Abstract
Clinical enterococcal resistance to linezolid is defined by the presence of the G2576T mutation. We evaluated the incidence of genetically proven linezolid resistance among vancomycin-resistant Enterococcus faecium strains and linezolid consumption for a possible association. A relationship was found (r2 = 0.73, P = 0.03) and predicts increasing resistance with current trends of linezolid use.
Linezolid, a member of the oxazolidinone class of antibiotics, exerts antibacterial activity by inhibiting the formation of the 70S initiation complex. This ultimately prevents the translation and replication of bacterial proteins. Enterococcal resistance to linezolid (MIC, >4 μg/ml) among clinical strains has been linked to the presence of a G2576T mutation in domain V of the 23S rRNA and may be accurately detected via PCR (8, 10, 11). Additionally, the number of genes containing this mutation has been correlated to the level of linezolid resistance detected (5, 6, 11). While the presence of the mutation in low copy levels has been found in enterococci that are susceptible and intermediate to linezolid (MIC, 1 to 4) (15), the presence of the mutation in more than one copy generally predicts some level of reduced susceptibility to linezolid (5, 9). The results of initial experiments seem to suggest that a single copy of the G2576T mutation imparts a proclivity to develop resistance (15). To date, no other mutation associated with clinical linezolid resistance among enterococci has been identified, to our knowledge.
The results of recent work in our laboratory (9) and the laboratories of others (3, 14) have cast doubt on the positive predictive value of several popular phenotypic detection methods for linezolid-intermediate or -resistant, vancomycin-resistant Enterococcus faecium (LIRVREF), and the gold standard for genotypic linezolid resistance determinants in enterococci is the detection and quantification of the G2576T mutation. Because classification of VREF as linezolid resistant may lead to unnecessary medical tests and treatments (12), it is important to characterize the emergence of genetically confirmed resistance. In this report, we screened clinical isolates of VREF for decreased phenotypic linezolid susceptibility, tested those identified as LIRVREF by phenotype for genetic resistance mutations, analyzed the prevalence of LIRVREF by genotype, and assessed the antimicrobial pressure of linezolid during the same time frame.
Isolate identification.
Isolates were obtained from clinical cultures or rectal surveillance cultures from 2002 to 2007 and were stored at −80°C. All isolates of VRE were identified to the species level by using a Vitek 2 system (Vitek Systems; bioMerieux, St. Louis, MO). When the species could not be identified with Vitek 2, isolates were identified with manual biochemical reactions (1).
Susceptibility testing.
Susceptibility testing of all isolates was performed with the Vitek 2 system using the GP61 card and following the manufacturer's instructions (Vitek Systems; bioMerieux, St. Louis, MO). Isolates with a linezolid MIC of 4 mg/liter or more were assessed for genetic resistance. As the automated system Vitek 2 has been shown to overcall resistant results (3, 9, 14), documentation of the clinical resistance mutation was required for the definition of linezolid resistance. Results for all LIRVREF isolates were confirmed by PCR for the G2576T mutation as previously described (9).
Molecular epidemiology.
All LIRVREF isolates underwent molecular typing using pulsed-field gel electrophoresis according to previously published methodologies (9). The similarities between isolates were determined by visual comparison of DNA banding patterns using the criteria of Tenover et al. (13), and isolates with a difference of more than six bands were considered genetically distinct. Isolates with a difference of three or fewer bands were considered closely related. Isolates that were closely related and had a clinical overlap (close proximity or shared medical services) were considered to be possible horizontal transfers. These isolates were excluded from calculations.
Antibiotic consumption.
Antibiotic consumption was characterized as defined daily doses (DDDs) per 1,000 patient days. Linezolid doses of 1,200 mg per day were considered one DDD (16). Linezolid consumption was obtained from antibiotic purchase data. Consumption was tallied for each fiscal year from 2002 to 2007. Each fiscal year was defined by internal hospital standards and spanned from September through August of the following calendar year.
LIRVREF incidence.
One LIRVREF isolate per patient per year was considered for inclusion. Events were standardized to the data for patient days per fiscal year to obtain rates. The definition of fiscal year remained constant throughout the study and represented the same time period for antibiotic utilization and LIRVREF incidence. The first distinct isolate identified was used in all calculations.
Statistics.
