Abstract
The development of effective therapies for hepatitis C virus (HCV) must take into account genetic variation among HCV strains. Response rates to interferon-based treatments, including the current preferred treatment of pegylated alpha interferon administered with ribavirin, are genotype specific. Of the numerous HCV inhibitors currently in development as antiviral drugs, nucleoside analogs that target the conserved NS5B active site seem to be quite effective against diverse HCV strains. To test this hypothesis, we examined the effects of a panel of nucleotide analogs, including ribavirin triphosphate (RTP) and several chain-terminating nucleoside triphosphates, on the activities of purified HCV NS5B polymerases derived from genotype 1a, 1b, and 2a strains. Unlike the genotype-specific effects on NS5B activity reported previously for nonnucleoside inhibitors (F. Pauwels, W. Mostmans, L. M. Quirynen, L. van der Helm, C. W. Boutton, A. S. Rueff, E. Cleiren, P. Raboisson, D. Surleraux, O. Nyanguile, and K. A. Simmen, J. Virol. 81:6909-6919, 2007), only minor differences in inhibition were observed among the various genotypes; thus, nucleoside analogs that are current drug candidates may be more promising for treatment of a broader variety of HCV strains. We also examined the effects of RTP on the HCV NS3 helicase/ATPase. As with the polymerase, only minor differences were observed among 1a-, 1b-, and 2a-derived enzymes. RTP did not inhibit the rate of NS3 helicase-catalyzed DNA unwinding but served instead as a substrate to fuel unwinding. NS3 added to RNA synthesis reactions relieved inhibition of the polymerase by RTP, presumably due to RTP hydrolysis. These results suggest that NS3 can limit the incorporation of ribavirin into viral RNA, thus reducing its inhibitory or mutagenic effects.
Hepatitis C virus (HCV) infects more than 170 million people worldwide, causing chronic hepatitis, cirrhosis, hepatocellular carcinoma, and/or liver failure (12). Clinical HCV isolates are categorized into different genotypes that display up to 30% sequence variation. Response rates to all interferon-based HCV therapies are genotype dependent, with patients infected with HCV genotype 1 having about one-half the chance of a sustained virological response as patients infected with other genotypes (37). Current HCV therapies combine pegylated alpha interferon and ribavirin (1-β-d-ribofuranosyl-1,2,4-triazole-3-carboxamide). A number of viral factors have been proposed to contribute to the different response rates to interferon (reviewed in reference 59), but less is known about the effects of viral variation on sensitivity to ribavirin. Here, we employ biochemical methods to examine the simple hypothesis that as a nucleoside analog, ribavirin may affect the enzymes encoded by HCV, and that variation among the enzymes from different genotypes could affect ribavirin response rates. We also examine whether HCV genetic variation influences the sensitivities of HCV enzymes to other nucleoside analogs similar to those currently in development as HCV-specific drugs.
The ∼9,600-nucleotide HCV genome harbors a single open reading frame that encodes a polyprotein, which is processed into 10 distinct proteins by both host and viral proteases (see reference 52 for a recent review). The two HCV proteins that interact most intimately with nucleoside triphosphates (NTPs) and their analogs are the nonstructural (NS) proteins NS5B (8) and NS3 helicase (18). NS3 is a bifunctional protein consisting of a protease at the N terminus and a helicase at the C terminus. The NS3 serine protease is responsible for processing the viral proteins from NS3 to NS5B, while the ATP-dependent NS3 helicase is presumably required for unwinding the duplex RNA and/or resolving secondary RNA structures. NS5B is an RNA-dependent RNA polymerase responsible for replicating the viral RNA.
The purine analog ribavirin inhibits the replication of a variety of RNA and DNA viruses (49). Compared to interferon monotherapy, pegylated alpha interferon in combination with ribavirin has led to substantially improved rates of sustained virological response in hepatitis C patients; however, the mechanism of ribavirin antiviral activity is still under debate. Four different mechanisms have been proposed (reviewed in reference 59). One proposed mechanism is the enhancement of the host adaptive antiviral immune response. A second is inhibition of host IMP dehydrogenase. In cells, ribavirin is converted into a nucleotide by adenosine kinase (58). Ribavirin monophosphate (RMP) then acts to lower GTP pools in ribavirin-treated cells because RMP is a competitive inhibitor of IMP dehydrogenase, which catalyzes the rate-limiting step of GTP synthesis (39). A third proposed antiviral mechanism is direct inhibition of the NS5B polymerase. Cellular kinases convert RMP to ribavirin diphosphate (RDP) and ribavirin triphosphate (RTP) (22). RTP acts as a competitive inhibitor of viral replication; once incorporated, it presents a significant block to RNA elongation and is a poor template for RNA synthesis. The fourth proposed mechanism is viral-RNA mutagenesis and “error catastrophe.” Ribavirin that has been incorporated into viral RNA can act as a mutagen because its base (1,2,4-triazole-3-carboxamide) can pair with either cytosine or uracil (15). As demonstrated with poliovirus, the addition of ribavirin-induced mutations to an already error-prone viral replication system leads to error catastrophe and the elimination of a viable infecting population (14). HCV NS5B is capable of using RTP as a substrate to incorporate ribavirin into viral RNA (36, 55); however, whether ribavirin acts to eliminate HCV in patients through error catastrophe is still debated (51).
Numerous NS5B polymerase inhibitors are presently being tested in patients. NS5B inhibitors can be divided into four different classes based on where they bind the enzyme. NS5B has the right-handed shape of a typical polymerase with a palm region containing the active site and finger and thumb domains. Potent inhibitors have been found that bind the palm (either nucleoside analogs or nonnucleoside inhibitors) and to two different sites on the thumb domain. There is some evidence that nucleoside inhibitors may be more broadly effective than nonnucleoside inhibitors. The results of one comparative study utilizing HCV replicons indicated that the nucleoside analog 2′-C-methyladenosine was similarly effective against diverse HCV isolates, in contrast to the variation observed with nonnucleoside inhibitors (35). In agreement with these cell-based assays are the results of a study of NS5B proteins derived from several HCV genotypes by Pauwels et al., which helped elucidate the role of variable NS5B residues on nonnucleoside inhibitor binding (43). We extend these studies with a biochemical analysis examining the influence of genotypic variation on NS5B sensitivity to nucleotide analogs.
Nucleotide-based NS5B inhibitors include mainly NTPs modified at the 2′ position, which are efficient nonobligate chain-terminating inhibitors of HCV replication. Such compounds include 2′-C-methyladenosine triphosphate, 2′-O-methylcytidine (10), and 2′-C-methylcytidine-3′-O-l-valine (valopicitabine; NM283) (46). NM283 is a prodrug that is cellularly converted to 2′-C-methyl-CTP, which is used as a substrate by NS5B to terminate a growing RNA chain. NS5B does not efficiently elongate chains terminated with 2′-C-methyl-C, even though they still possess a 3′-OH. Thus, in contrast to obligate chain terminators lacking a 3′-OH (e.g., zidovudine), these new NS5B inhibitors function as “nonobligate” chain terminators (16, 17). Molecular modeling suggests that the 3′-OH of such compounds is misaligned in the NS5B active site so that it is unable to accept the attack of an incoming NTP (38). Other NTP analogs have also been shown to inhibit HCV replication via the NS5B protein and are in various stages of development. They include 2′-deoxy-2′-fluorocytidine, which inhibits replication of subgenomic replicons in Huh-7 cells (50). Similarly, R7128, a prodrug of β-d-2′-deoxy-2′-fluoro-2′-C-methyl-cytidine, is being tested in combination with interferon and ribavirin (41), and 4′-azidocytidine is in preclinical trials (28).
In this study, we report the characterization of the NS5B polymerase from four isolates of HCV representing the 1a, 1b, and 2a genotypes. Although activities differed in steady-state RNA synthesis assays, each polymerase interacted similarly with NTP analogs. In agreement with previous reports, our results show that RTP is a poor substrate for NS5B (36, 55). The interaction of RTP with the NS3 helicase was also examined. NS3 helicase proteins derived from the 1a, 1b, and 2a genotypes all used RTP to fuel unwinding, and the resulting hydrolysis of RTP to RDP was found to abrogate RTP effects on NS5B. Thus, we uncover here a new mechanism whereby HCV can reduce the inhibitory effect of ribavirin by preventing its incorporation into viral RNA.
MATERIALS AND METHODS
Reagents.
Poly(U), poly(C), and ribavirin were purchased from Sigma (St. Louis, MO). RTP was purchased from Moravek Biochemicals, Inc. (Brea, CA), or Axxora (San Diego, CA). GTP, 3′-dGTP, 2′-O-methyl-GTP, 2′-O-methyl-ITP, ITP, 2′-dGTP, and 2′,3′-ddGTP were purchased from TriLink Biotechnologies (San Diego, CA). All oligonucleotides were purchased from Integrated DNA Technology (Coralville, IA), and their concentrations were determined from extinction coefficients provided by the manufacturers.
Proteins.
