGGCTCATTGTCC | — | 1.23 ± 0.2 |
| 27v_del6 | ACGTACACTAGTGGTCCGGGCGGGGCTCATTG--- | 109.5 | — |
| 27v_del7 | ACGTACACTAGTGGTCCGGGCGGGG--CATTG--- | 192.3 ± 10 | 3.21 ± 0.3 |
The Kd values of the mutant ODNs were determined as described in the legend to Fig. 3. IC50s were determined as described in Material and Methods. The data correspond to the means of three independent experiments ± SDs.
—, no binding or no inhibitory activity.
To analyze the role of the 3′ sequence, we used three additional mutants: ODN 27v_del3, which has a 5′-end 19-nt deletion which was able to fold as a stem-loop structure, according to the prediction of the mfold program, and ODNs 27v_del6 and 27v_del7, which harbor short deletions at the 3′ end. ODN 27v_del3 bound very poorly to NS5B; i.e., 20% was bound at an NS5B concentration of 1 μM, and it induced weak, albeit significant, inhibition of RNA synthesis (IC50, 1.23 ± 0.2 μM). The ODN 27v_del6 and 27v_del7 mutants bound to NS5B with an affinity similar to that of ODN 27v but did not inhibit or only poorly inhibited NS5B activity. Altogether, the data obtained with the five 27v deletion mutants demonstrated that two domains could be distinguished in ODN 27v. The 5′ region seemed to be essentially involved in polymerase binding, and the 3′ region seemed to be essentially involved in the inhibition of RNA synthesis.
By use of the same approach described above, several truncated mutants of ODN 127v were generated and tested. Deletion of any of the two stem-loops led to a loss in affinity and inhibition, suggesting that the 35 nt was necessary for binding and inhibition (data not shown).
Inhibition of JFH1 RNA synthesis and viral particle production.
Research efforts in the study of the multiplication of HCV particles have been hampered by the lack of a cellular system able to sustain the efficient replication of HCV. In this respect, the development of the replicon system, which mimics some intracellular virus steps, was an important breakthrough (30). More recently, Wakita and coworkers described a virus strain, termed JFH1, that productively replicates in Huh7 cells without cell culture adaptative mutations (47). To test whether aptamers 27v and 127v inhibit viral RNA synthesis in cell culture, a virus suspension (5 × 104 infectious particles/500 μl) containing three different concentrations of each aptamer was added to Huh7 cell cultures. Forty-eight hours later, the cellular RNAs were extracted and the viral RNA was quantified by real-time RT-PCR. Figure 8A depicts the results of three independent infection experiments. The HCV RNA copy number in cells infected with virus alone, i.e., from 7.8 × 106 to 22 × 106 HCV RNA copies/well following the experiments, was taken as 100%. This corresponded to the 156- to 440-fold amplification of input genomes. As described previously, the addition of interferon completely abolished HCV replication in Huh7 cells (13). When we added ODN 27v at the onset of infection, we observed 90%, 68%, and 19% reductions in RNA levels at concentrations of 5 μM, 1 μM, and 100 nM, respectively. Aptamer 127v reduced the HCV RNA level in Huh7 cells by 50% at a concentration of 5 μM, whereas no effect could be observed with control aptamer IV-04. By using the CellTiTer 96 Aqueous One solution, we determined that these three aptamers had no effect on cell proliferation, indicating that the decrease in the HCV RNA level observed in the presence of the ODN 27v was not due to a cytotoxic effect of this molecule (data not shown).
FIG. 8.
(A) Inhibition of JFH1 virus replication. The culture supernatant of Huh7-QR cells producing JFH1 virus (5 × 104 infectious viruses/500 μl) was used to infect naïve Huh7-QR cells. Viral particles were added either alone; with interferon at 150 U·ml−1; or with the various ODNs at 100 nM, 1 μM, or 5 μM. After a 48-h incubation, cellular RNAs were extracted from infected cells and the viral RNA copies were quantified by real-time RT-PCR. The results are expressed as a percentage of those obtained with cells infected with the virus alone. They correspond to the means of three independent infection experiments. Error bars represent the standard deviations. (B) Titration of HCV particles produced in the presence of DNA aptamers. A total of 5 × 104 infectious viruses/250 μl was used to infect naïve Huh7-QR cells in the presence or the absence of the DNA aptamers. After a 48-h incubation, the supernatants were used to infect Huh7-QR cells. HCV-infected cells were revealed after 48 h of culture with an anti-E2 antibody and Alexa Fluor 594-conjugated secondary antibodies, as described in Materials and Methods. The results are expressed as a percentage of those obtained with virus produced in the absence of aptamers. They correspond to the means of three independent infection experiments. Error bars represent the standard deviations. The left inset shows a typical focus of infected cells observed under fluorescent light. The right inset corresponds to the field observed under a phase-contrast microscope.
