Abstract
We have discovered a novel class of compounds active against hepatitis C virus (HCV), using a surrogate cellular system, HCV replicon cells. The leading compound in the series, ACH-806 (GS-9132), is a potent and specific inhibitor of HCV. The selection of resistance replicon variants against ACH-806 was performed to map the mutations conferring resistance to ACH-806 and to determine cross-resistance profiles with other classes of HCV inhibitors. Several clones emerged after the addition of ACH-806 to HCV replicon cells at frequencies and durations similar to that observed with NS3 protease inhibitors and NS5B polymerase inhibitors. Phenotypic analyses of these clones revealed that they are resistant to ACH-806 but remain sensitive to other classes of HCV inhibitors. Moreover, no significant change in the susceptibility to ACH-806 was found when the replicon cellular clones resistant to NS3 protease inhibitors and NS5B polymerase inhibitors were examined. Sequencing of the entire coding region of ACH-806-resistant replicon variants yielded several consensus mutations. Reverse genetics identified two single mutations in NS3, a cysteine-to-serine mutation at amino acid 16 and an alanine-to-valine mutation at amino acid 39, that are responsible for the resistance of the replicon variants to ACH-806. Both mutations are located at the N terminus of NS3 where extensive interactions with the central hydrophobic region of NS4A exist. These data provide evidence that ACH-806 inhibits HCV replication by a novel mechanism.
Hepatitis C virus (HCV) is the leading cause of liver disease worldwide. It is estimated that 170 million individuals are infected with HCV (56). The current therapeutic combination of pegylated alpha interferon (IFN-α) and ribavirin has a sustained viral response rate of ∼50% in genotype 1 HCV-infected patients and is limited by the adverse effects of both agents (8, 13). Therefore, the development of oral anti-HCV agents with improved efficacy and better tolerance is urgently needed.
HCV is an enveloped virus with a positive-stranded RNA genome of 9.6 kb. The viral genome encodes a large polyprotein that is cleaved cotranslationally and/or posttranslationally into at least 10 mature viral proteins: C, E1, E2, p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B (3, 26). Specific functions have been attributed to most of these viral proteins. For example, NS5B protein has an RNA-dependent RNA polymerase activity, the amino-terminal domain of NS3 carries serine protease activity, and NS4A is a cofactor of NS3 which enhances NS3 protease activity by forming a stable heterodimeric complex with NS3 (4, 5, 24, 26, 28, 30). Both NS5B polymerase and NS3 protease have been the prime targets for the development of HCV-specific agents. To date, multiple small molecules against the two targets have been reported (7, 17, 18, 48), and some of them have shown antiviral activity in HCV-infected patients (9, 20, 42, 43, 44, 54).
Among HCV-specific inhibitors discovered so far, NS5B nucleoside analogs target the polymerase catalytic site of NS5B. These inhibitors block nascent viral RNA synthesis by preventing further elongation after they are incorporated into nascent RNA chains (7, 18). On the other hand, NS5B nonnucleoside inhibitors, which belong to a number of different chemotypes, block the early steps of viral RNA replication by binding to four distinctive allosteric sites away from the active site of NS5B (17, 18). Different from NS5B inhibitors, NS3 protease inhibitors are substrate-based peptidomimetic compounds. They bind to the active site of the enzyme and competitively inhibit NS3 protease activity (48). Selections of resistance variants with many of these agents, using HCV replicon cells, have been reported (21, 22, 23, 31, 32, 36, 37, 39, 49, 50, 52). As expected, the signature-resistant mutations of most of the inhibitors are located around specific inhibitor-binding pockets and the cross-resistance exists among the inhibitors which bind to the same pocket.
The development of resistance to anti-human immunodeficiency virus (HIV) drugs has been a major factor that limits the efficacy of virus-specific therapies for treating HIV patients. Given the lack of a proofreading mechanism for HCV NS5B polymerase and the high replication rate of HCV in patients, it is well recognized that the emergence of resistant HCV variants is inevitable (34, 38). In fact, the appearance of viruses resistant to anti-HCV drug candidates has already been observed for clinical trials (45, 54). These resistant HCV viruses may exist as prior variants due to the presence of quasispecies in HCV patients, or they may be generated during the treatment period. Based on findings from the treatment of HIV patients, combination therapies with agents which act differently and are therefore not cross-resistant to each other are believed to be necessary to achieve the sustained suppression of HCV replication. Hence, efforts have been invested in the identification of anti-HCV compounds that act on targets other than HCV NS3 protease and NS5B polymerase.
Here, we report studies of the selection and characterization of HCV replicon variants with resistance to ACH-806 (or GS-9132), a novel and potent HCV inhibitor (Fig. 1A). ACH-806 was discovered by using HCV replicon cells. Mechanism-of-action studies have revealed that ACH-806 prevents the proper formation of replication complexes by selectively binding to NS4A (14; W. Yang, Y. Zhao, J. Fabrycki, X. Hou, X. Nie, A. Sanchez, A. Phadke, M. Deshpande, A. Agarwal, and M. Huang, unpublished data). Furthermore, ACH-806 has been confirmed to inhibit HCV replication in genotype 1 HCV-infected patients in a proof-of-concept clinical trial, although the reversible nephrotoxicity observed with the trial precludes its further clinical development (41). In this report, we describe the identification of key amino acid mutations responsible for the resistance by nucleotide sequence mapping of resistant replicon variants emerging under ACH-806 selection and the confirmation of their roles in replicon variants, through the use of reverse genetics. We also present evidence for the lack of cross-resistance between ACH-806 and other classes of HCV inhibitors, including nucleoside and nonnucleoside HCV NS5B inhibitors and HCV NS3 protease inhibitors. Finally, insight into the mechanism of resistance of this class of inhibitors is discussed.
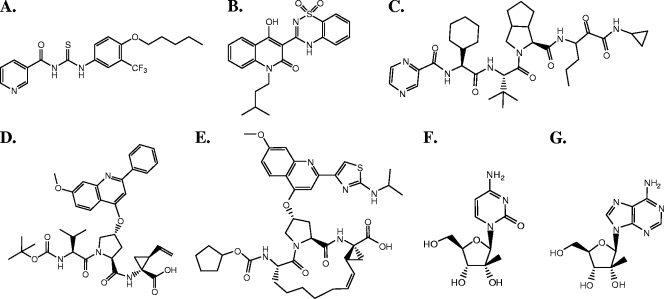
FIG. 1.
Chemical structures of HCV inhibitors used in this study. (A) ACH-806. (B) Nonnucleoside NS5B polymerase inhibitor, NNI-1, a benzothiadiazine derivative (47). (C) NS3 protease inhibitor, VX-950 (23). (D) NS3 protease inhibitor, PI-1 (6). (E) NS3 protease inhibitor, BILN 2061 (20). (F) Nucleoside NS5B polymerase inhibitor, Idenix NM-107 (2′-C-methylcytosine) (15). (G) Nucleoside NS5B polymerase inhibitor, NI-1 (2′-C-methyladenosine) (10).
MATERIALS AND METHODS
HCV inhibitors.
The structures of all inhibitors used are shown in Fig. 1. The synthesis of ACH-806 is published elsewhere. HCV NS5B polymerase nonnucleoside inhibitor (NNI-1; a benzothiadiazine derivative), HCV NS5B polymerase nucleoside inhibitors [NI-1; (2′-C-methyladenosine) and NM-107 (2′-C-methylcytosine)], and NS3 protease inhibitor (PI-1) were synthesized according to procedures described previously (6, 10, 15, 47) and were fully characterized by 1H nuclear magnetic resonance, high-performance liquid chromatography, and mass spectroscopy. IFN-α2b (intron A) was purchased from Schering-Plough Corporation.
Plasmids and HCV replicon-containing cell lines.
Subgenomic 1b (Con-1) replicon-encoding plasmids, pFK-I389neo/NS3-3′/wt and pFK-I341PI-Luc/NS3-3′/ET, and the subgenomic 1b (Con-1) replicon cell line Huh-9-13 were described previously (27, 29, 55). Huh-7 and its derivative HCV replicon cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), nonessential amino acids, penicillin (100 IU/ml), and streptomycin (100 μg/ml) at 37°C in an atmosphere of 5% CO2. Culture medium of replicon cells was additionally supplemented with 0.25 to 0.5 mg/ml G418, unless specified otherwise.
Selection of colonies resistant to HCV inhibitors.