NS5BΔ21, NS3 helicase fragment (NS3 amino acids 166 to 631) (NS3h), and NS3 were expressed and purified in Escherichia coli as recombinant proteins using DNA subcloned from cDNAs of HCV isolates from genotype 1a_H77 (pCV-H77c) (61), genotype 1b_J4 (pJ4L6S) (64), genotype 1b_con1 (pBDL429P+S+) (34), and genotype 2a (pHC-J6CF) (63).
For expression of the various HCV polymerases, a set of plasmids was constructed based on the scheme described by O'Farrell et al. (42). The coding sequences for NS5B lacking 21 C-terminal residues were PCR amplified from the HCV isolates indicated above. The following primer pairs, containing an NheI or SpeI restriction site in the upstream primer and an XhoI site in the downstream primer, were used: 1a(H77), 5′-CTA GCT AGC ATG TCT TAT TCC TGG ACA-3′ and 5′-CCG CTC GAG GCG GGG CCG GGC ATG AGA C-3′; 1b(J4), 5′-CTA GCT AGC ATG TCC TAT ACG TGG ACA-3′ and 5′-CCG CTC GAG GCG GGG TCG GGC ACG AGA-3′; 1b(con1), 5′-CTA GCT AGC ATG TCC TAC ACA TGG ACA-3′ and the same downstream primer as J4; and 2a(J6), 5′-CTA ACT AGT ATG TCA TAC TCC TGG ACC GG-3′ and 5′-CCG CTC GAG GCG GGG TCG GGC ACG CGA CA-3′ (restriction sites are underlined). The purified PCR products were digested with the appropriate restriction enzymes and ligated to the NheI/XhoI-digested pET23a vector (Novagen) to generate C-terminally His-tagged expression constructs. Each recombinant protein contained an additional Met and Ala at the N terminus and the sequence LEHHHHHH fused to the C terminus.
The NS5B expression plasmids were transformed into E. coli Rosetta(DE3) (EMD Chemicals, Inc., Gibbstown, NJ). The same protocol was used to express and purify each polymerase. Cells harboring the expression plasmid were grown overnight at 30°C. Cells from 20 ml of the starter culture were harvested and used to inoculate 2 liters of LB medium. The cells were grown at 37°C to an absorbance at 600 nm of 0.8 and induced with 0.4 mM isopropyl-β-d-thiogalactopyranoside. After growth at 25°C for 4 h, cells were harvested by centrifugation and resuspended in 50 mM sodium phosphate (pH 7.8), 400 mM NaCl, 10% glycerol, 0.25% Triton X-100, 5 mM β-mercaptoethanol, and 20 mM imidazole (buffer A) supplemented with protease inhibitor cocktail for His-tagged proteins (Sigma). The cells were lysed by sonication, and the extract was cleared by centrifugation at 18,500 × g for 20 min (fraction I). Fraction I was loaded onto an Ni-nitrilotriacetic acid column (Novagen) equilibrated with buffer A. The column was washed and eluted with buffer A containing a gradient of imidazole from 20 mM to 1 M. Fractions containing polymerase were combined and dialyzed (fraction II) overnight against 50 mM HEPES (pH 7.5), 200 mM NaCl, 10% glycerol, 0.25% Triton X-100, 0.1 mM dithiothreitol, 0.1 mM EDTA (buffer B). The sample was then applied to a Fractogel EMD COO− column (EMD Biosciences) equilibrated with buffer B, and the protein was eluted with a gradient of 200 to 500 mM NaCl (fraction III). Fraction III was loaded onto a Blue Sepharose 6 Fast Flow column (Amersham) equilibrated with buffer B containing 500 mM NaCl. The column was washed with this buffer, and then the protein was eluted with buffer B containing 2.5 M NaCl. The combined fractions were dialyzed against buffer B containing 30% glycerol for storage (fraction IV). The purified protein was concentrated in a Centricon-10 spin column (Amicon), and the protein concentration was measured by sodium dodecyl sulfate-polyacrylamide gel electrophoresis with bovine serum albumin as a standard.
The three truncated NS3 proteins (NS3h) lacking the protease domain have been described previously (30). Each NS3h contained NS3 residues 166 to 631, followed by a C-terminal PNSSSVDKLAAALEHHHHHH tag. Previously, NS3h_1a(H77) was called Hel-1a (30-32) and Hel-His (20), NS3h_1b(J4) was called Hel-1b (19, 30, 32), and NS3h_2a(J6) was called Hel-2a (19, 30, 32).
The full-length NS3 protein [NS3_1b(con1)] was isolated as an N-terminally His-tagged protein by amplifying cDNA from the plasmid pBDL429P+S+ using PCR primers NS3(+) (5′-GCG CGC GCT AGC GCG CCT ATT ACG GCC TAC-3′) and NS3(−) (5′-GCG CGC GAA TTC GGT CAC GTG ACG ACC TCC AG-3′). The PCR product was purified, digested with NheI and EcoRI (the restriction sites are underlined), and ligated into a similarly treated pET28 vector (Novagen). The NS3 protein was expressed and purified as described previously for truncated NS3, except that the final DEAE column was replaced with a Fractogel EMD COO- column (EMD Biosciences).
The concentrations of the purified proteins were determined by absorbance at 280 nm using extinction coefficients calculated using the program “Sequence Analysis” (Informagen, Greenland, NH).
RNA-dependent RNA polymerase assays.
RNA-dependent RNA polymerase activity was assayed using poly(C) as a template for incorporation of [α-32P]GTP. Unless otherwise noted, standard reactions were performed in a total volume of 10 μl containing 20 mM HEPES-KOH (pH 7.5), 10 mM NaCl, 10 mM MgCl2, 1 mM MnCl2, 4 U Superase·In (Ambion), 3 mM dithiothreitol, 25 μg/ml poly(C), 50 μM [α-32P]GTP, and 0.2 μM NS5B. The reaction mixtures were incubated at room temperature for 30 min, and the reactions were terminated with 5 μl 0.5 M EDTA. The reaction mixtures were spotted onto DE81 filter papers, which were washed three times for 15 min each time in 0.5 M sodium phosphate (pH 7) and washed twice in 70% ethanol before the radioactive product was counted in a liquid scintillation counter.
ATPase assays.
NTP hydrolysis was monitored by measuring NS3-catalyzed phosphate release from [γ-32P]ATP as previously described (21). These assays exploit the fact that nucleotides adsorb onto activated charcoal (Norit), whereas inorganic phosphate does not. The reactions were run at 37°C in 50 mM Tris-Cl (pH 7.5), 10 mM MgCl2, ATP, and helicase at the indicated concentrations in the presence or absence of RNA and were terminated with the addition of 0.1 ml of 16% Norit in 5% trichloroacetic acid. After centrifugation at 16,000 × g for 10 min, phosphate in the supernatant was measured by scintillation counting.
DNA-unwinding assays.
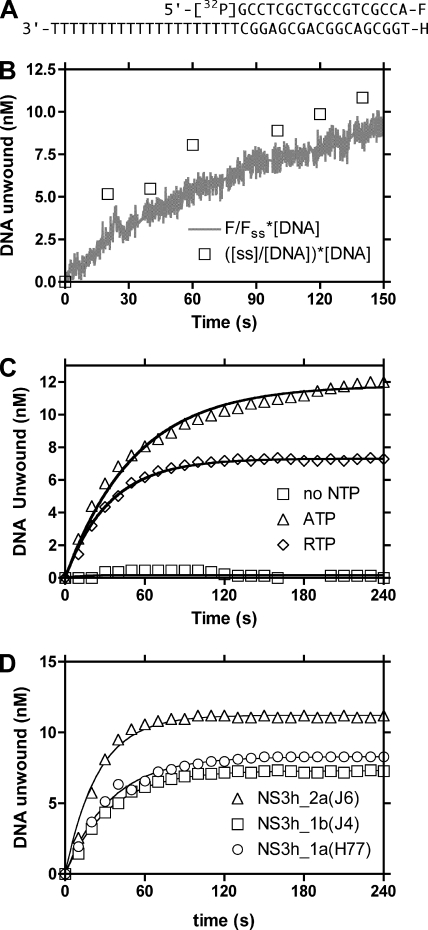
DNA helicase assays were performed using a labeled duplex (see Fig. 5A for sequence) with a single-stranded 3′ tail using a method based on one previously described (2). The short DNA strand contained a fluorescein (F) moiety covalently attached to the 3′ end, while the longer strand possessed a hexachlorofluorescein (H) moiety covalently attached to the 5′ end (bottom strand). The short strand was radiolabeled using [γ32P]ATP and polynucleotide kinase (Roche, IN). To anneal, 20 μM of each oligonucleotide was mixed in 10 mM Tris (pH 7.5), heated for 5 min at 90°C, and cooled slowly to room temperature over several hours.
FIG. 5.
Effect of RTP on NS3-catalyzed DNA unwinding. (A) DNA substrate used in the unwinding reaction. (B) DNA (100 nM) was preincubated with NS3h_1a(H77) (150 nM) for 10 min. Reactions were initiated by the addition of 2 mM ATP and 10 μM trap DNA. Unwinding reactions were monitored by measuring the increase in fluorescence (F) upon DNA substrate separation (solid line). The DNA unwound was calculated by using the fluorescence obtained for a sample of DNA fully unwound by being boiled in the presence of a DNA trap (Fss). At the indicated time points (squares), aliquots were withdrawn for gel analysis, where [ss] is the single-strand-product concentration and [DNA] is the substrate concentration. (C) DNA substrate (100 nM) was incubated with 150 nM NS3h_2a(J6) for 10 min, followed by addition of no NTP, 2 mM ATP, or 2 mM RTP. (D) Comparison of the abilities of RTP to support the unwinding catalyzed by NS3h_1a(H77), NS3h_1b(J4), or NS3h_2a(J6). The reactions were performed as for panel C.