Data analysis was performed using Intercooled Stata, version 9 (Statacorp, College Station, TX) (the Pearson correlation coefficient was used to assess for a possible correlation between antibiotic resistance data (nonstandardized LIRVREF incidence) and antibiotic resistance data standardized for patient days (standardized LIRVREF incidence). The Pearson correlation coefficient was also used to assess for a possible correlation between the number of LIRVREF isolates per 1,000 patient days (linezolid resistance) and linezolid DDD per 1,000 patient days (linezolid consumption). Simple linear regression was employed, with the number of LIRVREF isolates per 1,000 patient days as the dependent variable and linezolid DDD per 1,000 patient days as the independent variable. All tests were two tailed, and P values of less than 0.05 were considered statistically significant.
Results.
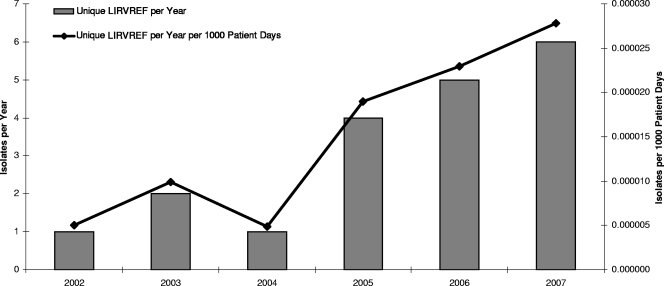
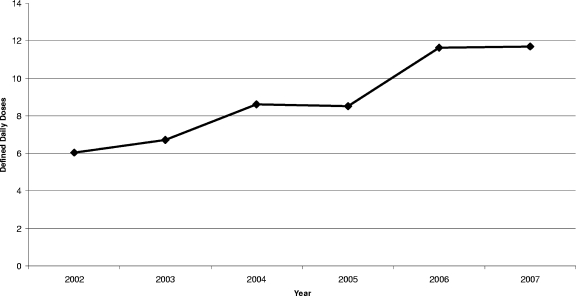
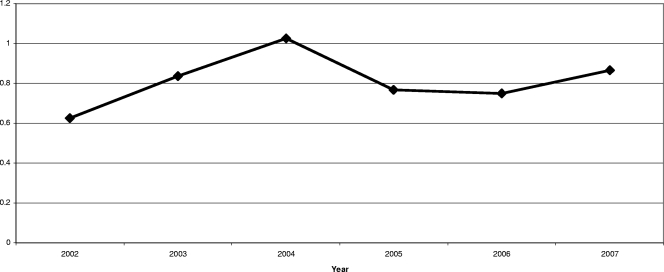
A total of 39 LIRVREF isolates were identified based on phenotype. Twenty-three of the isolates displayed the G2576T mutation. Of these, 19 isolates were genetically distinct according to the pulsed-field gel electrophoresis classification. The 19 LIRVREF-mutation-positive isolates were obtained from various clinical cultures (eight blood, five wound, three rectal swab, two urine, and one respiratory). The incidence of LIRVREF increased from one to six isolates per year in a linear trend from 2002 to 2007 (Fig. 1). The results were isometric when LIRVREF incidence per year was compared to LIRVREF incidence per year standardized to hospitalized-patient-days data by Pearson's correlation coefficient (r = 0.99, P < 0.001) (Fig. 1). Linezolid consumption ranged from 1.3 to 11.7 DDDs/1,000 patient days (Fig. 2), and consumption increased linearly from April of 2000 when linezolid was FDA approved. A correlation was noted between linezolid consumption (measured as DDD/1,000 patient days) and LIRVREF incidence per 1,000 patient days (r = 0.85), and 73% of the variability in LIRVREF incidence per 1,000 patient days was described by linezolid consumption (r2 = 0.73, P = 0.03) (Fig. 3).
FIG. 1.
Number of unique LIRVREF isolates per year. Asterisks indicate that a high correlation between the nonstandardized LIRVREF incidence and the patient day-standardized LIRVREF inicidence was observed (r = 0.99, P < 0.001).
FIG. 2.
DDDs of linezolid per 1,000 patient days.
FIG. 3.
Relationship between linezolid use and linezolid resistance. The variables were highly correlated (r = 0.85, P = 0.03), and the linear trend was significant (r2 = 0.73, P = 0.03).