The selection of resistant replicon cellular colonies was performed as described by Trozzi et al. (52), with the following modifications. HCV replicon cells (Huh-9-13 cells) were seeded in 100-mm-diameter tissue culture dishes at a density of 105 cells per dish. The next day, the culture medium was removed, and the selection medium containing 0.5 mg/ml G418 and 1 μM ACH-806 (a concentration approximately 25-fold greater than the 50% effective concentration [EC50]) was added. To select replicon cellular colonies resistant to other HCV inhibitors, specific compounds at a concentration of 10- to 50-fold their EC50 values were included in the selection medium. The selection medium was changed twice a week without passaging the cells. After approximately 3 weeks of selection, replicon cellular colonies resistant to ACH-806 and the other inhibitors became visible. Several resistant replicon cellular colonies were randomly picked up and propagated in the corresponding selection medium for subsequent characterizations. Huh-9-13 cells incubated in the selection medium without compound were used as the parental cell controls. To define the mutation(s) responsible for the resistant phenotypes, total RNA was extracted from individual resistant replicon cell lines with TRIzol reagent (Invitrogen, Carlsbad, CA). Reverse transcription-PCR (RT-PCR) was used to amplify either part of or the entire coding region of the HCV nonstructural proteins (NS3 to NS5B). Nucleotide sequences of PCR products were determined by Keck Foundation Biotechnology Resource Lab at Yale University, with an ABI model 377 sequencer unit according to the manufacturer's instructions. Sequences were aligned with Lasergene sequence analysis software (DNAStar, Inc., Madison, WI). All sequence numbers used in this report refer to the position of the corresponding nucleotide (nt) or amino acid of the complete HCV genome (HCV Con-1; GenBank accession number AJ238799).
Construction of molecular clones containing specific mutations.
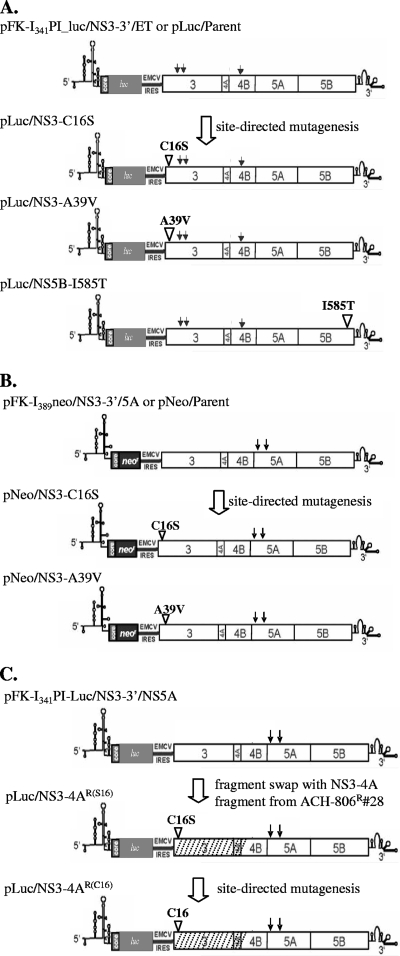
Three sets of plasmids containing replicon variants were constructed (Fig. 2). The first set of plasmids were constructed by introducing point mutations into the construct pFK-I341PI-Luc/NS3-3′/ET (in short, pLuc/Parent), which encodes a bicistronic nonselectable HCV replicon (Fig. 2A) (27). Point mutations were generated by PCR amplification of the target sequence, with primers containing the desired mutation. The resulting plasmids included (i) pLuc/NS3-C16S, replacement of the triplet UGC (nt 3465 to 3467) coding for cysteine 1042 in the polyprotein (residue 16 of NS3), with AGC coding for serine; (ii) pLuc/NS3-A39V, replacement of the triplet GCA (nt 3534 to 3436) coding for alanine 1065 in the polyprotein (residue 39 of NS3), with GUA coding for valine; and (iii) pLuc/NS5B-I585T, replacement of the triplet AUC (nt 9351 to 9353) coding for isoleucine 3004 in the polyprotein (residue 585 of NS5B), with ACC coding for threonine. The second set of plasmids was constructed by introducing point mutations into pFK-I389neo/NS3-3′/5A (in short, pNeo/Parent), which encodes a bicistronic selectable HCV replicon (Fig. 2B). The pNeo/Parent construct is the same as the prototype construct pFK-I389neo/NS3-3′/wt (29), except that it contains two adaptive mutations in NS5A, as shown in Fig. 2B (12). These two adaptive mutations were found to be present in all replicons isolated from the Huh-9-13 cell line. The resulting plasmids included (i) pNeo/NS3-C16S, replacement of the triplet UGC (nt 3465 to 3467) coding for cysteine 1042 in the polyprotein (residue16 of NS3), with AGC coding for serine; and (ii) pNeo/NS3-A39V, replacement of the triplet GCA (nt 3534 to 3436) coding for alanine 1065 in the polyprotein (residue 39 of NS3), with GUA coding for valine. The third set of plasmids was constructed in the backbone of pFK-I341PI-Luc/NS3-3′/NS5A, which encodes a bicistronic nonselectable replicon. This plasmid is identical to pI341PI-luc/NS3-3′/ET but contains different adaptive mutations, as shown in Fig. 2C. The resulting plasmids included (i) pLuc/NS3-4AR(S16), which contains a fragment encoding NS3, 4A, and part of NS4B (nt 3420 to 6530), from one of the ACH-806 resistant clones (no. 28). This plasmid was constructed as follows. Total RNA was isolated from the ACH-806-resistant clone 28 with TRIzol reagent (Invitrogen, Carlsbad, CA). RT-PCR was used to amplify the fragment encoding NS3, 4A, and a part of 4B, with a set of primers containing unique restriction enzyme sites (5′, HindIII; 3′, MluI). Following restriction enzyme digestion, the RT-PCR fragment was used to replace the corresponding region in pFK-I341PI-luc/NS3-3′/NS5A. The resulting plasmid contains a cysteine-to-serine mutation at amino acid residue 16 in NS3, in addition to several other nonconsensus mutations that are absent in the majority of other resistant clones but are present in clone no. 28; and (ii) pLuc/NS3-4AR(C16), which is identical to pLuc/NS3-4AR(S16) except that the serine mutation at amino acid residue 16 of NS3 was reverted back to cysteine by using PCR-mediated site-directed mutagenesis. All plasmid constructs were verified by DNA sequencing.
FIG. 2.
Construction of plasmids encoding replicon variants. (A) A single mutation of either cysteine to serine at position 16 of NS3, alanine to valine at position 39 of NS3, or isoleucine to threonine at position 585 of NS5B was introduced into the parental construct pFK-I341PI-Luc/NS3-3′/ET (pLuc/Parent). The resulting plasmids were designated pLuc/NS3-C16S, pLuc/NS3-A39V, and pLuc/NS5B-I585T. EMCV IRES, encephalomyocarditis virus internal ribosome entry site. (B) A single mutation of either cysteine to serine at position 16 of NS3 or alanine to valine at position 39 of NS3 was introduced into the parental construct pFK-I389neo/NS3-3′/5A (pNeo/Parent). The resulting plasmids were designated pNeo/NS3-C16S and pNeo/NS3-A39V. (C) The RT-PCR fragment (shaded boxes) amplified from total RNA of ACH-806-resistant clone no. 28 was used to replace the NS3 and 4A and part of 4B encoding region in pFK-I341PI_Luc/NS3-3′/5A, and the resulting plasmid was designated pLuc/NS3-4BR(S16) because it contained a cysteine-to-serine mutation at amino acid residue 16 of NS3. The construct pLuc/NS3-4BR(C16) was identical to pLuc/NS3-4BR(S16), except that the serine mutation at amino acid residue 16 of NS3 reverted back to cysteine. Abbreviation: #28, a clone cell line, no. 28. All the mutations were confirmed by sequencing analysis. Arrows represent adaptive mutations, which are ET (E1202G, T1280I, and K1846T) (55) in all replicons in panel A and S2204I and D2254E in all replicons in panels B and C (12).
RNA transcription and electroporation.
To generate run-off transcripts of HCV replicons, plasmid DNA was first linearized with AseI, followed by ScaI. After DNA was extracted with phenol-chloroform and ethanol precipitation, it was used as the template in an in vitro T7 transcription reaction (Megascript T7 kit; Ambion, Austin, TX). Transcripts were extracted with acidic phenol, and the concentration of RNA was determined by measuring the optical density at 260 nm.. The RNA was transfected into Huh-7 cells by electroporation, as described previously (27, 29). In brief, single-cell suspensions of Huh-7 cells were prepared at a density of 107 cells per ml in Cytomix solution supplemented with 2 mM ATP and 5 mM glutathione. After 1 μg (colony formation assay) or 5 μg (transient transfection assay) of in vitro-transcribed RNA was mixed with 400 μl of the cell suspension in a Gene Pulser cuvette (0.4-cm gap), electroporation was immediately performed at 270 V and 960 μF, with a Gene Pulser system (Bio-Rad, Hercules, CA) (27, 29, 53). Electroporated cells were immediately diluted with 10 ml of DMEM supplemented with 10% FBS.