The indicated amounts of HCV helicase and duplex DNA were preincubated in reaction buffer (50 mM Tris, pH 7.5, 4 mM MgCl2) for 10 min at room temperature before initiation by adding 1 mM ATP (or RTP) and 1 μM of a trap (an unmodified 18-mer DNA oligonucleotide bearing the same sequence as the short strand). The trap served to sequester excess enzyme, to prevent enzyme recycling, and to prevent unwound DNA from reannealing to the template DNA.
For the gel-based analysis, the reaction was terminated by the addition of stopping buffer (0.25% bromophenol blue, 0.25% xylenecyanol FF, 30% glycerol, 200 mM EDTA, 2% sodium dodecyl sulfate) and separated by a 12% native polyacrylamide gel. After running for 30 min at 200 V, the gel was dried and subsequently visualized using a PhosphorImager and quantified by using ImageQuant software from Molecular Dynamics.
For the fluorescence-based analyses, separation of the oligonucleotides was monitored by an increase in the fluorescein fluorescence, which was excited at 492 nm, with fluorescence emission monitored at 522 nm. Such assays were performed in 500 μl in a Varian Cary Eclipse fluorescence spectrophotometer (Walnut Creek, CA) or in 10-μl volumes using a Lightcycler PCR machine equipped with a fluorescence detection system from Roche (Indianapolis, IN). Complete unwinding of the annealed substrate was determined by boiling the substrates for 5 min, cooling them in the presence of a DNA trap (unlabeled short strand), and reading the fluorescence.
Data analysis.
Nonlinear least-square analysis was performed using GraphPad Prism (GraphPad Software, San Diego, CA). Inhibitor concentrations leading to 50% inhibition (IC50s) were determined by fitting the data to a sigmoid dose-response curve. Kms were determined by fitting the data directly to the Michaelis-Menten equation. Kis were determined by fitting the data to a standard model for competitive inhibition:
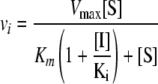 |
(1) |
where Vmax is the maximum rate of metabolism, vi is the velocity with inhibitor, [S] is the substrate concentration, and [I] is the inhibitor concentration. The error bars in the figures represent standard deviations obtained in repeated independent reactions (3 ≤ n ≤ 5), and uncertainties in Table 1 represent standard deviations of data obtained for three to five independent titrations with substrate. Errors in Kis in Table 1 reflect the same percent errors as the Kms used for Ki calculation in equation 1, because Kms had larger uncertainties than Kis calculated from repeat titrations using the same Km.
TABLE 1.
Steady-state kinetic parameters describing the interaction of GTP and various NTP analogs with polymerases isolated from HCV strains of various genotypes
| Parameter | Valuea
|
|||
|---|---|---|---|---|
| NS5BΔ21_1a(H77) | NS5BΔ21_1b(con1) | NS5BΔ21_1b(J4) | NS5BΔ21_2a(J6) | |
| GTP | ||||
| kcat (min−1) | 0.58 ± 0.50 | 4.4 ± 2.2 | 2.5 ± 0.8 | 2.7 ± 0.4 |
| Km (μM) | 900 ± 500 | 57 ± 11 | 65 ± 12 | 190 ± 40 |
| kcat/Km (min−1 mM−1) | 0.6 ± 0.3 | 77 ± 38 | 38 ± 12 | 14 ± 2 |
| Ki (μM) | ||||
| 3′-dGTP | 0.42 ± 0.23 | 0.23 ± 0.04 | 0.12 ± 0.02 | 0.18 ± 0.04 |
| 2′-O-methyl-GTP | 2.4 ± 1.3 | 1.0 ± 0.2 | 1.5 ± 0.3 | 2.9 ± 0.6 |
| 2′-O-methyl-ITP | 6.4 ± 3.6 | 3.4 ± 0.7 | 2.6 ± 0.5 | 3.6 ± 0.8 |
| ITP | 250 ± 140 | 84 ± 16 | 61 ± 11 | 54 ± 11 |
| RTP | 140 ± 80 | 190 ± 40 | 230 ± 40 | 270 ± 60 |
| 2′-dGTP | 280 ± 160 | 150 ± 30 | 280 ± 50 | 110 ± 20 |
| 2′,3′-ddGTP | 390 ± 220 | 380 ± 70 | 260 ± 50 | 150 ± 30 |
Mean values obtained from three to five independent titrations.
RESULTS
Expression and purification of HCV NS5B polymerase from various HCV strains.
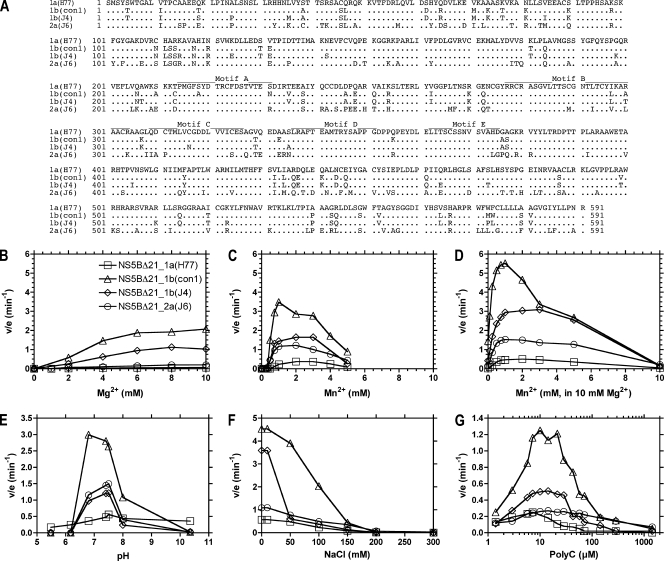
We chose to study recombinant NS5B proteins derived from the 1a(H77), 1b(con1), 1b(J4), and 2a(J6) strains. All four isolates are infectious to chimpanzees (7, 62-64); the 1b(con1) isolate also forms the backbone of many commonly used HCV replicons (3). Each protein has a C-terminal His tag and lacks the C-terminal 21 amino acids. Deletion of the final 21 amino acids, which most likely tether the polymerase to cellular membranes, aids in the production of a soluble recombinant protein in E. coli (1, 40, 53, 54, 60). The four polymerases were expressed similarly, despite the fact that they possess numerous sequence differences even in known motifs characteristic of RNA-dependent RNA polymerases (29) (Fig. 1A).
FIG. 1.
(A) Alignment of the NS5B amino acid sequences from the 1a(H77), 1b(con1), 1b(J4), and 2a(J6) strains used in this study. RNA-dependent RNA polymerase motifs A through E are indicated (29). (B to G) Characterization of conditions for optimal RNA synthesis by the four NS5BΔ21 polymerases. The ability of each polymerase to synthesize poly(G) from [α-32P]GTP using poly(C) template was assessed as specific activity (v/e), where v is nmoles RNA/min and e is nmoles of enzyme. To test metal ion requirements, NS5B reactions were titrated with MgCl2 (B), MnCl2 (C), and MnCl2 (D) in the presence of 10 mM MgCl2. The effects of pH (E), salt concentration (F), and template concentration (G) were also determined. All reaction mixtures contained 500 μM GTP. The reaction mixtures in panel E contained various buffers at a concentration of 50 mM and at their respective pKas: citric acid (5.4), MES (morpholineethanesulfonic acid) (6.15), PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] (6.8), MOPS (morpholinepropanesulfonic acid) (7.2), HEPES (7.5), Tris (8.1), and CAPS (N-cyclohexyl-3-aminopropanesulfonic acid) (10.4). The reactions in panel F were carried out in the absence of MnCl2. All other assay conditions were as described in Materials and Methods.
Characterization of NS5BΔ21 variants.
The abilities of the four NS5BΔ21 proteins to synthesize [32P]RNA from [α-32P]NTPs was measured using a template made of RNA transcribed from HCV replicon cDNA, RNA oligonucleotide, or homopolymer [poly(C)]. Products were analyzed either by measuring [32P]RNA bound to DEAE filters after washing them or by analyzing 32P-labeled oligonucleotide using denaturing polyacrylamide gels (data not shown). Polymerases derived from all four genotypes were found to be active in all of the assays. An assay using only [α-32P]GTP and poly(C) was chosen for further analysis for simplicity and so that our results could be directly compared with those of the recent comparative study of nonnucleoside NS5B inhibitors by Pauwels et al. (43).