Discussion.
The results of this study establish an ecological link between linezolid consumption and increasing incidence of enterococci with decreased susceptibility to linezolid. Previous studies have identified exposure to linezolid as a risk factor for linezolid resistance among enterococci (4, 7). However, these studies used phenotypic testing methods to detect linezolid resistance, and as such, the rates of nonsusceptibility and the relationship between linezolid use and resistance reported in these studies may not be an accurate reflection of the incidence of genotypically confirmed resistance (4, 7). This is the first report to assess the incidence of G2576T mutants in the setting of increasing linezolid consumption and demonstrate a correlation between linezolid consumption and genotypically proven linezolid resistance among VREF strains.
At our institution, linezolid use has increased since the introduction of the drug in 2000. For several years, persistent use of this agent did not appear to translate into the emergence of resistant organisms; however, since 2002 an increase in LIRVREF isolates possessing the G2576T mutation has been observed. We standardized our resistance data for increasing patient numbers in recent years and verified an increasing rate of LIRVREF incidence despite these increases in patient census. Additionally, we confirmed that our VRE burden did not increase over the study period. The results shown in Fig. 4 demonstrate a stable rate of clinical VRE isolation. We previously demonstrated that commonly utilized phenotypic tests, such as E-test and disk diffusion, do not accurately reflect the true incidence of linezolid resistance compared to the rate of isolation of G2576T mutants (9). These findings were further corroborated through the work of Tenover and colleagues, who also described significant error rates for the detection of linezolid resistance among enterococci when phenotypic methods were utilized, with very major errors reaching rates between 20% and 40% for some automated systems (9, 14). Additionally, disk diffusion tests and E-tests have shown 7% major error rates (13). Only agar dilution and broth microdilution methods appear to have a high correlation with the incidence of genotypic resistance (9, 14). Data such as these highlight the attractiveness of genetic confirmation for resistance. It is especially convenient when only a single mutation is associated with resistance, such as with the G2576T mutation for linezolid resistance among clinical enterococci. A reliable means of identifying linezolid resistance is critical, since false reporting of resistance may lead to unnecessary increases in resource utilization and subsequent patient morbidity (12).
FIG. 4.
Clinical VRE rate (one isolate per patient per year per 1,000 patient days).
Potential limitations of our analysis follow. First, this study used antibiotic purchase data as a method of quantifying antimicrobial usage; however, since each compartmentalized time period (1 year) was relatively long and the period analyzed (fiscal year) remained constant throughout the study, the purchase data are thought to correlate well with actual antimicrobial consumption. Second, horizontal spread of resistant isolates may potentially be implicated in outbreaks of LIRVREF (2). This is unlikely to have accounted for the increase in resistance at our institution, since only genetically distinct isolates were used for the calculations. As such, it appears that the increasing isolation of genotypically confirmed isolates of LIRVREF has followed an increase in linezolid usage at our institution. Third, these data are ecological and do not establish causality; however, we suggest that increased linezolid consumption leads to an increase in the incidence of LIRVREF. This presumption is strengthened by the facts that the data are biologically plausible, such an association is concordant with results of in vitro experiments, and linezolid use and resistance are temporally linked.
Conclusion.
Linezolid consumption and LIRVREF incidence, as defined by the presence of the G2576T mutation, are correlated and increasing at our institution. Due to the ecological nature of the data, it is impossible to establish a causal link, but the two processes are likely related. Future work should attempt to discern this association.
Acknowledgments
This study was sponsored in part by a Chicago College of Pharmacy Faculty Research Stimulation Grant, Midwestern University, Downers Grove, IL.
Footnotes
Published ahead of print on 7 April 2008.
REFERENCES
- 1.Facklam, R., and J. A. Elliott. 1995. Identification, classification, and clinical relevance of catalase-negative, gram-positive cocci, excluding the streptococci and enterococci. Clin. Microbiol. Rev. 8:479-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herrero, I. A., N. C. Issa, and R. Patel. 2002. Nosocomial spread of linezolid-resistant, vancomycin-resistant Enterococcus faecium. N. Engl. J. Med. 346:867-869. [DOI] [PubMed] [Google Scholar]
- 3.Jones, R. N., G. J. Moet, L. Woosley, H. S. Sader, and T. R. Fritsche. 2007. Critical evaluation of linezolid susceptibility testing using two automated systems (Vitek, Vitek2), abstr. D-225. Abstr. 47th Intersci. Conf. Antimicrob. Agents Chemother., Chicago, IL.