Transient HCV replication assay.
Following transfection of nonselectable replicon RNA molecules, which were synthesized in vitro, into Huh-7 cells by electroporation, cells were seeded into 96-well plates at a density of 8,000 cells per well in a final volume of 100 μl. One day after the cells were plated, 100 μl of DMEM-10% FBS containing a 2× concentration of a specific inhibitor was added, and the cells were further incubated for 72 h. The replication capacity of HCV replicon variants as well as the inhibition of HCV replicon replication was quantified by measurement of the luciferase activity, using a commercial kit (Britelite Ultra-high sensitive luminescence reporter gene assay system; Perkin Elmer, Wellesley, MA). Briefly, at the end of 72 h of treatment, cells were lysed, and lysates were transferred to an opaque white 96-well plate. Immediately after the Britelite substrate was added, the plate was read with a MicroBeta model liquid scintillation and luminescence counter (Perkin Elmer, Wellesley, MA). The replication capacity of replicon variants was assessed by the level of luciferase activity (in relative luminescence units [RLU]) measured at 4 days posttransfection in cells not treated with inhibitor. This level was normalized by the efficiency of transfection, which was expressed as the luciferase activity at 4 h posttransfection. The inhibition of replicon replication by an inhibitor was expressed as the concentration that caused a reduction of luciferase activity (RLU) by 50% (EC50) or 90% (EC90) in comparison to that of the no-inhibitor-treated controls.
Colony formation assay.
Huh-7 cells transfected by electroporation with selectable replicon RNA molecules were seeded in 10-cm-diameter tissue culture dishes at a density of 3 × 105 cells per dish. Initially, cells were cultured in DMEM supplemented with 10% FBS and ACH-806 at various concentrations (0 to 3.16 μM). The next day, the medium was replaced with DMEM supplemented with 10% FBS, 0.5 mg/ml G418, and ACH-806 at the same concentration range (0 to 3.16 μM). Cells were refed with the same media twice a week until colonies formed. The colonies were fixed with 10% formaldehyde and stained with 2% crystal violet in 20% ethanol.
Dot blot hybridization.
The level of replicon RNA was quantitatively measured by dot blot hybridization. Briefly, 8,000 replicon cells were seeded into each well of a 96-well plate. The next day, half-log dilutions of ACH-806 or control compounds were added to the cells. After 3 days of incubation, the cells were harvested with 70 μl of lysis buffer (50 mM Tris-HCl [pH 8.0], 140 mM NaCl, 1.5 mM MgCl2, 0.5% Nonidet P-40, 1,000 U/ml RNasin, and 1 mM dithiothreitol). Nuclei were removed from the lysates by centrifugation at 300 × g for 3 min. Fifty microliters of each supernatant was transferred to the wells of a 96-well manifold equipped with a nylon membrane (Schleicher & Schuell, Keene, NH). The membrane was subjected to hybridization with a 32P-labeled negative-sense riboprobe complementary to NS5B at 58°C overnight. Following hybridization, the membrane was washed and dried, and the radioactivity in each spot was counted with a MicroBeta model liquid scintillation and luminescence counter (Perkin Elmer). Anti-HCV activity was expressed as the concentration that caused a reduction of radioactivity (cpm) by 50% (EC50) in comparison to that of the untreated control. In most cases, the membrane was also exposed to X-ray film after counting. Additional details of this assay will be published elsewhere (Yang et al., unpublished).
Structural analysis and modeling.
Coordinates for the three-dimensional structure of the full-length NS3 protein were obtained from the Protein Data Bank entry 1CU1 (59). 1CU1 was analyzed using InsightII 2000 software (Accelrys), using an SGI Octane2 workstation. Distances between the nearest heavy atoms of Ala 39 and Cys 16 and the NS4A cofactor were measured.
RESULTS
By screening a diverse collection of small molecules in HCV replicon cells, we discovered leading compounds with EC50 values as low as 3 μM. Further systematic optimization of these leaders yielded analogs with greatly improved potency, including a clinical development candidate, ACH-806 (or GS-9132) (Fig. 1A). ACH-806 displays potent activity against both genotype 1a and 1b replicons with EC50 values between 10 and 50 nM, depending on the replicon cells and the end points. Mechanism-of-action studies have shown that ACH-806 does not inhibit the enzymatic activities of NS5B polymerase and NS3 protease per se (data not shown), suggesting that a novel mechanism is attributed to the anti-HCV activity of ACH-806.
Selection of replicon variants resistant to ACH-806.
Selection was initiated by culturing Huh-9-13 cells, genotype 1b replicon-containing cells (29), in the presence of ACH-806 and G418. Significant cell death was observed approximately 10 days after selection was initiated. Colonies of replicon cells resistant to both the inhibitor and the selecting antibiotic became visible 2 weeks later. Two independent selections were performed (selections 1 and 2 in Table 1), with an average frequency of ∼0.04% for the emergence of resistant colonies. The duration and the frequency of resistant colony formation to ACH-806 were in accordance with prior data reported for NS3 protease inhibitors and for NS5B polymerase inhibitors (21, 31, 32, 36, 37, 49, 52). It should be noted that passaging a resistant replicon cellular line (no. 28, see below) under higher concentrations (up to 4 μM) of ACH-806 failed to impose further selection, i.e., no apparent cell death ever occurred. No higher concentration was attempted due to the limitation in ACH-806 solubility (the compound crystallized after 3 days in cultures at concentrations of 10 μM and higher).
TABLE 1.
Phenotypes and genotypes of ACH-806-resistant replicon cell lines
| Cell line, clone (selection) | Fold change in EC50 (μM) of the indicated compounda
|
Codon (amino acid) of NS3 at residue:
|
||||
|---|---|---|---|---|---|---|
| ACH-806 | NNI-1 | PI-1 | NI-1 | 16 | 39 | |
| Parental | 1.0 | 1.0 | 1.0 | 1.0 | UGC (Cys) | GCA (Ala) |
| 806R, 22 (1) | 13.9 | 2.2 | 2.2 | 2.2 | AGC (Ser) | GCA (Ala) |
| 806R, 23 (1) | 18.6 | 0.3 | 2.5 | ND | AGC (Ser) | GCA (Ala) |
| 806R, 24 (1) | 11.2 | 1.2 | 1.2 | 2.0 | AGC (Ser) | GCA (Ala) |
| 806R, 25 (1) | 14.9 | 0.9 | 1.1 | ND | AGC (Ser) | GCA (Ala) |
| 806R, 28 (1) | 21.3 | 0.8 | 0.8 | 3.0 | AGC (Ser) | GCA (Ala) |
| 806R, 6 (2) | 18.6 | 2.6 | 4.2 | 2.0 | UGC (Cys) | GTA (Val) |
| 806R, 8 (2) | 9.5 | 1.2 | 1.1 | 1.9 | UGC (Cys) | GTA (Val) |
| 806R, 9 (2) | 11.5 | 3.0 | 2.7 | 2.4 | UGC (Cys) | GTA (Val) |
| Avg fold change | 14.9 | 1.3 | 1.7 | 2.7 | ||
Fold changes in EC50 were calculated by a comparison of that seen with the parental replicon cellular line Huh-9-13. The EC50 ± standard deviations (μM) from three independent experiments against Huh-9-13 were 0.04 ± 0.02 (ACH-806), 0.78 ± 0.2 (NNI-1), 0.79 ± 0.16 (PI-1), and 0.48 ± 0.05 (NI-1). ND, not determined.
For phenotypic and genotypic analyses, eight random colonies were picked and expanded. The susceptibility levels of these resistant replicon cellular lines to ACH-806 and to other classes of inhibitors were compared to that of their parental replicon cellular lines by measuring HCV replicon RNA with a dot blot assay as described in Materials and Methods. An exemplary result that includes a dot blot image and dose-response curves obtained from one ACH-806-resistant replicon cellular line is shown in Fig. 3. The summary of phenotypes of all eight cellular lines is presented in Table 1. The average increase of EC50 values to ACH-806 of all resistant replicon cellular lines was about 15-fold (ranging from 9.5 to 21.3). In contrast, the average increase of EC50 to other classes of inhibitors was less than twofold (ranging from 0.3 to 4.2). These results suggested that the resistant phenotype shown in the cloned replicon cellular lines was specific to ACH-806.