To confirm that the decrease in viral RNA synthesis induced by aptamers 27v and 127v correlated with a decrease in virus particle production, the supernatants of cells infected with JFH1 in the presence or the absence of the aptamers were used to reinfect naïve Huh7 cells. The foci of the fluorescent cells were revealed with an anti-E2 antibody and were numbered under a fluorescent microscopy. We performed three independent infection experiments. The number of infectious particles in the control varied from 440/ml to 1,660/ml following the experiments. The results presented in Fig. 8B confirmed the inhibitory effects of aptamers 27v and 127v, since the number of infectious particles decreased in the presence of aptamer 27v at 5 μM and 1 μM and in the presence of aptamer 127v at 5 μM, whereas ODN IV-04 had no effect. However, the decrease in the proportion of infectious particles produced in the presence of 5 μM and 1 μM ODN 27v was less pronounced than the decrease in the HCV RNA copy number (66% and 90%, respectively, at 5 μM and 25% and 68%, respectively, at 1 μM). This could result (i) from the use of a more concentrated viral suspension to get a sufficient number of foci and (ii) from the induction of new foci by surviving virus resulting from incomplete inhibition.
Entry of DNA aptamers into Huh7 cells.
To verify that these aptamers were able to enter Huh7-QR cells during HCV infection, we performed confocal microscopy experiments (see Fig. S2 in the supplemental material). Huh7-QR cells were infected with a dilution of JFH1 virus containing 1 μM of the ODN labeled at the 5′ end with Cy3. Twenty-four hours later, the cells were fixed with formaldehyde, the plasma membranes were labeled with biotinylated concanavalin A bound to Alexa Fluor 488-streptavidin, and the nuclei were stained with 4′-6-diaminido-2-phenylindole. As shown in Fig. S2 in the supplemental material, an intense red fluorescence corresponding to the three aptamers was observed in the cytoplasms of the infected cells (see Fig. S2, panels 2, 3, and 4, in the supplemental material). These results show unequivocally that the three aptamers used in this experiment were able to enter HCV-infected cells in the absence of transfection agents. Moreover, the cellular uptake of ODN 27v appeared to be more efficient than that of the other ODNs. This could explain, at least in part, the lower level of inhibition observed with ODN 127v.
DISCUSSION
Here we report on the characterization of two DNA aptamers that specifically bind to HCV RNA polymerase. ODNs 27v and 127v inhibited HCV NS5B activity with IC50s of 196 ± 16 nM and 322 ± 48 nM, respectively, values similar to or twice as high as the NS5B concentration used in our experiment (150 nM). The affinities of ODNs 27v and 127v for HCV NS5B were determined by gel shift experiments. The Kd values of 132.3 ± 20 nM and 320.6 ± 21 nM for ODNs 27v and 127v, respectively, showed that they have a relatively lower affinity for their target than other recently described DNA aptamers selected for the inhibition of HCV NS5B (23). This difference might be attributed to the selection conditions and the technique used, i.e., the surface plasmon resonance assay used in the study of Jones et al. (23) versus the filter binding assay used here (3). Moreover, differences in affinity may be ascribed to the source of NS5B, since our aptamers were selected by using the NS5B from subtype 1a, whereas those obtained by Jones et al. (23) were isolated by using a subgenotype 3a NS5B.
Studies challenging the specificity of the inhibition of HCV NS5B by ODNs 27v and 127v showed that these agents were not able to inhibit other polymerases, like HIV-1 RT and poliovirus 3Dpol, by using RNA as templates. Interestingly, these aptamers significantly inhibited the polymerase activity of the GBV-B NS5B. This enzyme shares 37% identity and 52% similarity with the HCV NS5B, and their polymerase activities also have common features; they may thus share ODN binding domains (35, 50).