- 4.Kainer, M. A., R. A. Devasia, T. F. Jones, B. P. Simmons, K. Melton, S. Chow, J. Broyles, K. L. Moore, A. S. Craig, and W. Schaffner. 2007. Response to emerging infection leading to outbreak of linezolid resistant enterococci. Emerg. Infect. Dis. 13:1024-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lobritz, M., R. Hutton-Thomas, S. Marshall, and L. B. Rice. 2003. Recombination proficiency influences frequency and locus of mutational resistance to linezolid in Enterococcus faecalis. Antimicrob. Agents Chemother. 47:3318-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marshall, S. H., C. J. Donskey, R. Hutton-Thomas, R. A. Salata, and L. B. Rice. 2002. Gene dosage and linezolid resistance in Enterococcus faecium and Enterococcus faecalis. Antimicrob. Agents Chemother. 46:3334-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pai, M. P., K. A. Rodvold, P. C. Schreckenberger, R. D. Gonzales, J. M. Petrolatti, and J. P. Quinn. 2002. Risk factors associated with the development of infection with linezolid- and vancomycin-resistant Enterococcus faecium. Clin. Infect. Dis. 35:1269-1272. [DOI] [PubMed] [Google Scholar]
- 8.Prystowsky, J., F. Siddiqui, J. Chosay, D. L. Shinabarger, J. Millichap, L. R. Peterson, and G. A. Noskin. 2001. Resistance to linezolid: characterization of mutations in rRNA and comparison of their occurrences in vancomycin-resistant enterococci. Antimicrob. Agents Chemother. 45:2154-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qi, C., X. Zheng, A. Obias, M. H. Scheetz, M. Malczynski, and J. R. Warren. 2006. Comparison of testing methods for detection of decreased linezolid susceptibility due to G2576T mutation of the 23S rRNA gene in Enterococcus faecium and Enterococcus faecalis. J. Clin. Microbiol. 44:1098-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raad, I. I., H. A. Hanna, R. Y. Hachem, T. Dvorak, R. B. Arbuckle, G. Chaiban, and L. B. Rice. 2004. Clinical-use-associated decrease in susceptibility of vancomycin-resistant Enterococcus faecium to linezolid: a comparison with quinupristin-dalfopristin. Antimicrob. Agents Chemother. 48:3583-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruggero, K. A., L. K. Schroeder, P. C. Schreckenberger, A. S. Mankin, and J. P. Quinn. 2003. Nosocomial superinfections due to linezolid-resistant Enterococcus faecalis: evidence for a gene dosage effect on linezolid MICs. Diagn. Microbiol. Infect. Dis. 47:511-513. [DOI] [PubMed] [Google Scholar]
- 12.Scheetz, M. H., C. Qi, G. A. Noskin, J. R. Warren, M. J. Postelnick, M. Malczynski, J. Huang, and T. R. Zembower. 2006. The clinical impact of linezolid susceptibility reporting in patients with vancomycin-resistant enterococci. Diagn. Microbiol. Infect. Dis. 56:407-413. [DOI] [PubMed] [Google Scholar]
- 13.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tenover, F. C., P. P. Williams, S. Stocker, A. Thompson, L. A. Clark, B. Limbago, R. B. Carey, S. M. Poppe, D. Shinabarger, and J. E. McGowan, Jr. 2007. Accuracy of six antimicrobial susceptibility methods for testing linezolid against staphylococci and enterococci. J. Clin. Microbiol. 45:2917-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Werner, G., M. Bartel, N. Wellinghausen, A. Essig, I. Klare, W. Witte, and S. Poppert. 2007. Detection of mutations conferring resistance to linezolid in Enterococcus spp. by fluorescence in situ hybridization. J. Clin. Microbiol. 45:3421-3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.World Health Organization Collaborating Centre for Drug Statistics Methodology. 2007. Anatomic therapeutic chemical/defined daily dose (ATC/DDD) index. www.whocc.no/atcddd/. Accessed 4 November 2007.