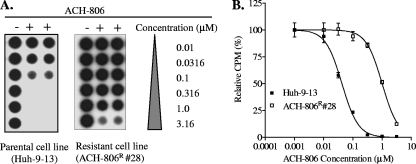
FIG. 3.
Comparison of the susceptibility level of the parental cell line with that of the cell line resistant to ACH-806. The dose responses of ACH-806 in Huh-9-13 cells, a parental cell line, and in ACH-806R, a clone (no. 28) derived from Huh-9-13 under the selection of ACH-806. Both cell lines were treated in duplicate with ACH-806 at the concentrations indicated for 3 days. The HCV replicon level following the treatment was quantified with a dot blot assay using a 32P-labeled riboprobe as described in Materials and Methods. (A) Autoradiograph of the membrane. (B) Dose-response curve. The fitting was performed based on the radioactivity (cpm) in each spot on the membrane relative to that of the untreated controls (100%) by using nonlinear regression with Prism software (GraphPad Software, Inc., San Diego, CA).
Genotypic analysis of resistant replicon variants.
To ascertain if the reduced ACH-806 susceptibility of the resistant replicon cellular lines was due to a distinctive replicon mutation(s), we determined the sequences of the entire nonstructural protein-coding regions of replicons extracted from the three resistant replicon cellular lines (no. 22, no. 24 and no. 28 from selection 1) and aligned them with the sequences of the parental replicon. Besides some nonconserved mutations randomly scattering throughout the replicons, we found two sense mutations that were conserved in all three replicons isolated from resistant replicon cellular lines. One mutation resulted in the replacement of cysteine 16 of NS3 (residue 1042 of the polyprotein) with serine (C16S) (Table 1), and the other mutation resulted in the replacement of isoleucine 585 of NS5B (residue 3004 of the polyprotein) with threonine (I585T). Since the C16S mutation in NS3 was the mutation that conferred resistance to ACH-806 (see below), the subsequent genetic analysis focused on the NS3 region. After sequencing the NS3 region of replicons extracted from the remaining five resistant replicon cellular lines, we found that two of them (no. 23 and no. 25 from selection 1) carried the C16S mutation; however, the other three lines (no. 6, no. 8, and no. 9 from selection 2) carried a different mutation that replaced alanine 39 of NS3 (residue 1065 of the polyprotein) with valine (A39V) (Table 1). The same A39V mutation was previously detected in resistant replicon variants selected with several ACH-806 analogs and was identified as the mutation responsible for the reduced susceptibility of the resistant replicon cellular clones to those analogs (data not shown).
Determination of mutations responsible for the resistant phenotype.
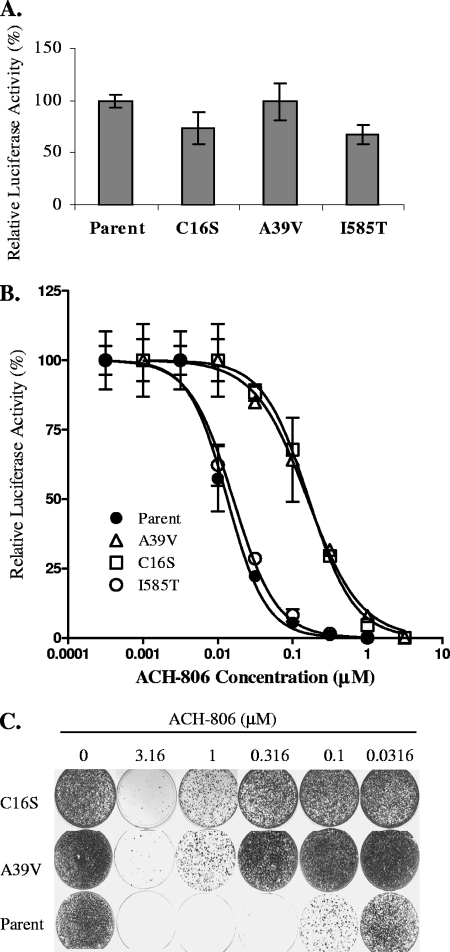
To investigate the genetic basis of resistance to ACH-806 in the resistant replicon cellular lines, specifically the roles of C16S and A39V in NS3 and I585T in NS5B, we introduced these mutations separately into a nonselectable replicon containing a luciferase reporter gene as shown in Fig. 2A. Following the transfection of the parental replicon RNA molecule and the C16S, A39V, and I585T variant replicon RNA molecules into Huh-7 cells, the replication capacity of all replicons was determined as described in Materials and Methods. Replicon RNA carrying the A39V mutation replicated as efficiently as its parent, whereas the C16S and I585T mutations caused a slight decrease in the replication capacity (about 75% of that of the parental replicon) (Fig. 4A). The susceptibility of each of these four replicons to ACH-806 as well as to other classes of inhibitors was compared side by side, and the results are summarized in Table 2 and Fig. 4B. The I585T mutation did not significantly affect the potency of any compound. The C16S and A39V mutations, on the other hand, increased the EC50 values of ACH-806 12- and 14-fold, respectively. In contrast, neither of the mutations significantly affected the potency of other control inhibitors. In addition, a replicon variant carrying both the C16S and the A39V mutations was also made but failed to replicate efficiently in Huh-7 cells.
FIG. 4.
Reverse genetic studies. (A) Comparison of the replication capacity of the parental cell line with that of the mutant replicon variants in transiently transfected cells. Following transfection into the Huh-7 cells of replicon RNAs synthesized in vitro, the cells were plated in 96-well plates and cultured in the absence of any HCV inhibitors. Luciferase activities of each replicon-transfected cells were measured at 4 h and on day 4 posttransfection, and the values were normalized by the efficiency of transfection, i.e., luciferase activities at 4 h posttransfection. Parent, C16S, A39V and I585T are replicons made from pLuc/parent, pLuc/NS3-C16S, pLuc/NS3-A39V, and pLuc/NS5B-I585T, respectively. Error bars indicate standard derivations from two replicates. (B) Dose-response curves of the parental and mutant replicons to ACH-806 treatment in transiently transfected cells. Following transfection of replicon RNAs synthesized in vitro into Huh-7 cells, the cells were plated in 96-well plates and cultured in the presence of ACH-806. Luciferase activities of each replicon-transfected cells were measured on day 4 posttransfection. The curve fitting was performed, based on luciferase activity in each well relative to that of the untreated controls (100%), by nonlinear regression with Prism software (GraphPad Software, Inc., San Diego, CA). Parent, C16S, A39V and I585T are replicons made from pLuc/parent, pLuc/NS3-C16S, pLuc/NS3- A39V, and pLuc/NS5B-I585T, respectively. Error bars indicate standard deviations from two replicates. (C) Comparison of the susceptibilities of parental cells with those of mutant replicons to ACH-806 by colony formation assay. Naïve Huh-7 cells were transfected with the replicons synthesized in vitro. After cells were treated with G418 (0.5 mg/ml) and various concentrations of ACH-806 as indicated at the top for ∼4 weeks, they were fixed, and colonies were stained with crystal violet solution. Parent, C16S, and A39V are replicons made from pNeo/Parent, pNeo/C16S, and pNeo/NS3-A39V, respectively.
TABLE 2.
Susceptibility of the parental and mutant replicon variants to ACH-806 and control inhibitorsa
| Compound | Susceptibility ± SD (μM) of the Luc/Parent replicon at:
|
Fold change in susceptibility ± SD (μM) of the indicated variant repliconb
|
||||||
|---|---|---|---|---|---|---|---|---|
| Luc/NS3-A39V
|
Luc/NS3-C16S
|
Luc/NS5B-I585T
|
||||||
| EC50 | EC90 | EC50 | EC90 | EC50 | EC90 | EC50 | EC90 | |
| ACH-806 | 0.01 ± 0.01 | 0.08 ± 0.01 | 14.0 | 13.7 | 11.1 ± 1.0 | 11.3 ± 0.9 | 0.7 ± 0.1 | 1.1 ± 0.1 |
| NNI-1 | 1.76 ± 0.56 | 7.50 ± 1.00 | 1.0 | 1.1 | 1.1 ± 0.2 | 1.0 ± 0.1 | 1.3 ± 0.4 | 1.1 ± 0.2 |
| PI-1 | 0.90 ± 0.88 | 4.77 ± 2.10 | 1.3 | 0.7 | 0.8 ± 0.4 | 1.1 ± 0.6 | 0.8 ± 0.4 | 1.0 + 0.0 |
| NI-1 | 0.14 ± 0.06 | 1.25 ± 0.44 | 0.9 | 1.2 | 1.2 ± 0.6 | 0.9 ± 0.2 | 1.4 ± 0.8 | 1.1 ± 0.0 |
Data shown are averages ± standard deviations of the results from two independent experiments, except for those for the replicon Luc/NS3-A39V, where only one experiment was conducted.