The results of single-round-synthesis experiments as well as gel-based initiation and elongation assays showed that ODN 127v inhibits the initiation and transition steps of RNA synthesis but does not inhibit elongation, whereas ODN 27v inhibits both initiation and elongation. Although both aptamers inhibited initiation, our data strongly suggest that they interfere with this step by different mechanisms. ODN 27v competes with the RNA template, as shown by the data from competition gel shift assays indicating that it efficiently dissociates template RNA in complex with NS5B. More importantly, NS5B was able to initiate RNA synthesis at the 3′ end of ODN 27v in the presence of both Mn2+ and Mg2+ as divalent cations. In the presence of Mn2+, the molecules initiated were efficiently elongated (Fig. 6E), while in the presence of Mg2+, a very small amount of the full-length copy of ODN 27v was synthesized and only short abortive products were observed. These data were obtained in assays demanding high concentrations of the NS5B enzyme (1.13 μM) and template (1 μM). This could explain why, under the conditions routinely used for NS5B inhibition (150 nM NS5B and 87 nM template RNA) in the presence of Mg2+, no 35-nt RNA was synthesized when ODN 27v was used as the template (Fig. 1C). In the latter case, ODN 27v could displace template RNA from its NS5B binding site, leading to an abortive initiation. An RNA aptamer, R20-25, has been also shown to disrupt the 3′ UTR-NS5B complex, suggesting that it binds to the same domain as the natural template (46). The novelty of our observation is that this can be achieved with a short DNA molecule, even though the natural template of NS5B is RNA. Competitive inhibition of RNA-directed synthesis by DNA aptamers has been shown for HIV-1 RT. One example is RT1t49, which has been studied extensively (26). Other NS5B inhibitors have been shown to inhibit de novo initiation prior to the elongation step. This is the case of some nonnucleoside analogues, such as benzimidazoles (43) and benzothiadiazines (11, 18, 44), which have been reported to inhibit RNA synthesis by their direct action on the enzyme before the formation of the elongation complex. A major problem with antiviral molecules is the rapid appearance of resistant viral strains. In this regard, it has been shown that the inhibitory potential of RT1t49 on RT activity is relatively insensitive to sequence variations in the enzyme (26). In addition, the results obtained with the same aptamer indicated that RT mutants resistant to such an inhibitor that overlaps the template binding site have severe defects in HIV replication, suggesting that they could not be isolated in vivo (15).
In addition to its effect on the initiation step, ODN 27v also interfered with elongation. Indeed, when ODN 27v was added after the formation of the initiation complex, an additional inhibitory effect was observed (Fig. 5A). This was directly demonstrated in a gel-based assay (Fig. 6E). The lack of a visible arrest band indicates that ODN 27v decreases the elongation of RNA template by competing for and/or blocking a postinitiation event.
In contrast, ODN 127v appeared to block the initiation step by another mechanism. First, it only partially dissociated the NS5B-RNA complex at a concentration as high as 10 μM. Second, gel-based initiation assays showed that ODN 127v prevents the formation of the 3-nt initiation product but cannot be used as a template (Fig. 6B to D). Thus, it is likely that ODN 127v binding locks NS5B in a form that prevents initiation. The results of both the single-round-synthesis and the gel-based assays showed that ODN 127v did not inhibit elongation from the RNA template (Fig. 5B and 6E). However, this aptamer apparently decreases the fidelity of RNA replication, as shown by the appearance of new products slightly larger (Fig. 1C, lanes 11 to 13) or smaller (Fig. 6E, lane 4) than the template products. The fact that these two aptamers inhibit RNA synthesis by different mechanisms suggests that they bind to different domains of NS5B that are important for the activity of the enzyme. Determination of the residues involved in the interaction between NS5B and the aptamers may provide information about essential functional regions of NS5B. RNA aptamers that inhibit HCV NS5B were shown to bind to different sites on HCV NS5B. Aptamer R20-25, described by Vo et al. (46), binds to the same NS5B domain as the HCV 3′ UTR. On the contrary, the RNA aptamer selected by Biroccio and coworkers (4) binds to a basic patch near the C-terminal domain of NS5B.