RNA synthesis assays were optimized using each NS5BΔ21 (Fig. 1B to G). In all assays, the rates of RNA synthesis were linearly dependent on both time and enzyme concentrations, demonstrating that the reactions were performed under a “steady state.” Although specific activities (rate/enzyme) varied among the polymerases (1b > 2a > 1a), they all displayed similar basic properties. All NS5BΔ21 proteins require divalent metal cations to synthesize RNA, and although Mg2+ is likely the physiological metal of choice (Fig. 1B), all four enzymes synthesize RNA somewhat faster in the presence of Mn2+ (Fig. 1C). Mn2+ concentrations higher than 3 mM inhibited all enzymes, and optimal RNA synthesis took place when 10 mM Mg2+ was supplemented with 1 mM Mn2+ (Fig. 1B, C, and D). Likewise, all four genotypes were most active at physiological pH (Fig. 1E), and like the NS3 helicase (24), they were all most active in the absence of added salts (Fig. 1F).
For each NS5BΔ21 protein, the maximal RNA synthesis rate was achieved when there were roughly 200 to 300 template nucleotides for each molecule of polymerase (Fig. 1G). As reported previously, excess template RNA inhibits steady-state RNA synthesis rates (57). We always performed poly(C) assays with template concentrations exceeding those of enzymes by 300-fold. All subsequent assays were performed using 20 mM HEPES-KOH (pH 7.5), 10 mM NaCl, 10 mM MgCl2, 1 mM MnCl2, and 25 μg/ml poly(C) (71.5 μM C).
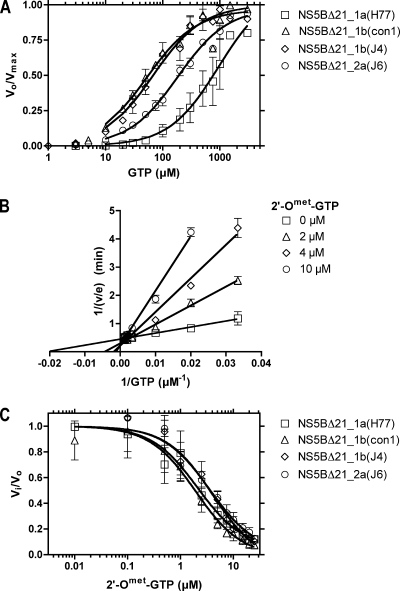
To better understand the molecular basis for the dramatic differences in activity, standardized steady-state assays were performed at various [α-32P]GTP concentrations (Fig. 2A). As seen before, the concentrations of GTP needed to achieve half-maximal velocities (Km) were notably different for the various polymerases (43). Turnover rates (kcat) were calculated for each enzyme (Table 1) assuming that the polymerase functions as a 100% active monomer (Vmax/Et, where Vmax is the maximum rate of catalysis and Et is total enzyme), although it must be noted that these two assumptions likely oversimplify the experimental conditions. There is growing evidence that NS5B functions as an oligomer (48, 56), and it is also possible that low activity levels are due to a partially denatured enzyme (9). Active-site titrations and structural studies are currently under way in our laboratory to test these hypotheses, but no final data are available at this time. It may therefore be preferable to interpret the kcats in Table 1 as “specific activities” expressed in terms of moles RNA synthesized/min/mole of protomer. Nevertheless, it is clearly evident that the less active polymerases from genotypes 1a_H77 and 2a_J6 also display a higher Km for the GTP substrate. As a result, the catalytic efficiency of the “worst” enzyme, NS5BΔ21_1a(H77), is 130-fold lower than the catalytic efficiency of the “best,” NS5BΔ21_1b(con1) (Table 1).
FIG. 2.
Interaction of NS5Bs from various genotypes with GTP and 2′-O-methyl GTP. (A) GTP concentration dependence of RNA synthesis rates by HCV genotype 1a, 1b, and 2a polymerases. The rates (Vo) of [α-32P]GTP incorporation into poly(G) were determined for different concentrations of GTP. Analysis of the data yielded the steady-state kinetic constants summarized in Table 1. (B) Inhibition of NS5B RNA synthesis by 2′-O-methyl GTP. Shown is a Lineweaver-Burke plot of RNA synthesis by NS5BΔ21_1b(J4) in the presence of 0, 2, 4, and 10 μM 2′-O-methyl (Omet) GTP and increasing concentrations of GTP. (C) Comparison of the effects of 2′-O-methyl GTP on RNA synthesis by the four polymerases. The points represent mean values of fraction of activity remaining (Vi/Vo) obtained from three to five independent reactions. The error bars represent standard deviations.
Inhibition of NS5B RNA-dependent RNA polymerase activity.
To compare the abilities of the four NS5B enzymes to synthesize RNA in the presence of different nucleotide analogs, we analyzed commercially available analogs of GTP and of ITP, which can be inserted into an RNA duplex, forming two Watson-Crick hydrogen bonds with C, rather than three. These compounds included canonical 2′-dGTP, obligate chain terminators (3′-dGTP and 2′,3′-ddGTP), nonobligate chain terminators (2′-O-methyl GTP and 2′-O-methyl ITP), and the mutagen RTP. Using NS5BΔ21_1b(J4), RNA synthesis assays were performed at various GTP concentrations in the presence of various inhibitor concentrations. The data were analyzed by using double-reciprocal (Lineweaver-Burke) plots and by fitting velocity data directly to equations describing competitive (where the inhibitor binds the enzyme only), uncompetitive (where the inhibitor binds the enzyme-substrate complex only), mixed (where the inhibitor binds the enzyme and the enzyme-substrate complex), and noncompetitive (where the inhibitor binds the enzyme and the enzyme-substrate complex with the same affinity) inhibition using nonlinear regression analysis. All data fit best to models for competitive inhibition, as shown in a double-reciprocal plot (Fig. 2B) with 2′-O-methyl GTP as the inhibitor. The data revealed that 2′-O-methyl GTP and 2′-O-methyl ITP behaved like the corresponding 2′-O-methyl CTP derivative studied previously (10). Each of the four polymerases studied responded similarly to various concentrations of 2′-O-methyl GTP (Fig. 2C).
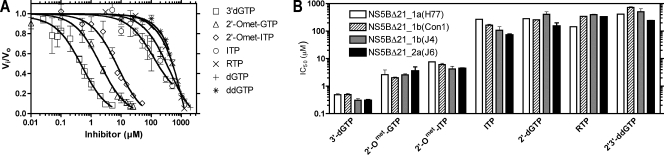
To compare the various enzymes using the panel of inhibitors, the concentration of GTP was held constant at 50 μM while the reactions catalyzed by each enzyme were titrated with each inhibitor (Fig. 3A). Each titration was performed in triplicate, and the average IC50 was calculated for each compound/enzyme combination (Fig. 3B). IC50 values were generally similar among the four enzymes. The same data were then used to calculate a Ki for each compound by fitting it to equation 1 using the Kms for GTP listed in Table 1. Interestingly, despite the large differences in apparent GTP binding affinities, overall the Kis among the enzymes showed little variation. Although statistical analysis yielded a number of significant differences, these two- to threefold differences in equilibrium constants represent only minor differences in free energy. The few differences of potential interest are noted below.
FIG. 3.
Inhibition of NS5B-catalyzed RNA synthesis by seven NTP analogs. (A) Fraction of RNA synthesis by NS5BΔ21_1b(con1) remaining (Vi/Vo) in the presence of 50 μM GTP and increasing concentrations of the various inhibitors, including 2′-O-methyl (2′-Omet) derivatives. The points represent mean values obtained from three to five independent reactions. (B) Data from inhibitor titrations were used to calculate IC50 values for each compound and all four polymerases. Inhibitors are shown in order of decreasing potency. Mean values were obtained from three to five independent titrations. In panels A and B, the error bars represent standard deviations.
For each enzyme, 3′-dGTP was the most potent inhibitor, followed by the 2′-O-methyl (Omet) derivatives. ITP behaved as an alternate substrate, with Kis that paralleled substrate Kms. NS5B appeared to select against the substrates for DNA synthesis, and as a result, both canonical 2′-dGTP and the chain terminator ddGTP were poor inhibitors of all enzymes, although the 2a(J6) enzyme showed somewhat less discrimination against them. RTP was likewise a poor inhibitor of all enzymes, with Kis determined from these steady-state assays reflective of dissociation contents previously determined using transient-state kinetics and an engineered NS5B capable of using a stably annealed primer-template substrate (36). Comparison of Kms with Kis for RTP indicated that, in the case of the 1a(H77) enzyme, RTP competed better with GTP (Table 1).
Effects of ribavirin and RTP on NS3-catalyzed ATP hydrolysis.
We next asked whether the panel of NTP analogs could influence another enzyme in the HCV replicase, the NS3 helicase, which uses the energy from NTP hydrolysis to unwind duplex RNA (or DNA). We found that the helicase was capable of hydrolyzing each of the various NTP analogs to fuel unwinding (D. N. Frick and C. A. Belon, unpublished data). These findings are in agreement with previous studies indicating that the helicase indiscriminately hydrolyzes all canonical NTPs (19, 47). However, for RTP, this result was somewhat surprising, given that RTP was previously shown to be an inhibitor of both hydrolysis and unwinding, indicating that it might not be a substrate for the helicase (4). For this reason, and because of the clear clinical relevance of examining the mechanism of ribavirin, we carried out a more detailed analysis of the interactions of NS3 helicase with ribavirin and RTP.