Fold changes at EC50 and EC90 were calculated by comparison with those seen with the Luc/Parent replicon.
To further confirm the roles of C16S and A39V in conferring resistance to ACH-806, we introduced both mutations separately into a selectable replicon which carries the neo gene and the adaptive mutations that differ from the nonselectable replicon (Fig. 2B). Following transfection of the parental replicon RNA, the C16S replicon RNA and the A39V replicon RNA, into Huh-7 cells, the susceptibility of each of these three replicons to ACH-806, as evaluated by the number of colonies formed under the treatment of both G418 and ACH-806, was compared side by side (Fig. 4C). Again, an approximately 10-fold reduction in the susceptibility of C16S or the A39V replicon variant to ACH-806 was observed.
A third approach, fragment cloning, was used to reinforce the role of the C16S mutation and to exclude the role of other nonconsensus mutations in conferring resistance to ACH-806. An ∼3-kb fragment covering the coding regions of NS3, NS4A, and part of NS4B was amplified by RT-PCR from total RNA isolated from the ACH-806-resistant clone no. 28. The PCR product was cloned into the nonselectable replicon containing a luciferase reporter gene (Fig. 2C). Replicon RNA molecules were made from eight individual clones and were transfected into Huh-7 cells to determine the replication capacity by luciferase activity. From the eight clones, two were able to replicate efficiently, and sequencing of the entire cloned fragment in these two clones confirmed the existence of the C16S mutation in addition to other nonconsensus mutations. The serine at amino acid residue 16 of NS3 in one of these two clones was then converted back to cysteine by using site-directed mutagenesis, leaving other nonconsensus mutations unchanged (Fig. 2C). The replication capacity of this resulting replicon [Luc/N3-4AR(C16)] was 95% of that of its parent [Luc/N3-4AR(S16)], determined as previously described. After they were transfected into Huh-7 cells, the susceptibility of both replicons was compared. The reversion of serine back to cysteine at amino acid residue 16 of NS3 increased the susceptibility of the replicon to ACH-806 about 10-fold and maintained the susceptibility to other classes of HCV inhibitors (Table 3). Similar results were also obtained with a fragment containing NS3, 4A, and part of 4B derived from a resistant cell line carrying the A39V mutation (data not shown). In summary, all three approaches yielded a 10- to 15-fold decrease in the susceptibility to ACH-806 in the replicon variants carrying either the C16S or the A39V mutation, a value similar to that observed for the resistant cell clones emerging under the selection of ACH-806 (Table 1). Hence, we conclude that either of the C16S or the A39V mutations in the HCV replicon is sufficient to confer the replicon resistance to ACH-806.
TABLE 3.
Susceptibility of the mutant replicon variants with and without the C16S mutation to ACH-806 and control inhibitorsa
| Compound | Susceptibility ± SD (μM) of the indicated repliconsa
|
Fold change (μM) atb
|
||||
|---|---|---|---|---|---|---|
| Luc/NS3-4AR(S16)
|
Luc/NS3-4AR(C16)
|
EC50 | EC90 | |||
| EC50 | EC90 | EC50 | EC90 | |||
| ACH-806 | 0.27 ± 0.04 | 2.35 ± 0.24 | 0.02 ± 0.00 | 0.24 ± 0.01 | 13.5 | 9.8 |
| NNI-1 | 0.44 ± 0.13 | 2.84 ± 0.06 | 0.63 ± 0.03 | 3.04 ± 0.05 | 0.7 | 0.9 |
| PI-1 | 0.38 ± 0.10 | 1.88 ± 0.08 | 0.64 ± 0.05 | 2.52 ± 0.03 | 0.6 | 0.7 |
| NI-1 | 0.19 ± 0.00 | 0.86 ± 0.01 | 0.24 ± 0.01 | 0.90 ± 0.01 | 0.8 | 1.0 |
Data shown are averages ± standard deviations of results from two independent experiments.
Data represent fold changes in EC50 and EC90 relative to that seen with the Luc/NS3-4AR(C16) replicon.
No cross-resistance between ACH-806 and other classes of HCV inhibitors.
In the study described above, we showed that there were no significant differences between the susceptibility to ACH-806-resistant replicon variants and that of their parental replicons to the other classes of HCV inhibitors (Table 2 and 3). In addition, the susceptibility of ACH-806-resistant replicon cellular lines to IFN and ribavirin was also similar to that of the parent cell line (data not shown). The lack of cross-resistance between ACH-806 and other classes of HCV inhibitors was further confirmed by the evaluation of the susceptibility to ACH-806 of replicon cellular lines resistant to other classes of HCV inhibitors. The replicon cellular lines resistant to each of the HCV inhibitors were obtained by selection with the Huh-9-13 cell line. These resistant replicon cell lines were highly resistant to their inducing agent (a change in EC50 values of >20-fold) and carried the signature mutations reported previously by others (23, 31, 36, 39, 49, 52) (Table 4). No significant differences were observed when the susceptibility levels to ACH-806 of these resistant replicon cellular lines and the parental replicon cellular line were compared side by side (Table 4). Hence, we conclude that ACH-806 is not cross-resistant with the other classes of inhibitors that we have tested, including nucleoside and nonnucleoside NS5B polymerase inhibitors and NS3 protease inhibitors.
TABLE 4.
Susceptibility of other resistant cell lines to ACH-806
| Cell line (resistance mutation) | Fold change in EC50 (μM) for the indicated compounda
|
|||||
|---|---|---|---|---|---|---|
| ACH-806 | VX-950 | BILN 2061 | NM107 | NI-1 | NNI-1 | |
| Parental | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| VX-950R (A156T) | 0.6 | 43.7 | >100 | ND | 0.6 | ND |
| BILN 2061R (D168V) | 1.1 | 2.9 | >100 | ND | 1.4 | 0.8 |
| NM107R (S282T) | 0.5 | 0.3 | 1.1 | 30.7 | 11.5 | 2.9 |
| NI-1R (S282T) | 0.8 | ND | 1.4 | >68.7 | 30.3 | 5.0 |
| NNI-1R (M414T) | 0.9 | 2.3 | 2.8 | ND | 0.6 | >28.4 |
The fold change in EC50 values was calculated by comparison with those seen with the parent replicon cell line Huh-9-13. The EC50 values ± standard deviations from two independent experiments against Huh-9-13 cells are 0.04 ± 0.03 μM (ACH-806), 0.61 ± 0.14 μM (VX-950), 1.2 ± 0.61 nM (BILN 2061), 1.16 ± 0.1.22 μM (NM 107), 0.56 ± 0.17 μM (NI-1), and 1.37 ± 0.56 μM (NNI-1). ND, not determined.
Modeling of NS3 mutations.
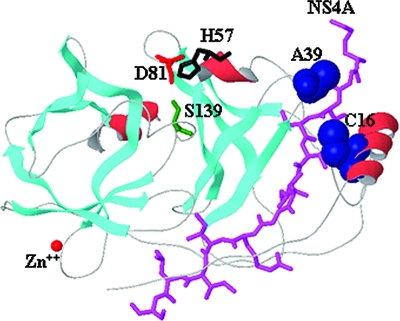
Structural and biochemical analyses of NS3 protease have shown that the very N-terminal region of NS3 interacts with NS4A to form the NS3-NS4A complex (4, 11, 24, 46, 59). In the absence of an NS4A cofactor, folding of the N-terminal region of NS3 was incomplete (2, 16, 35). Both the C16S and the A39V mutations are located in the N-terminal region of NS3. The three-dimensional locations of Cys 16 and Ala 39 are shown in Fig. 5, along with the binding position of the NS4A cofactor. The distances between Cys 16 (Cβ) of NS3 and Val 26 (CG1) of NS4A and Ala 39 (Cβ) of NS3 and Gly 21 (Cα) of NS4A are 4.1 Å and 7.3 Å, respectively. Examination of the HCV sequence database (http://hcv.lanl.gov) revealed that both Cys 16 and Ala 39 are highly conserved in genotype 1 HCV.
FIG. 5.