In addition to their inhibitory potential, aptamers can be used with NS5B to understand the mechanisms involved in HCV RNA replication. Using two types of RNA aptamers specific for bacteriophage Qβ replicase, Brown and Gold have proposed a model for the replication of this bacteriophage (8). Kettenberger et al. have resolved the structure of the Saccharomyces cerevisiae RNA polymerase II-RNA aptamer FC* complex, allowing a better understanding of the transcription regulation mechanisms in this yeast (25). DNA aptamers directed against E. coli RNA polymerase have allowed the study of the mechanism of transcription in bacteria and the conformational flexibility of this enzyme (27). All these examples illustrate the potential of aptamers for use in the conduct of structure-function studies.
Finally, we determined that in addition to the in vitro inhibitory effect on RNA synthesis, aptamer 27v is also able to interfere with the RNA synthesis of HCV JFH1 in Huh7 cells. This was shown both by quantification of the HCV RNA by quantitative RT-PCR and by titration of the infectious particles produced in the presence of the aptamer. Although the efficient delivery of nucleic acids across the cell membrane remains a challenge, DNA or RNA aptamers acting as competitive inhibitors could represent useful drugs for the treatment of viral infections in the future. In this work we show that the best inhibitory aptamer, aptamer 27v, enters human cells during HCV infection and inhibits viral RNA synthesis. However, aptamer 27v is less efficient at the inhibition of JFH1 virus replication in Huh7 cells than it was in the in vitro experiments. Four main hypotheses can be proposed to explain this difference. First, the decrease in JFH1 genomic RNA could be due to innate immunity. Indeed, some CpG motifs or short DNA molecules could stimulate innate immunity pathways, but this seems unlikely, as no effect was observed with aptamer IV-04, used as a control. Second, the conditions of RNA synthesis in vitro and in infected cells were strictly different: in vitro we used a purified and soluble enzyme, whereas in cellulo, HCV NS5B works in complex with viral and cellular proteins in a specific membrane compartment (17, 42). Third, the aptamers used in our experiments were unmodified. Consequently, they were sensitive to cellular nucleases, even though DNA molecules have been shown to be more stable than RNA. Chemical modifications could improve their lifetimes in the cell. Finally, the results of the experiments reported here do not allow us to definitively establish that the mechanism of action of aptamer 27v within the cell is exclusively through the inhibition of polymerase. Further experiments must be performed to clarify this point.
In the case of aptamer 127v, this aptamer did not reduce or only slightly reduced the HCV RNA level even at a concentration of 5 μM. The lack of a significant effect of aptamer 127v on strain JFH1 RNA synthesis is in agreement with data obtained in the in vitro assay, showing that it only partially inhibits JFH1 NS5B activity (Fig. 2C).
Here, we have characterized in further detail two DNA aptamers able to inhibit specifically the activity of the HCV RNA polymerase in vitro. These results illustrate the power of the selective evolution of ligands by an exponential enrichment strategy for the isolation of specific ODNs able to inhibit HCV NS5B by different mechanisms. Moreover, the inhibitory effects observed for human cells infected with HCV show that aptamers may be useful tools for studying HCV RNA replication, thus becoming a very attractive and alternative approach to therapy against HCV.
Supplementary Material
Acknowledgments
This work was supported by the Agence Nationale de Recherche contre le Sida (ANRS), the Centre National de la Recherche Scientifique, and the University Victor Segalen Bordeaux 2. P.B. was supported by a Ph.D. fellowship from the ANRS.
We are grateful to S. Litvak for discussions and encouragement.