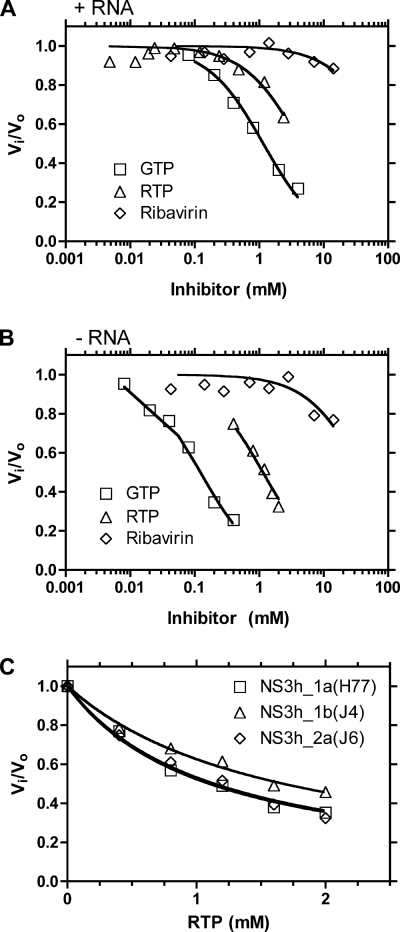
To examine the relative affinity of RTP for NS3, the abilities of RTP, ribavirin, and GTP to inhibit NS3-catalyzed [γ-32P]ATP hydrolysis were assessed. Reactions were first performed in the presence of saturating amounts of poly(U) RNA using the truncated NS3 lacking the protease domain derived from genotype 2a [NS3h_2a(J6)]. NS3h_2a(J6)-catalyzed ATP hydrolysis was titrated with ribavirin (0 to 14.2 mM), RTP (0 to 2.4 mM), or GTP (0 to 0.4 mM) in the presence of 50 μM [γ-32P]ATP. GTP was hydrolyzed by NS3 helicase at rates comparable to those for ATP, as reported previously (19, 30, 47). ATP hydrolysis was only weakly inhibited by ribavirin, with an IC50 more than 300 times that of GTP, while the IC50 for RTP was approximately 10-fold higher than that of GTP (Fig. 4). The data confirmed that ribavirin must be phosphorylated to interact with NS3 and that RTP interacts with the NS3 helicase, but as with NS5B, RTP interacts with the NS3 enzyme notably less strongly than typical cellular NTPs.
FIG. 4.
Effect of RTP on NS3-catalyzed ATP hydrolysis. (A) Inhibition of reactions (Vi/Vo) containing 50 μM [γ-32P]ATP by increasing concentrations of ribavirin, GTP, and RTP in the presence of 2.7 mg/ml poly(U) RNA. The data were fitted to equation 1 using the Km (270 μM) for ATP (30). (B) Inhibition of reactions containing 20 μM [γ-32P]ATP by increasing concentrations of ribavirin, GTP, and RTP in the absence of RNA. The data were fitted to equation 1 using the Km (10 μM). (C) RTP inhibition of ATP hydrolysis catalyzed by NS3h_1a(H77) (squares), NS3h_1b(J4) (triangles), or NS3h_2a(J6) (diamonds). Reaction mixtures contained 8 μM [γ-32P]ATP in the presence of the indicated concentrations of RTP in the absence of RNA. The data were fitted to equation 1 using a Km for ATP (10 μM) determined previously (30) and the following Kis for RTP: NS3h_1a(H77), 380 μM; NS3h_1b(J4), 560 μM; and NS3h_2a(J6), 380 μM.
The presence of RNA significantly reduces the affinity of the helicase for its NTP substrates (19, 30, 47). In order to better determine the apparent affinity of ribavirin and RTP, the experiments were repeated without RNA (Fig. 4B). Increasing concentrations of ribavirin, RTP, or GTP were included in reactions catalyzed by NS3h_2a(J6) in the presence of 20 μM [γ-32P]ATP. Again, the IC50 for ribavirin was more than 100-fold higher than the Km for ATP, and RTP again bound approximately 10-fold less tightly than GTP. It was interesting that even though the NS3 helicase has been shown to be nonselective toward its NTP substrates (19, 30), like NS5B, NS3 distinguishes between GTP and RTP.
Since RTP is a more potent inhibitor of the NS3 ATPase in the absence of RNA, the apparent affinity of RTP for NS3h_1a(H77), NS3h_1b(J4), or NS3h_2a(J6) was determined in the absence of nucleic acids (Fig. 4C). Assays monitoring ATP hydrolysis catalyzed by each helicase were titrated with increasing concentrations of RTP. Analysis of the data showed clearly that the IC50s for RTP for the three NS3 helicases were not significantly different (Fig. 4C).
RTP can fuel the NS3 helicase.
We next examined the ability of RTP to inhibit DNA unwinding catalyzed by HCV helicase, because a previous report mentioned that RTP inhibited the HCV helicase but that complete inhibition of unwinding could not be attained (4). Surprisingly, we found that RTP did not inhibit the rate of the reaction (data not shown), and we speculated that NS3 could instead use RTP as an alternate substrate to fuel unwinding. To examine if RTP was acting as an alternate substrate, we modified an existing fluorescence-based helicase assay so that it could be performed in microvolumes. This was necessary because the reactions required relatively high concentrations of RTP, which was in limited supply.
The helicase assay chosen (based on methods from reference 2) used a radiolabeled duplex containing a 3′ overhang and had a fluorophore and a quencher covalently attached to the 3′ and 5′ ends of the complementary strands (Fig. 5A). Unwinding reactions were initiated by addition of ATP to a DNA-helicase mixture, along with 100-fold excess trap DNA, which functions both to prevent reannealing and to prevent dissociated helicase from rebinding the substrate. To first confirm that the observed fluorescence correlated with unwound DNA, a 500-μl reaction was continually monitored in a fluorescence spectrophotometer, and periodically, 10-μl aliquots were withdrawn and analyzed on 12% nondenaturing polyacrylamide gels (Fig. 5B). After thus calibrating the assay, 10-μl microreactions were monitored instead using a Lightcycler equipped with a fluorescence detection system (Fig. 5C and D).
NS3h unwound duplex DNA substrate when ATP was replaced with RTP, although to a lesser extent (Fig. 5C). If one assumes that the reaction took place in a single turnover, the data can be interpreted to imply that when RTP is used as a substrate, the enzyme is somewhat less processive. In other words, less total DNA is unwound because the enzyme falls from the template (and binds the trap DNA) sooner when RTP is used as the reaction fuel. The effect of RTP on unwinding was then examined using helicases isolated from the three different genotypes (Fig. 5D). Unwinding reaction mixtures containing NS3h_1a(H77), NS3h_1b(J4), or NS3h_2a(J6) were mixed with DNA substrate prior to initiation by addition of either ATP or RTP and trap DNA. All helicases unwound the DNA similarly, but NS3h_2a was slightly faster and unwound a higher percentage of the duplex. The results were confirmed by repeating each assay three times and analyzing the final reaction products on polyacrylamide gels. These data agreed with our previous report that genotype 2a(J6) encodes a slightly more active helicase than genotype 1a(H77) or 1b(J4) (30).
The effect of NS3 helicase on RTP-NS5B interaction.
If NS3 helicase used RTP to fuel unwinding, converting RTP to RDP, the helicase would lower the RTP pool available as a potential substrate for the NS5B polymerase. NS5B and NS3 coordinate their actions in the HCV replicase (26) in an interaction that takes place via the protease domain of NS3 (66). Formation of an NS3-NS5B complex facilitates nucleic acid unwinding (66), and NS3 has been shown to stimulate NS5B-catalyzed RNA synthesis on HCV RNA templates (45).
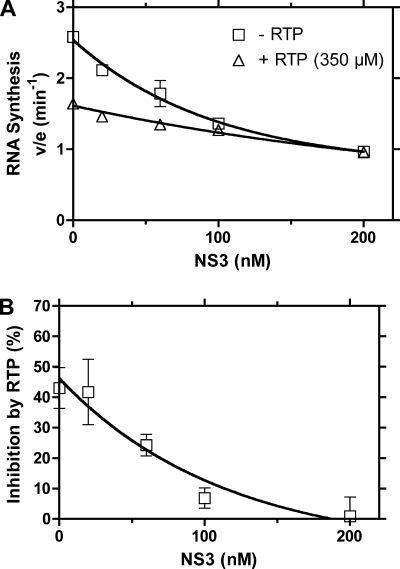
To first examine if we could recapitulate a stimulatory effect of NS3 and NS5B in our system using poly(C) template, RNA synthesis by NS5BΔ21_1b(con1) was monitored in reactions containing various amounts of purified NS3 from the same genotype [NS3_1b(con1)]. No stimulation was observed. In all titrations, NS3 inhibited total RNA synthesis, presumably because NS3 was competing for a limited supply of GTP needed for RNA synthesis (Fig. 6A). Similar inhibition was observed using other NS5B proteins, other helicase constructs, or templates made of HCV replicon RNA or synthetic RNA oligonucleotides (data not shown). When the experiment was repeated in the presence of 350 μM RTP, less inhibition by NS3 was observed (Fig. 6A), and when NS3 and NS5B were present at equal concentrations, RTP no longer appeared to inhibit NS5B (Fig. 6B). This experiment is noteworthy because it suggests that concentrations of RTP needed to exert a strong effect on pure NS5B may not be cellularly possible because of the action of the NS3 helicase.