Locations of NS3 C16 and A39. NS3 protease domain (PDB code 1NS3) is gray, where β strands (cyan) and α helices (red) are shown as ribbons; catalytic site residues are red (Asp 81), black (His 57), and green (Ser 139); the structural zinc ion is displayed as a red sphere; the NS4A cofactor is shown in magenta; and the CPK representation of the resistant mutants (Ala 39 and Cys 16) is blue. The distances between Cys 16 (Cb) of NS3 and Val 26 (CG1) of NS4A and Ala 39 (Cb) of NS3 and Gly 21 (Cα) of NS4A are 4.1 Å and 7.3 Å, respectively.
DISCUSSION
In this report, we present for the first time the resistant profile of ACH-806, a new class of HCV inhibitor. By using a selection procedure similar to the one that has been described for other classes of HCV inhibitors (23, 31, 36, 37, 39, 52), resistant replicon cell lines have been obtained, and eight of these cell lines have been further characterized (Table 1). All of the replicon cell lines exhibit a reduction in the susceptibility to ACH-806, an average EC50 value increase of 15-fold compared to the EC50 value of the parental replicon cell line to this inhibitor. These resistant replicon cell lines are divided into two groups, based on genotypic analyses, one group carrying a C16S mutation and the other an A39V mutation. Both mutations are mapped to the N terminus of the NS3 protease domain. Different approaches of reverse genetics have been employed to study the roles of the two mutations in conferring resistance to ACH-806. The results of those studies unequivocally demonstrate that either of the C16S or the A39V mutations is sufficient to confer resistance to ACH-806. This conclusion is based on the fact that reduction in the susceptibility of the replicons rendered by each of the C16S and A39V mutations in the reverse genetics is close to that found in the original ACH-806-resistant replicon cellular clones. In addition, we previously used IFN to cure a replicon cell line, which was derived from selection with an ACH-806 analog, was resistant to ACH-806 and its analogs and also contained replicon molecules with the A39V mutation in NS3 (data not shown). When we transfected the nonselectable naïve replicon into the cured cell line, we found no change in its susceptibility to ACH-806 and its analogs (data not shown). These data suggest that any cellular adaptation that may have occurred during selection with the ACH-806 analog plays a negligible role in conferring resistance to this class of inhibitors.
Unlike the resistant replicon variants selected with other classes of HCV-specific inhibitors, where a higher reduction in the susceptibility is often seen (23, 36, 52), all ACH-806-resistant replicon cell lines exhibit a moderate reduction (Table 1). Resistant replicon cell lines obtained with several analogs of ACH-806 display the same phenomenon (data not shown). We do not know the exact reason for this, as of now, although multiple scenarios could be used to interpret this difference. One attractive hypothesis is that the variants able to confer high resistance to ACH-806 do not replicate well, such as the C16S and A39V double mutant, so they cannot be selected out under the current selection condition. It is possible that the presence of viral quasispecies in HCV-infected patients, due to the error-prone nature of NS5B polymerase and high replication rate in vivo (34, 38), might provide more opportunities for the selection of resistant viruses, which display resistance levels that are different from those presented in this study. Nevertheless, the low-level resistance will be advantageous if it holds true in the clinical setting.
In addition to ACH-806, we have performed resistance selection studies with 10 analogs of ACH-806. The A39V mutation was detected in most of the replicon clones obtained from selection with these analogs. However, the C16S mutation was also detected in a few clones. Although we do not know the exact reason why this is, we suspect that it is related to how ACH-806 acts. Unlike viral enzyme inhibitors which inhibit enzymatic activities through direct binding to the viral enzymes, ACH-806 acts by interrupting the interaction between two viral proteins through binding to one of them. It is possible that a mutation conferring resistance may depend on changes in other amino acid residues involved in the interaction between NS3 and NS4A. Amino acid differences are not unusual among replicon cellular lines and are observed within a replicon cellular line under various passaging. For example, we have seen different nonconsensus mutations in replicon molecules isolated from the Huh-9-13 cell line at various times (data not shown). Consequently, replicons carrying different resistant mutations could emerge in one replicon cellular line from different selections with ACH-806. A similar phenomenon was observed for BMS-378806, an HIV entry inhibitor (25). BMS-378806 acts by interrupting the binding of the HIV-1 envelope protein, gp120, to cellular CD4 receptors. The major resistance mutation, in one selection, is at amino acid 426 of gp120 and, in another selection, at amino acid 475 of gp120, even though the two parental viruses do not differ significantly in their gp120 sequences (i.e., they are both derived from the same strain) (1, 40). This is in great contrast to 3TC (an HIV reverse transcriptase inhibitor). The mutation at amino acid 184 of reverse transcriptase is always found no matter what viral strains are used for the selection of variants resistant to 3TC.
Mechanism-of-action studies with ACH-806 have revealed that the inhibitor prevents the proper formation of replication complexes by binding selectively to NS4A (14; Yang et al., unpublished). Although no resistance mutation is found in NS4A, the location of both the C16 and the A39 resistance mutations is intriguing. The NS3 protease domain complexes with NS4A and adopts a chymotrypsin-like folding pattern composed of a C-terminal six-stranded β barrel and an N-terminal eight-stranded β barrel (16, 35, 58). The catalytic triad histidine 57, aspartate 81, and serine 139 is located between the two domains. One of the β strands in the N-terminal barrel is composed of the central region of NS4A (21 to 32 amino acids), which interacts extensively with the A1 and A0 β strands of NS3 (16). These structural observations are in agreement with the results generated by deletion mutagenesis that indicate the interaction domain of NS4A and NS3 lies in the N terminus of NS3 (11, 46). The location of Cys 16 is in the α0 helix that is involved in hydrophobic interactions with Val 24 and Val 26 of NS4A. The distance between Cys 16 (Cβ) and Val 26 (CG1) is 4.1 Å. The location of Ala 39 is in a loop between the A1 and B1 β strands of the NS3 N-terminal region. This loop is close to the N terminus of the cofactor part of NS4A (21 to 32 residues). Although modeling of the local structure shows that the spatial distance between Ala 39 (Cβ) and Gly 21 (Cα) of NS4A is 7.3 Å, it is unclear if Ala 39 could interact with other parts of NS4A since no structural information beyond the cofactor part of NS4A is currently available. Nevertheless, based on this information as well as the evidence that ACH-806 acts by binding to NS4A, we propose a model. When ACH-806 binds to NS4A, the binding affinity of NS4A to NS3 is reduced, resulting in several changes in replication complexes, including the impairment of certain NS4A-dependent cleavages, the reduced stability of NS3 and NS4A and other potential changes (14; Yang et al., unpublished). We believe that the mutations on the N terminus of NS3 (the C16S or the A39V mutation) alter the interaction between NS3 and NS4A so that the binding affinity of NS4A to NS3 is restored. We do not know the exact reason why no resistance mutation is identified in NS4A, where the inhibitor binds. There are precedents, though, where resistance mutations are not located in the protein to which the inhibitor binds. For example, resistance mutations are mapped to HIV envelope protein gp120, not to the CCR5 coreceptor where CCR5 antagonists bind (19, 33, 51, 57) because of the impossibility for the gene encoding CCR5 coreceptor, a host gene, to mutate. By a similar, though not the same, analogy, if the mutations in NS4A are not as tolerant as the mutations at the N terminus of NS3 to replicon growth, the replicons carrying the mutations at the N terminus of NS3 could be selected more readily under ACH-806 treatment. The lack of a significant growth disadvantage for either the A39V or the C16S replicon variant (Fig. 4) is supportive for this assumption.
This work has also demonstrated that ACH-806 and it analogs lack cross-resistance to other HCV inhibitors. ACH-806-resistant replicon cellular lines are susceptible to other HCV inhibitors, including HCV NS5B inhibitors, NS3 protease inhibitors, and IFN and ribavirin. Replicon cellular lines resistant to other classes of HCV inhibitors are as susceptible to ACH-806 as their parental replicon cellular line. No cross-resistance between ACH-806 and other HCV inhibitors provides further support for the conclusion that ACH-806 employs a novel mechanism for inhibiting HCV replication as well as a rationale for combining an agent of this class with NS5B polymerase inhibitors, NS3 protease inhibitors, and IFN and/or ribavirin in the treatment of HCV-infected patients.
Acknowledgments
We thank Jane Thanassi for help with manuscript preparation.