Footnotes
Published ahead of print on 27 March 2008.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Ago, H., T. Adachi, A. Yoshida, M. Yamamoto, N. Habuka, K. Yatsunami, and M. Miyano. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Structure 7:1417-1426. [DOI] [PubMed] [Google Scholar]
- 2.Astier-Gin, T., P. Bellecave, S. Litvak, and M. Ventura. 2005. Template requirements and binding of hepatitis C virus NS5B polymerase during in vitro RNA synthesis from the 3′-end of virus minus-strand RNA. FEBS J. 272:3872-3886. [DOI] [PubMed] [Google Scholar]
- 3.Bellecave, P., M. L. Andreola, M. Ventura, L. Tarrago-Litvak, S. Litvak, and T. Astier-Gin. 2003. Selection of DNA aptamers that bind the RNA-dependent RNA polymerase of hepatitis C virus and inhibit viral RNA synthesis in vitro. Oligonucleotides 13:455-463. [DOI] [PubMed] [Google Scholar]
- 4.Biroccio, A., J. Hamm, I. Incitti, R. De Francesco, and L. Tomei. 2002. Selection of RNA aptamers that are specific and high-affinity ligands of the hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 76:3688-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biswal, B. K., M. M. Cherney, M. Wang, L. Chan, C. G. Yannopoulos, D. Bilimoria, O. Nicolas, J. Bedard, and M. N. G. James. 2005. Crystal structures of the RNA-dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors. J. Biol. Chem. 280:18202-18210. [DOI] [PubMed] [Google Scholar]
- 6.Boiziau, C., E. Dausse, L. Yurchenko, and J. J. Toulme. 1999. DNA aptamers selected against the HIV-1 trans-activation-responsive RNA element form RNA-DNA kissing complexes. J. Biol. Chem. 274:12730-12737. [DOI] [PubMed] [Google Scholar]
- 7.Bressanelli, S., L. Tomei, A. Roussel, I. Incitti, R. L. Vitale, M. Mathieu, R. De Francesco, and F. A. Rey. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl. Acad. Sci. USA 96:13034-13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, D., and L. Gold. 1996. RNA replication by Q beta replicase: a working model. Proc. Natl. Acad. Sci. USA 93:11558-11562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheney, I. W., S. Naim, V. C. Lai, S. Dempsey, D. Bellows, M. P. Walker, J. H. Shim, N. Horscroft, Z. Hong, and W. Zhong. 2002. Mutations in NS5B polymerase of hepatitis C virus: impacts on in vitro enzymatic activity and viral RNA replication in the subgenomic replicon cell culture. Virology 297:298-306. [DOI] [PubMed] [Google Scholar]
- 10.Deval, J., C. M. D'Abramo, Z. Zhao, S. McCormick, D. Coutsinos, S. Hess, M. Kvaratskhelia, and M. Gotte. 2007. High resolution footprinting of the hepatitis C virus polymerase NS5B in complex with RNA. J. Biol. Chem. 282:16907-16916. [DOI] [PubMed] [Google Scholar]
- 11.Dhanak, D., K. J. Duffy, V. K. Johnston, J. Lin-Goerke, M. Darcy, A. N. Shaw, B. Gu, C. Silverman, A. T. Gates, M. R. Nonnemacher, et al. 2002. Identification and biological characterization of heterocyclic inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 277:38322-38327. [DOI] [PubMed] [Google Scholar]
- 12.Dufour, E., J. Reinbolt, M. Castroviejo, B. Ehresmann, S. Litvak, L. Tarrago-Litvak, and M. L. Andreola. 1999. Cross-linking localization of a HIV-1 reverse transcriptase peptide involved in the binding of primer tRNALys3. J. Mol. Biol. 285:1339-1346. [DOI] [PubMed] [Google Scholar]
- 13.Dumas, E., C. Masante, T. Astier-Gin, D. Lapaillerie, and M. Ventura. 2007. The hepatitis C virus minigenome: a new cellular model for studying viral replication. J. Virol. Methods 142:59-66. [DOI] [PubMed] [Google Scholar]
- 14.Dutartre, H., J. Boretto, J.-C. Guillemot, and B. Canard. 2004. A relaxed discrimination of 2′-O-methyl GTP relative to GTP between de novo and elongative RNA synthesis by the hepatitis C RNA-dependent RNA polymerase NS5B. J. Biol. Chem. 280:6359-6368. [DOI] [PubMed] [Google Scholar]