FIG. 6.
NS3 blocks inhibition of NS5B by RTP. (A) The ability of NS5BΔ21_1b(con1) to synthesize RNA in the presence and absence of RTP and the presence of increasing concentrations of NS3_1b(con1) was measured. The IC50 (350 μM) for RTP was used for the reactions containing RTP. While NS3 and RTP each inhibit the polymerase separately, the presence of NS3 mitigates the inhibitory effect of RTP. (B) Percent inhibition by RTP. The error bars indicate standard deviations.
DISCUSSION
The significance of the studies performed here is threefold. First, we showed that genetic variation among HCV isolates can lead to polymerases that interact with NTP substrates with up to 100-fold differences but that this variation does not lead to significant differences in the effects of nucleotide-based inhibitors. Second, the work here addressed the question of whether RTP is an “inhibitor” of HCV helicase. Previously, Borowski et al. found that RTP partially inhibits HCV helicase-catalyzed DNA unwinding, with about 35% of the original activity remaining at 0.6 mM RTP (4). We showed here that this remaining activity is explained by the fact that RTP acts as an alternate substrate for the NS3 helicase. We showed that RTP fuels helicase, and as a consequence, helicase can effectively reduce the incorporation of ribavirin into viral RNA. This observation does not directly support or contradict the hypothesis that ribavirin exerts its action as a lethal viral mutagen, but it indicates that the activity of the helicase would make such a mechanism less effective.
In addition to ribavirin, which forms the basis for all current HCV therapies in combination with various types of interferon, several nucleoside analogs are promising HCV drug candidates, some of which have already entered into clinical trials. Given the genotype-specific response rates to current treatments and the wide range of circulating genotypes, it is surprising that few systematic comparative biochemical studies have been performed. We have found only one such study published to date, which examined the effects of nonnucleoside NS5B inhibitors on genotypic variants (43). Compared with the relative effects of various nonnucleosides on NS5B from various genotypes, the differences among the relative IC50s and Kis reported here are quite minor (43). Thus, nonobligate chain terminators appear more promising for eliminating a broader variety of HCV strains. We included in our study nucleotide analogs that are commercially available. Although not under clinical investigation, the 2′-O-methyl NTPs studied here are similar to compounds that have entered clinical trials. Our results are in agreement with replicon and infectious cell culture studies demonstrating that 2′-C-methyladenosine is active against multiple genotypes (23, 33, 35). Our findings with multiple NTP analogs suggest that other nucleotide-based drug candidates are likely to be broadly effective as well. Resistance to nonobligate chain terminators has already been documented (38), but these mutations add significant costs in terms of viral fitness (17).
Like other NTP analogs, RTP seems to affect the genotypes studied here similarly. The ability of NS5B to use RTP as a substrate was first analyzed using a modified version of NS5B that can efficiently extend RNA primers (36). That study showed that NS5B enzyme incorporates RTP even more poorly than our estimates, with an apparent Kd (dissociation constant) 100-fold higher than that of GTP, and that ribavirin is incorporated at a rate about 400-fold lower than that of GTP (36). A later study using wild-type [1b(NIH1)] full-length recombinant NS5B confirmed that RTP is worse than GTP as an NS5B substrate and also revealed that templates containing ribavirin were transcribed by NS5B up to 3,000-fold more slowly than natural RNA (55). Our data show that RTP binds NS5B up to five times more weakly than GTP. Interestingly, NS5B from the 1a(H77) strain, the only strain of the four examined here that contains a phenylalanine instead of a tyrosine at NS5B position 415, seems to bind RTP the best (Fig. 3 and Table 1). An F415Y substitution has previously been linked with ribavirin resistance, both in genotype 1a-infected patients and in replicons (44, 65); in other genotypes, tyrosine is the consensus residue. Although the data with NS5BΔ21_1a(H77) have a high error rate due to the enzyme's low specific activity, the basic trend suggests that with this genotype 1a strain, RTP competes better with GTP (compare IC50s [Fig. 3B], Kms, and Kis [Table 1]). This finding is consistent with 415F conferring greater sensitivity to RTP.
Since RTP is clearly such a poor substrate for NS5B, its effects on other HCV enzymes may provide other clues to its mechanism of action. For example, other studies have previously examined the effects of ribavirin and RTP on HCV NS3 DNA unwinding (4) and NTPase activity (5). This report confirms the previous observation that RTP acts as a competitive inhibitor of ATP hydrolysis (5), but our data do not support the earlier conclusion that RTP inhibits the helicase-unwinding activity (4). Instead, we demonstrated that RTP can act as an alternate fuel for the unwinding reaction. The fact that RTP competes with ATP for binding to the helicase is not surprising, because the NS3 helicase is nonselective, hydrolyzing all eight canonical (d)NTPs, NTP analogs, and even tripolyphosphate (19, 47). The more important finding here is that the presence of RTP actually fuels nucleic acid unwinding. Genetic variation appears to have little effect on the RTP-NS3 interaction.
The fact that NS3 can hydrolyze RTP indicates a mechanism by which HCV can avoid the effects of a viral mutagen like ribavirin. By rapidly hydrolyzing RTP to RDP, NS3 can effectively prevent the incorporation of ribavirin into viral RNA (Fig. 6). Although RDP could be converted back to RTP by cellular kinases, it is likely that the close association of the helicase with the polymerase (27) would keep the pool of RTP immediately available to the polymerase at low levels. The hydrolysis of RTP by NS3 could explain why some ribavirin-resistant mutants have been mapped outside NS5B (44). It is possible that mutations elsewhere in the viral replicase could enhance or diminish the ability of NS3 to hydrolyze potentially inhibitory nucleotides.
Several theories have been invoked to explain the antiviral effect of ribavirin (as outlined in the introduction). The theory that RTP exerts its inhibitory effect as a mutagen is still controversial (51), and the results of two recent clinical studies have (25) and have not (11) supported this hypothesis. While cell culture studies have indicated that ribavirin increases the rate of viral RNA mutations (6, 13, 15), it remains unclear whether or to what degree the resulting “error catastrophe” of the virus accounts for the antiviral effect of ribavirin in patients. Our data suggest that HCV helicase could significantly reduce the impact of RTP as a direct inhibitor of the NS5B polymerase and as a viral mutagen and that NTP hydrolysis by the helicase should be taken into account in the development of NTP analogs as HCV drugs.
Acknowledgments
This work was supported by National Institutes of Health grant AI052395.
We are grateful to Fred Jaffe for valuable technical support.
Footnotes
Published ahead of print on 7 April 2008.