Footnotes
Published ahead of print on 14 April 2008.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbato, G., D. O. Cicero, M. C. Nardi, C. Steinkuhler, R. Cortese, R. De Francesco, and R. Bazzo. 1999. The solution structure of the N-terminal proteinase domain of the hepatitis C virus (HCV) NS3 protein provides new insights into its activation and catalytic mechanism. J. Mol. Biol. 289:371-384. [DOI] [PubMed] [Google Scholar]
- 3.Bartenschlager, R., L. Ahlborn-Laake, J. Mous, and H. Jacobsen. 1994. Kinetic and structural analyses of hepatitis C virus polyprotein processing. J. Virol. 68:5045-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartenschlager, R., V. Lohmann, T. Wilkinson, and J. O. Koch. 1995. Complex formation between the NS3 serine-type proteinase of the hepatitis C virus and NS4A and its importance for polyprotein maturation. J. Virol. 69:7519-7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behrens, S. E., L. Tomei, and R. De Francesco. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15:12-22. [PMC free article] [PubMed] [Google Scholar]
- 6.Boehringer Ingelheim (Canada), Ltd. February 2000. Hepatitis C inhibitor tri-peptides. International patent WO 00/09543.
- 7.Carroll, S. S., and D. B. Olsen. 2006. Nucleoside analog inhibitors of hepatitis C virus replication. Infect. Disord. Drug Targets 1:17-29. [DOI] [PubMed] [Google Scholar]
- 8.Di Bisceglie, A. M., and J. H. Hoofnagle. 2002. Optimal therapy of hepatitis C. Hepatology 36:S121-S127. [DOI] [PubMed] [Google Scholar]
- 9.Dieterich, D., E. Lawitz, T. Nguyen, Z. Younes, J. Santoro, N. Gitlin, D. McEniry, R. Chasen, J. Goff, S. Knox, K. Kleber, B Belanger, and N. Brown. 2006. Early clearance of HCV RNA with valopicitabine (NM283) plus peg-interferon in treatment-naive patients with HCV-1 infection: first results from a Phase IIb trial. J. Hepatol. 44(Suppl. 2):S271. [Google Scholar]
- 10.Eldrup, A. B., C. R. Allerson, C. F. Bennett, S. Bera, B. Bhat, N. Bhat, M. R. Bosserman, J. Brooks, C. Burlein, S. S. Carroll, P. D. Cook, K. L. Getty, M. MacCoss, D. R. McMasters, D. B. Olsen, T. P. Prakash, M Prhavc, Q. Song, J. E. Tomassini, and J. Xia. 2004. Structure-activity relationship of purine ribonucleosides for inhibition of hepatitis C virus RNA-dependent RNA polymerase. J. Med. Chem. 47:2283-2295. [DOI] [PubMed] [Google Scholar]
- 11.Failla, C., L. Tomei, and R. De Francesco. 1995. An amino-terminal domain of the hepatitis C virus NS3 protease is essential for interaction with NS4A. J. Virol. 69:1769-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replication. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoofnagle, J. H., and L. B. Seeff. 2006. Peginterferon and ribavirin for chronic hepatitis C. N. Engl. J. Med. 355:2444-2451. [DOI] [PubMed] [Google Scholar]
- 14.Huang, M., Y. Sun, W. Yang, X. Hou, J. Fabrycki, X. Nie, A. Sanchez, Y. Zhao, A. Phadke, and M. Deshpande. 2007. ACH-806: A potent inhibitor of HCV replication with a novel mechanism of action. J. Hepatol. 46(Suppl. 1):A583. [Google Scholar]
- 15.Idenix (Cayman) Limited. June 2003. Modified 2′ and 3′-nucleoside prodrugs for treating flaviridae infections. International patent WO 2004/002999 A2.
- 16.Kim, J. L., K. A. Morgenstern, C. Lin, T. Fox, M. D. Dwyer, J. A. Landro, S. P. Chambers, W. Markland, C. A. Lepre, E. T. O'Malley, S. L. Harbeson, C. M. Rice, M. A. Murcko, P. R. Caron, and J. A. Thomson. 1996. Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell 87:343-355. [DOI] [PubMed] [Google Scholar]
- 17.Koch, U., and F. Narjes. 2006. Allosteric inhibition of the hepatitis C virus NS5B RNA dependent RNA polymerase. Infect. Disord. Drug Targets 6:31-41. [DOI] [PubMed] [Google Scholar]
- 18.Koch, U., and F. Narjes. 2007. Recent progress in the development of inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. Curr. Top. Med. Chem. 7:1302-1329. [DOI] [PubMed] [Google Scholar]
- 19.Kuhmann, S. E., P. Pugach, K. J. Kunstman, J. Taylor, R. L. Stanfield, A. Snyder, J. M. Strizki, J. Riley, B. M. Baroudy, I. A. Wilson, B. T. Korber, S. M. Wolinsky, and J. P. Moore. 2004. Genetic and phenotypic analyses of human immunodeficiency virus type 1 escape from a small-molecule CCR5 inhibitor. J. Virol. 78:2790-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamarre, D., P. C. Anderson, M. Bailey, P. Beaulieu, G. Bolger, P. Bonneau, M. Bos, D. R. Cameron, M. Cartier, M. G. Cordingley, A. M. Faucher, N. Goudreau, S. H. Kawai, G. Kukolj, L. Lagace, S. R. LaPlante, H. Narjes, M. A. Poupart, J. Rancourt, R. E. Sentjens, R. St. George, B. Simoneau, G. Steinmann, D. Thibeault, Y. S. Tsantrizos, S. M. Weldon, C. L. Yong, and M. Llinas-Brunet. 2003. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature 426:186-189. [DOI] [PubMed] [Google Scholar]
- 21.Le Pogam, S., H. Kang, S. F. Harris, V. Leveque, A. M. Giannetti, S. Ali, W. R. Jiang, S. Rajyaguru, G. Tavares, C. Oshiro, T. Hendricks, K. Klumpp, J. Symons, M. F. Browner, N. Cammack, and I. Najera. 2006. Selection and characterization of replicon variants dually resistant to thumb- and palm-binding nonnucleoside polymerase inhibitors of the hepatitis C virus. J. Virol. 80:6146-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le Pogam, S., W. R. Jiang, V. Leveque, S. Rajyaguru, H. Ma, H. Kang, S. Jiang, M. Singer, S. Ali, K. Klumpp, S. Smith, J. Symons, N. Cammack, and I. Najera. 2006. In vitro selected Con1 subgenomic replicons resistant to 2′-C-methyl-cytidine or to R1479 show lack of cross resistance. Virology 351:349-359. [DOI] [PubMed] [Google Scholar]
- 23.Lin, C., K. Lin, Y-P. Luong, B. G. Rao, Y. Y. Wei, D. L. Brennan, J. R. Fulghum, H. M. Hsiao, S. Ma, J. P. Maxwell, K. M. Cottrell, R. B. Perni, C. A. Gates, and A. D. Kwong. 2004. In vitro resistance studies of hepatitis C virus serine protease inhibitors, VX-950 and BILN 2061: structural analysis indicates different resistance mechanisms. J. Biol. Chem. 279:17508-17514. [DOI] [PubMed] [Google Scholar]
- 24.Lin, C., J. A. Thomson, and C. M. Rice. 1995. A central region in the hepatitis C virus NS4A protein allows formation of an active NS3-NS4A serine proteinase complex in vivo and in vitro. J. Virol. 69:4373-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin, P. F., W. Blair, T. Wang, T. Spicer, Q. Guo, N. Zhou, Y. F. Gong, H. G. Wang, R. Rose, G. Yamanaka, B. Robinson, C. B. Li, R. Fridell, C. Deminie, G. Demers, Z. Yang, L. Zadjura, N. Meanwell, and R. Colonno. 2003. A small molecule HIV-1 inhibitor that targets the HIV-1 envelope and inhibits CD4 receptor binding. Proc. Natl. Acad. Sci. USA 100:11013-11018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol 1. Lippincott Williams & Wilkins Co., Philadelphia, PA.