- 15.Fisher, T. S., P. Joshi, and V. R. Prasad. 2002. Mutations that confer resistance to template-analog inhibitors of human immunodeficiency virus (HIV) type 1 reverse transcriptase lead to severe defects in HIV replication. J. Virol. 76:4068-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flint, M., C. Maidens, L. D. Loomis-Price, C. Shotton, J. Dubuisson, P. Monk, A. Higginbottom, S. Levy, and J. A. McKeating. 1999. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative receptor, CD81. J. Virol. 73:6235-6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gosert, R., D. Egger, V. Lohmann, R. Bartenschlager, H. E. Blum, K. Bienz, and D. Moradpour. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 77:5487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu, B., V. K. Johnston, L. L. Gutshall, T. T. Nguyen, R. R. Gontarek, M. G. Darcy, R. Tedesco, D. Dhanak, K. J. Duffy, C. C. Kao, et al. 2003. Arresting initiation of HCV RNA synthesis using heterocyclic derivatives. J. Biol. Chem. 278:16602-16607. [DOI] [PubMed] [Google Scholar]
- 19.Hong, Z., C. E. Cameron, M. P. Walker, C. Castro, N. Yao, J. Y. Lau, and W. Zhong. 2001. A novel mechanism to ensure terminal initiation by hepatitis C virus NS5B polymerase. Virology 285:6-11. [DOI] [PubMed] [Google Scholar]
- 20.Hoofnagle, J. H. 2002. Course and outcome of hepatitis C. Hepatology 36:S21-S29. [DOI] [PubMed] [Google Scholar]
- 21.Huang, D. B., D. Vu, L. A. Cassiday, J. M. Zimmerman, L. J. Maher, and G. Ghosh. 2003. Crystal structure of NF-kappaB (p50)2 complexed to a high-affinity RNA aptamer. Proc. Natl. Acad. Sci. USA 100:9268-9273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jayasena, S. D. 1999. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 45:1628-1650. [PubMed] [Google Scholar]
- 23.Jones, L. A., L. E. Clancy, W. D. Rawlinson, and P. A. White. 2006. High-affinity aptamers to subtype 3a hepatitis C virus polymerase display genotypic specificity. Antimicrob. Agents Chemother. 50:3019-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kao, C. C., X. Yang, A. Kline, Q. M. Wang, D. Barket, and B. A. Heinz. 2000. Template requirements for RNA synthesis by a recombinant hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 74:11121-11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kettenberger, H., A. Eisenfuhr, F. Brueckner, M. Theis, M. Famulok, and P. Cramer. 2006. Structure of an RNA polymerase II-RNA inhibitor complex elucidates transcription regulation by noncoding RNAs. Nat. Struct. Mol. Biol. 13:44-48. [DOI] [PubMed] [Google Scholar]
- 26.Kissel, J. D., D. M. Held, R. W. Hardy, and D. H. Burke. 2007. Single-stranded DNA aptamer RT1t49 inhibits RT polymerase and RNase H functions of HIV type 1, HIV type 2, and SIVCPZ RTs. AIDS Res. Hum. Retrovir. 23:699-708. [DOI] [PubMed] [Google Scholar]
- 27.Kulbachinskiy, A., A. Feklistov, I. Krasheninnikov, A. Goldfarb, and V. Nikiforov. 2004. Aptamers to Escherichia coli core RNA polymerase that sense its interaction with rifampicin, sigma-subunit and GreB. Eur. J. Biochem. 271:4921-4931. [DOI] [PubMed] [Google Scholar]
- 28.Lesburg, C. A., M. B. Cable, E. Ferrari, Z. Hong, A. F. Mannarino, and P. C. Weber. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6:937-943. [DOI] [PubMed] [Google Scholar]
- 29.Liu, Y., W. W. Jiang, J. Pratt, T. Rockway, K. Harris, S. Vasavanonda, R. Tripathi, R. Pithawalla, and W. M. Kati. 2006. Mechanistic study of HCV polymerase inhibitors at individual steps of the polymerization reaction. Biochemistry 45:11312-11323. [DOI] [PubMed] [Google Scholar]
- 30.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 31.Luo, G., R. K. Hamatake, D. M. Mathis, J. Racela, K. L. Rigat, J. Lemm, and R. J. Colonno. 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol. 74:851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manns, M. P., J. G. McHutchison, et al. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958-965. [DOI] [PubMed] [Google Scholar]
- 33.Mergny, J. L., A. T. Phan, and L. Lacroix. 1998. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 435:74-78. [DOI] [PubMed] [Google Scholar]