REFERENCES
- 1.Adachi, T., H. Ago, N. Habuka, K. Okuda, M. Komatsu, S. Ikeda, and K. Yatsunami. 2002. The essential role of C-terminal residues in regulating the activity of hepatitis C virus RNA-dependent RNA polymerase. Biochim. Biophys. Acta 1601:38-48. [DOI] [PubMed] [Google Scholar]
- 2.Bjornson, K. P., M. Amaratunga, K. J. Moore, and T. M. Lohman. 1994. Single-turnover kinetics of helicase-catalyzed DNA unwinding monitored continuously by fluorescence energy transfer. Biochemistry 33:14306-14316. [DOI] [PubMed] [Google Scholar]
- 3.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 4.Borowski, P., M. Lang, A. Niebuhr, A. Haag, H. Schmitz, J. S. zur Wiesch, J. Choe, M. A. Siwecka, and T. Kulikowski. 2001. Inhibition of the helicase activity of HCV NTPase/helicase by 1-beta-d-ribofuranosyl-1,2,4-triazole-3-carboxamide-5′-triphosphate (ribavirin-TP). Acta Biochim. Pol. 48:739-744. [PubMed] [Google Scholar]
- 5.Borowski, P., O. Mueller, A. Niebuhr, M. Kalitzky, L. H. Hwang, H. Schmitz, M. A. Siwecka, and T. Kulikowsk. 2000. ATP-binding domain of NTPase/helicase as a target for hepatitis C antiviral therapy. Acta Biochim. Pol. 47:173-180. [PubMed] [Google Scholar]
- 6.Brochot, E., G. Duverlie, S. Castelain, V. Morel, C. Wychowski, J. Dubuisson, and C. Francois. 2007. Effect of ribavirin on the hepatitis C virus (JFH-1) and its correlation with interferon sensitivity. Antivir. Ther. 12:805-813. [PubMed] [Google Scholar]
- 7.Bukh, J., T. Pietschmann, V. Lohmann, N. Krieger, K. Faulk, R. E. Engle, S. Govindarajan, M. Shapiro, M. St Claire, and R. Bartenschlager. 2002. Mutations that permit efficient replication of hepatitis C virus RNA in Huh-7 cells prevent productive replication in chimpanzees. Proc. Natl. Acad. Sci. USA 99:14416-14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carroll, S. S., and D. B. Olsen. 2006. Nucleoside analog inhibitors of hepatitis C virus replication. Infect. Disord. Drug Targets 6:17-29. [DOI] [PubMed] [Google Scholar]
- 9.Carroll, S. S., V. Sardana, Z. Yang, A. R. Jacobs, C. Mizenko, D. Hall, L. Hill, J. Zugay-Murphy, and L. C. Kuo. 2000. Only a small fraction of purified hepatitis C RNA-dependent RNA polymerase is catalytically competent: implications for viral replication and in vitro assays. Biochemistry 39:8243-8249. [DOI] [PubMed] [Google Scholar]
- 10.Carroll, S. S., J. E. Tomassini, M. Bosserman, K. Getty, M. W. Stahlhut, A. B. Eldrup, B. Bhat, D. Hall, A. L. Simcoe, R. LaFemina, C. A. Rutkowski, B. Wolanski, Z. Yang, G. Migliaccio, R. De Francesco, L. C. Kuo, M. MacCoss, and D. B. Olsen. 2003. Inhibition of hepatitis C virus RNA replication by 2′-modified nucleoside analogs. J. Biol. Chem. 278:11979-11984. [DOI] [PubMed] [Google Scholar]
- 11.Chevaliez, S., R. Brillet, E. Lazaro, C. Hezode, and J. M. Pawlotsky. 2007. Analysis of ribavirin mutagenicity in human hepatitis C virus infection. J. Virol. 81:7732-7741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 13.Contreras, A. M., Y. Hiasa, W. He, A. Terella, E. V. Schmidt, and R. T. Chung. 2002. Viral RNA mutations are region specific and increased by ribavirin in a full-length hepatitis C virus replication system. J. Virol. 76:8505-8517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crotty, S., C. E. Cameron, and R. Andino. 2001. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc. Natl. Acad. Sci. USA 98:6895-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crotty, S., D. Maag, J. J. Arnold, W. Zhong, J. Y. Lau, Z. Hong, R. Andino, and C. E. Cameron. 2000. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 6:1375-1379. [DOI] [PubMed] [Google Scholar]
- 16.Dutartre, H., J. Boretto, J. C. Guillemot, and B. Canard. 2005. A relaxed discrimination of 2′-O-methyl-GTP relative to GTP between de novo and elongative RNA synthesis by the hepatitis C RNA-dependent RNA polymerase NS5B. J. Biol. Chem. 280:6359-6368. [DOI] [PubMed] [Google Scholar]
- 17.Dutartre, H., C. Bussetta, J. Boretto, and B. Canard. 2006. General catalytic deficiency of hepatitis C virus RNA polymerase with an S282T mutation and mutually exclusive resistance towards 2′-modified nucleotide analogues. Antimicrob. Agents Chemother. 50:4161-4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frick, D. N. 2007. The hepatitis C virus NS3 protein: a model RNA helicase and potential drug target. Curr. Issues Mol. Biol. 9:1-20. [PMC free article] [PubMed] [Google Scholar]
- 19.Frick, D. N., S. Banik, and R. S. Rypma. 2007. Role of divalent metal cations in ATP hydrolysis catalyzed by the hepatitis C virus NS3 helicase: magnesium provides a bridge for ATP to fuel unwinding. J. Mol. Biol. 365:1017-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frick, D. N., R. S. Rypma, A. M. Lam, and B. Gu. 2004. The nonstructural protein 3 protease/helicase requires an intact protease domain to unwind duplex RNA efficiently. J. Biol. Chem. 279:1269-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frick, D. N., D. J. Weber, J. R. Gillespie, M. J. Bessman, and A. S. Mildvan. 1994. Dual divalent cation requirement of the MutT dGTPase. Kinetic and magnetic resonance studies of the metal and substrate complexes. J. Biol. Chem. 269:1794-1803. [PubMed] [Google Scholar]
- 22.Gallois-Montbrun, S., Y. Chen, H. Dutartre, M. Sophys, S. Morera, C. Guerreiro, B. Schneider, L. Mulard, J. Janin, M. Veron, D. Deville-Bonne, and B. Canard. 2003. Structural analysis of the activation of ribavirin analogs by NDP kinase: comparison with other ribavirin targets. Mol. Pharmacol. 63:538-546. [DOI] [PubMed] [Google Scholar]
- 23.Graham, D. J., M. Stahlhut, O. Flores, D. B. Olsen, D. J. Hazuda, R. L. Lafemina, and S. W. Ludmerer. 2006. A genotype 2b NS5B polymerase with novel substitutions supports replication of a chimeric HCV 1b:2b replicon containing a genotype 1b NS3-5A background. Antivir. Res. 69:24-30. [DOI] [PubMed] [Google Scholar]
- 24.Gwack, Y., D. W. Kim, J. H. Han, and J. Choe. 1996. Characterization of RNA binding activity and RNA helicase activity of the hepatitis C virus NS3 protein. Biochem. Biophys. Res. Commun. 225:654-659. [DOI] [PubMed] [Google Scholar]
- 25.Hofmann, W. P., A. Polta, E. Herrmann, U. Mihm, B. Kronenberger, T. Sonntag, V. Lohmann, B. Schonberger, S. Zeuzem, and C. Sarrazin. 2007. Mutagenic effect of ribavirin on hepatitis C nonstructural 5B quasispecies in vitro and during antiviral therapy. Gastroenterology 132:921-930. [DOI] [PubMed] [Google Scholar]
- 26.Ishido, S., T. Fujita, and H. Hotta. 1998. Complex formation of NS5B with NS3 and NS4A proteins of hepatitis C virus. Biochem. Biophys. Res. Commun. 244:35-40. [DOI] [PubMed] [Google Scholar]
- 27.Jennings, T. A., Y. Chen, D. Sikora, M. K. Harrison, B. Sikora, L. Huang, E. Jankowsky, M. E. Fairman, C. E. Cameron, and K. D. Raney. 2008. RNA unwinding activity of the hepatitis C virus NS3 helicase is modulated by the NS5B polymerase. Biochemistry 47:1126-1135. [DOI] [PubMed] [Google Scholar]
- 28.Klumpp, K., V. Leveque, S. Le Pogam, H. Ma, W. R. Jiang, H. Kang, C. Granycome, M. Singer, C. Laxton, J. Q. Hang, K. Sarma, D. B. Smith, D. Heindl, C. J. Hobbs, J. H. Merrett, J. Symons, N. Cammack, J. A. Martin, R. Devos, and I. Najera. 2006. The novel nucleoside analog R1479 (4′-azidocytidine) is a potent inhibitor of NS5B-dependent RNA synthesis and hepatitis C virus replication in cell culture. J. Biol. Chem. 281:3793-3799. [DOI] [PubMed] [Google Scholar]
- 29.Koonin, E. V. 1991. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J. Gen. Virol. 72:2197-2206. [DOI] [PubMed] [Google Scholar]
- 30.Lam, A. M., D. Keeney, P. Q. Eckert, and D. N. Frick. 2003. Hepatitis C virus NS3 ATPases/helicases from different genotypes exhibit variations in enzymatic properties. J. Virol. 77:3950-3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lam, A. M., D. Keeney, and D. N. Frick. 2003. Two novel conserved motifs in the hepatitis C virus NS3 protein critical for helicase action. J. Biol. Chem. 278:44514-44524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lam, A. M., R. S. Rypma, and D. N. Frick. 2004. Enhanced nucleic acid binding to ATP-bound hepatitis C virus NS3 helicase at low pH activates RNA unwinding. Nucleic Acids Res. 32:4060-4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 34.Lindenbach, B. D., B. M. Pragai, R. Montserret, R. K. Beran, A. M. Pyle, F. Penin, and C. M. Rice. 2007. The C terminus of hepatitis C virus NS4A encodes an electrostatic switch that regulates NS5A hyperphosphorylation and viral replication. J. Virol. 81:8905-8918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ludmerer, S. W., D. J. Graham, E. Boots, E. M. Murray, A. Simcoe, E. J. Markel, J. A. Grobler, O. A. Flores, D. B. Olsen, D. J. Hazuda, and R. L. LaFemina. 2005. Replication fitness and NS5B drug sensitivity of diverse hepatitis C virus isolates characterized by using a transient replication assay. Antimicrob. Agents Chemother. 49:2059-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maag, D., C. Castro, Z. Hong, and C. E. Cameron. 2001. Hepatitis C virus RNA-dependent RNA polymerase (NS5B) as a mediator of the antiviral activity of ribavirin. J. Biol. Chem. 276:46094-46098. [DOI] [PubMed] [Google Scholar]