- 34.Mergny, J. L., J. Li, L. Lacroix, S. Amrane, and J. B. Chaires. 2005. Thermal difference spectra: a specific signature for nucleic acid structure. Nucleic Acids Res. 33:e138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muerhoff, A. S., T. P. Leary, J. N. Simons, T. J. Pilot-Matias, G. J. Dawson, J. C. Erker, M. L. Chalmers, G. G. Schlauder, S. M. Desai, and I. K. Mushahwar. 1995. Genomic organization of GB viruses A and B: two new members of the Flaviviridae associated with GB agent hepatitis. J. Virol. 69:5621-5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nimjee, S. M., C. P. Rusconi, and B. A. Sullenger. 2005. Aptamers: an emerging class of therapeutics. Annu. Rev. Med. 56:555-583. [DOI] [PubMed] [Google Scholar]
- 37.Phan, A. T., V. Kuryavyi, J. B. Ma, A. Faure, M. L. Andreola, and D. J. Patel. 2005. An interlocked dimeric parallel-stranded DNA quadruplex: a potent inhibitor of HIV-1 integrase Proc. Natl. Acad. Sci. USA 102:634-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reigadas, S., M. Ventura, L. Sarih-Cottin, M. Castroviejo, S. Litvak, and T. Astier-Gin. 2001. HCV RNA-dependent RNA polymerase replicates in vitro the 3′ terminal region of the minus strand viral RNA more efficiently than the 3′ terminal region of the plus RNA. Eur. J. Biochem. 268:5857-5867. [DOI] [PubMed] [Google Scholar]
- 39.Saito, I., T. Miyamura, A. Ohbayashi, H. Harada, T. Katayama, S. Kikuchi, Y. Watanabe, S. Koi, M. Onji, Y. Ohta, et al. 1990. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma Proc. Natl. Acad. Sci. USA 87:6547-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanchez-Pescador, R., M. D. Power, P. J. Barr, K. S. Steimer, M. M. Stempien, S. L. Brown-Shimer, W. W. Gee, A. Renard, A. Randolph, J. A. Levy, D. Dina, and P. A. Luciw. 1985. Nucleotide sequence and expression of an AIDS-associated retrovirus (ARV-2). Science 227:484-492. [DOI] [PubMed] [Google Scholar]
- 41.SantaLucia, J., Jr. 1998. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. USA 95:1460-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi, S. T., K. J. Lee, H. Aizaki, S. B. Hwang, and M. M. C. Lai. 2003. Hepatitis C virus RNA replication occurs on a detergent-resistant membrane that cofractionates with caveolin-2. J. Virol. 77:4160-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tomei, L., S. Altamura, L. Bartholomew, A. Biroccio, A. Ceccacci, L. Pacini, F. Narjes, N. Gennari, M. Bisbocci, I. Incitti, et al. 2003. Mechanism of action and antiviral activity of benzimidazole-based allosteric inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 77:13225-13231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tomei, L., S. Altamura, L. Bartholomew, M. Bisbocci, C. Bailey, M. Bosserman, A. Cellucci, E. Forte, I. Incitti, L. Orsatti, et al. 2004. Characterization of the inhibition of hepatitis C virus RNA replication by nonnucleosides. J. Virol. 78:938-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toulme, J. J. 2000. Aptamers: selected oligonucleotides for therapy. Curr. Opin. Mol. Ther. 2:318-324. [PubMed] [Google Scholar]
- 46.Vo, N. V., J. W. Oh, and M. M. Lai. 2003. Identification of RNA ligands that bind hepatitis C virus polymerase selectively and inhibit its RNA synthesis from the natural viral RNA templates. Virology 307:301-316. [DOI] [PubMed] [Google Scholar]
- 47.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H.-G. Krausslich, M. Mizokami, et al. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu, J. Z., N. Yao, M. Walker, and Z. Hong. 2005. Recent advances in discovery and development of promising therapeutics against hepatitis C virus NS5B RNA-dependent RNA polymerase. Mini Rev. Med. Chem. 5:1103-1112. [DOI] [PubMed] [Google Scholar]
- 49.Zhong, W., A. S. Uss, E. Ferrari, J. Y. Lau, and Z. Hong. 2000. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase J. Virol. 74:2017-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhong, W., P. Ingravallo, J. Wright-Minogue, A. S. Uss, A. Skelton, E. Ferrari, J. Y. Lau, and Z. Hong. 2000. RNA-dependent RNA polymerase activity encoded by GB virus-B non-structural protein 5B. J. Viral Hepat. 7:335-342. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.