- 37.Manns, M. P., J. G. McHutchison, S. C. Gordon, V. K. Rustgi, M. Shiffman, R. Reindollar, Z. D. Goodman, K. Koury, M. Ling, and J. K. Albrecht. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958-965. [DOI] [PubMed] [Google Scholar]
- 38.Migliaccio, G., J. E. Tomassini, S. S. Carroll, L. Tomei, S. Altamura, B. Bhat, L. Bartholomew, M. R. Bosserman, A. Ceccacci, L. F. Colwell, R. Cortese, R. De Francesco, A. B. Eldrup, K. L. Getty, X. S. Hou, R. L. LaFemina, S. W. Ludmerer, M. MacCoss, D. R. McMasters, M. W. Stahlhut, D. B. Olsen, D. J. Hazuda, and O. A. Flores. 2003. Characterization of resistance to non-obligate chain-terminating ribonucleoside analogs that inhibit hepatitis C virus replication in vitro. J. Biol. Chem. 278:49164-49170. [DOI] [PubMed] [Google Scholar]
- 39.Miller, J. P., L. J. Kigwana, D. G. Streeter, R. K. Robins, L. N. Simon, and J. Roboz. 1977. The relationship between the metabolism of ribavirin and its proposed mechanism of action. Ann. N. Y. Acad. Sci. 284:211-229. [DOI] [PubMed] [Google Scholar]
- 40.Moradpour, D., V. Brass, E. Bieck, P. Friebe, R. Gosert, H. E. Blum, R. Bartenschlager, F. Penin, and V. Lohmann. 2004. Membrane association of the RNA-dependent RNA polymerase is essential for hepatitis C virus RNA replication. J. Virol. 78:13278-13284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murakami, E., H. Bao, M. Ramesh, T. R. McBrayer, T. Whitaker, H. M. Micolochick Steuer, R. F. Schinazi, L. J. Stuyver, A. Obikhod, M. J. Otto, and P. A. Furman. 2006. Mechanism of activation of β-d-2′-deoxy-2′-fluoro-2′-C-methylcytidine and inhibition of hepatitis C virus NS5B RNA polymerase. Antimicrob. Agents Chemother. 51:503-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Farrell, D., R. Trowbridge, D. Rowlands, and J. Jager. 2003. Substrate complexes of hepatitis C virus RNA polymerase (HC-J4): structural evidence for nucleotide import and de-novo initiation. J. Mol. Biol. 326:1025-1035. [DOI] [PubMed] [Google Scholar]
- 43.Pauwels, F., W. Mostmans, L. M. Quirynen, L. van der Helm, C. W. Boutton, A. S. Rueff, E. Cleiren, P. Raboisson, D. Surleraux, O. Nyanguile, and K. A. Simmen. 2007. Binding site identification and genotypic profiling of hepatitis C virus polymerase inhibitors. J. Virol. 81:6909-6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pfeiffer, J. K., and K. Kirkegaard. 2005. Ribavirin resistance in hepatitis C virus replicon-containing cell lines conferred by changes in the cell line or mutations in the replicon RNA. J. Virol. 79:2346-2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Piccininni, S., A. Varaklioti, M. Nardelli, B. Dave, K. D. Raney, and J. E. McCarthy. 2002. Modulation of the hepatitis C virus RNA-dependent RNA polymerase activity by the non-structural (NS) 3 helicase and the NS4B membrane protein. J. Biol. Chem. 277:45670-45679. [DOI] [PubMed] [Google Scholar]
- 46.Pierra, C., A. Amador, S. Benzaria, E. Cretton-Scott, M. D'Amours, J. Mao, S. Mathieu, A. Moussa, E. G. Bridges, D. N. Standring, J. P. Sommadossi, R. Storer, and G. Gosselin. 2006. Synthesis and pharmacokinetics of valopicitabine (NM283), an efficient prodrug of the potent anti-HCV agent 2′-C-methylcytidine. J. Med. Chem. 49:6614-6620. [DOI] [PubMed] [Google Scholar]
- 47.Preugschat, F., D. R. Averett, B. E. Clarke, and D. J. Porter. 1996. A steady-state and pre-steady-state kinetic analysis of the NTPase activity associated with the hepatitis C virus NS3 helicase domain. J. Biol. Chem. 271:24449-24457. [DOI] [PubMed] [Google Scholar]
- 48.Qin, W., H. Luo, T. Nomura, N. Hayashi, T. Yamashita, and S. Murakami. 2002. Oligomeric interaction of hepatitis C virus NS5B is critical for catalytic activity of RNA-dependent RNA polymerase. J. Biol. Chem. 277:2132-2137. [DOI] [PubMed] [Google Scholar]
- 49.Sidwell, R. W., J. H. Huffman, G. P. Khare, L. B. Allen, J. T. Witkowski, and R. K. Robins. 1972. Broad-spectrum antiviral activity of Virazole: 1-beta-d-ribofuranosyl-1,2,4-triazole-3-carboxamide. Science 177:705-706. [DOI] [PubMed] [Google Scholar]
- 50.Stuyver, L. J., T. R. McBrayer, T. Whitaker, P. M. Tharnish, M. Ramesh, S. Lostia, L. Cartee, J. Shi, A. Hobbs, R. F. Schinazi, K. A. Watanabe, and M. J. Otto. 2004. Inhibition of the subgenomic hepatitis C virus replicon in huh-7 cells by 2′-deoxy-2′-fluorocytidine. Antimicrob. Agents Chemother. 48:651-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Summers, J., and S. Litwin. 2006. Examining the theory of error catastrophe. J. Virol. 80:20-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tellinghuisen, T. L., M. J. Evans, T. von Hahn, S. You, and C. M. Rice. 2007. Studying hepatitis C virus: making the best of a bad virus. J. Virol. 81:8853-8867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tomei, L., R. L. Vitale, I. Incitti, S. Serafini, S. Altamura, A. Vitelli, and R. De Francesco. 2000. Biochemical characterization of a hepatitis C virus RNA-dependent RNA polymerase mutant lacking the C-terminal hydrophobic sequence. J. Gen. Virol. 81:759-767. [DOI] [PubMed] [Google Scholar]
- 54.Vo, N. V., J. R. Tuler, and M. M. Lai. 2004. Enzymatic characterization of the full-length and C-terminally truncated hepatitis C virus RNA polymerases: function of the last 21 amino acids of the C terminus in template binding and RNA synthesis. Biochemistry 43:10579-10591. [DOI] [PubMed] [Google Scholar]
- 55.Vo, N. V., K. C. Young, and M. M. Lai. 2003. Mutagenic and inhibitory effects of ribavirin on hepatitis C virus RNA polymerase. Biochemistry 42:10462-10471. [DOI] [PubMed] [Google Scholar]
- 56.Wang, Q. M., M. A. Hockman, K. Staschke, R. B. Johnson, K. A. Case, J. Lu, S. Parsons, F. Zhang, R. Rathnachalam, K. Kirkegaard, and J. M. Colacino. 2002. Oligomerization and cooperative RNA synthesis activity of hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 76:3865-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, Y. K., K. L. Rigat, J. H. Sun, M. Gao, and S. B. Roberts. 2007. RNA template-mediated inhibition of hepatitis C virus RNA-dependent RNA polymerase activity. Arch. Biochem. Biophys. 470:146-152. [DOI] [PubMed] [Google Scholar]
- 58.Willis, R. C., D. A. Carson, and J. E. Seegmiller. 1978. Adenosine kinase initiates the major route of ribavirin activation in a cultured human cell line. Proc. Natl. Acad. Sci. USA 75:3042-3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wohnsland, A., W. P. Hofmann, and C. Sarrazin. 2007. Viral determinants of resistance to treatment in patients with hepatitis C. Clin. Microbiol. Rev. 20:23-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamashita, T., S. Kaneko, Y. Shirota, W. Qin, T. Nomura, K. Kobayashi, and S. Murakami. 1998. RNA-dependent RNA polymerase activity of the soluble recombinant hepatitis C virus NS5B protein truncated at the C-terminal region. J. Biol. Chem. 273:15479-15486. [DOI] [PubMed] [Google Scholar]
- 61.Yanagi, M., R. H. Purcell, S. U. Emerson, and J. Bukh. 1997. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc. Natl. Acad. Sci. USA 94:8738-8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yanagi, M., R. H. Purcell, S. U. Emerson, and J. Bukh. 1999. Hepatitis C virus: an infectious molecular clone of a second major genotype (2a) and lack of viability of intertypic 1a and 2a chimeras. Virology 262:250-263. [DOI] [PubMed] [Google Scholar]
- 63.Yanagi, M., M. St Claire, S. U. Emerson, R. H. Purcell, and J. Bukh. 1999. In vivo analysis of the 3′ untranslated region of the hepatitis C virus after in vitro mutagenesis of an infectious cDNA clone. Proc. Natl. Acad. Sci. USA 96:2291-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yanagi, M., M. St Claire, M. Shapiro, S. U. Emerson, R. H. Purcell, and J. Bukh. 1998. Transcripts of a chimeric cDNA clone of hepatitis C virus genotype 1b are infectious in vivo. Virology 244:161-172. [DOI] [PubMed] [Google Scholar]
- 65.Young, K. C., K. L. Lindsay, K. J. Lee, W. C. Liu, J. W. He, S. L. Milstein, and M. M. Lai. 2003. Identification of a ribavirin-resistant NS5B mutation of hepatitis C virus during ribavirin monotherapy. Hepatology 38:869-878. [DOI] [PubMed] [Google Scholar]
- 66.Zhang, C., Z. Cai, Y. C. Kim, R. Kumar, F. Yuan, P. Y. Shi, C. Kao, and G. Luo. 2005. Stimulation of hepatitis C virus (HCV) nonstructural protein 3 (NS3) helicase activity by the NS3 protease domain and by HCV RNA-dependent RNA polymerase. J. Virol. 79:8687-8697. [DOI] [PMC free article] [PubMed] [Google Scholar]