- 27.Lohmann, V., S. Hoffmann, U. Herian, F. Penin, and R. Bartenschlager. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77:3007-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lohmann, V., F. Korner, U. Herian, and R. Bartenschlager. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 30.Lohmann, V., A. Roos, F. Korner, J. O. Koch, and R. Bartenschlager. 1998. Biochemical and kinetic analyses of NS5B RNA-dependent RNA polymerase of the hepatitis C virus. Virology 249:108-118. [DOI] [PubMed] [Google Scholar]
- 31.Lu, L., T. J. Pilot-Matias, K. D. Stewart, J. T. Randolph, R. Pithawalla, W. He, P. P. Huang, L. L. Klein, H. Mo, and A. Molla. 2004. Mutations conferring resistance to a potent hepatitis C virus serine protease inhibitor in vitro. Antimicrob. Agents Chemother. 48:2260-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu, L., T. Dekhtyar, S. Masse, R. Pithawalla, P. Krishnan, W. He T. Ng, G. Koev, K. Stewart, D. Larson, T. Bosse, R. Wagner, T. Pilot-Matias, H. Mo, and A. Molla. 2007. Identification and characterization of mutations conferring resistance to an HCV RNA-dependent RNA polymerase inhibitor in vitro. Antiviral Res. 76:93-97. [DOI] [PubMed] [Google Scholar]
- 33.Marozsan, A. J., S. E. Kuhmann, T. Morgan, C. Herrera, E. Rivera-Troche, S. Xu, B. M. Baroudy, J. Strizki, and J. P. Moore. 2005. Generation and properties of a human immunodeficiency virus type 1 isolate resistant to the small molecule CCR5 inhibitor, SCH-417690 (SCH-D). Virology 338:182-199. [DOI] [PubMed] [Google Scholar]
- 34.Martell, M., J. L. Esteban, J. Quer, J. Genesca, A. Weiner, R. Esteban, J. Guardia, and J. Gomez. 1992. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J. Virol. 66:3225-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCoy, M. A., M. M. Senior, J. J. Gesell, L. Ramanathan, and D. F. Wyss. 2001. Solution structure and dynamics of the single-chain hepatitis C virus NS3 protease NS4A cofactor complex. J. Mol. Biol. 305:1099-1110. [DOI] [PubMed] [Google Scholar]
- 36.Migliaccio, G., J. E. Tomassini, S. S. Carroll, L. Tomei, S. Altamura, B. Bhat, L. Bartholomew, M. R. Bosserman, A. Ceccacci, L. F. Colwell, R. Cortese, R. De Francesco, A. B. Eldrup, K. L. Getty, X. S. Hou, R. L. LaFemina, S. W. Ludmerer, M. MacCoss, D. R. McMasters, M. W. Stahlhut, D. B. Olsen, D. J. Hazuda, and O. A. Flores. 2003. Characterization of resistance to non-obligate chain-terminating ribonucleoside analogs that inhibit hepatitis C virus replication in vitro. J. Biol. Chem. 278:49164-49170. [DOI] [PubMed] [Google Scholar]
- 37.Mo, H., L. Lu, T. Pilot-Matias, R. Pithawalla, R. Mondal, S. Masse, T. Dekhtyar, T. Ng, G. Koev, V. Stoll, K. D. Stewart, J. Pratt, P. Donner, T. Rockway, C. Maring, and A. Molla. 2005. Mutations conferring resistance to a hepatitis C virus (HCV) RNA-dependent RNA polymerase inhibitor alone or in combination with an HCV serine protease inhibitor in vitro. Antimicrob. Agents Chemother. 49:4305-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neumann, A. U., N. P. Lam, H. Dahari, D. R. Gretch, T. E. Wiley, T. J. Layden, and A. S. Perelson. 1998. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science 282:103-107. [DOI] [PubMed] [Google Scholar]
- 39.Nguyen, T. T., A. T. Gates, L. L. Gutshall, V. K. Johnston, B. Gu, K. J. Duffy, and R. T. Sarisky. 2003. Resistance profile of a hepatitis C virus RNA-dependent RNA polymerase benzothiadiazine inhibitor. Antimicrob. Agents Chemother. 47:3525-3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peden, K. W. 1992. Instability of HIV sequences in high copy number plasmids. J. Acquir. Immune Defic. Syndr. 5:313-315. [PubMed] [Google Scholar]
- 41.Pottage, J. C., E. Lawitz, D. Mazur, D. Wyles, H. Vargas, R. Ghalib, R. Gugliotti, M. Donohue, and H. Robison. 2007. Short-term antiviral activity and safety of ACH-806 (GS-9132), an NS4A antagonist, in HCV genotype 1 infected individuals. J. Hepatol. 46(Suppl. 1):A783. [Google Scholar]
- 42.Reesink, H. W., S. Zeuzem, C. J. Weegink, N. Forestier, A. van Vliet, J. van de Wetering de Rooij, L. McNair, S. Purdy, R. Kauffman, J. Alam, and P. L. Jansen. 2006. Rapid decline of viral RNA in hepatitis C patients treated with VX-950: a phase Ib, placebo-controlled, randomized study. Gastroenterology 131:997-1002. [DOI] [PubMed] [Google Scholar]
- 43.Roberts, S., G. Cooksley, D. Shaw, H. K. Berns, M. T. Brandl, S. H. Fettner, G. Hill, D. Ipe, K. Klumpp, M. Mannino, E. O'Mara, Y. Tu, and C. B. Washington. 2006. Interim results of a multiple ascending dose study of R1626, a novel nucleoside analog targeting HCV polymerase in chronic HCV patients. J. Hepatol. 44(Suppl. 2):S269. [Google Scholar]
- 44.Sarrazin, C., R. Rouzier, F. Wagner, N. Forestier, D. Larrey, S. K. Gupta, M. Hussain, A. Shah, D. Cutler, J. Zhang, and S. Zeuzem. 2007. SCH 503034, a novel hepatitis C virus protease inhibitor, plus pegylated interferon alpha-2b for genotype 1 nonresponders. Gastroenterology 132:1270-1278. [DOI] [PubMed] [Google Scholar]
- 45.Sarrazin, C., T. L. Kieffer, D. Bartels, B. Hanzelka, U. Muh, M. Welker, D. Wincheringer, Y. Zhou, H. M. Chu, C. Lin, C. Weegink, H. Reesink, S. Zeuzem, and A. D. Kwong. 2007. Dynamic hepatitis C virus genotypic and phenotypic changes in patients treated with the protease inhibitor telaprevir. Gastroenterology 132:1767-1777. [DOI] [PubMed] [Google Scholar]
- 46.Satoh, S., Y. Tanji, M. Hijikata, K. Kimura, and K. Shimotohno. 1995. The N-terminal region of hepatitis C virus nonstructural protein 3 (NS3) is essential for stable complex formation with NS4A. J. Virol. 69:4255-4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.SmithKline Beecham Corporation. May 2001. Novel anti-infectives. International patent no. WO 01/85172 A1.
- 48.Thomson, J. A., and R. B. Perni. 2006. Hepatitis C virus NS3-4A protease inhibitors: countering viral subversion in vitro and showing promise in the clinic. Curr. Opin. Drug Discov. Devel. 9:606-617. [PubMed] [Google Scholar]
- 49.Tomei, L., S. Altamura, L. Bartholomew, A. Biroccio, A. Ceccacci, L. Pacini, F. Narjes, N. Gennari, M. Bisbocci, I. Incitti, L. Orsatti, S. Harper, I. Stansfield, M. Rowley, R. De Francesco, and G. Migliaccio. 2003. Mechanism of action and antiviral activity of benzimidazole-based allosteric inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 77:13225-13231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tong, X., R. Chase, A. Skelton, T. Chen, J. Wright-Minogue, and B. A. Malcolm. 2006. Identification and analysis of fitness of resistance mutations against the HCV protease inhibitor SCH 503034. Antiviral Res. 70:28-38. [DOI] [PubMed] [Google Scholar]
- 51.Trkola, A., S. E. Kuhmann, J. M. Strizki, E. Maxwell, T. Ketas, T. Morgan, P. Pugach, S. Xu, L. Wojcik, J. Tagat, A. Palani, S. Shapiro, J. W. Clader, S. McCombie, G. R. Reyes, B. M. Baroudy, and J. P. Moore. 2002. HIV-1 escape from a small molecule, CCR5-specific entry inhibitor does not involve CXCR4 use. Proc. Natl. Acad. Sci. USA 99:395-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trozzi, C., L. Bartholomew, A. Ceccacci, G. Biasiol, L. Pacini, S. Altamura, F. Narjes, E. Muraglia, G. Paonessa, U. Koch, R. De Francesco, C. Steinkuhler, and G. Migliaccio. 2003. In vitro selection and characterization of hepatitis C virus serine protease variants resistant to an active-site peptide inhibitor. J. Virol. 77:3669-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van den Hoff, M. J., A. F. Moorman, and W. H. Lamers. 1992. Electroporation in “intracellular” buffer increases cell survival. Nucleic Acids Res. 20:2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Villano, S. A., D. Raible, D. Harper, J. Speth, P. Chandra, P. Shaw, and G. Bichier. 2007. Antiviral activity of the non-nucleoside polymerase inhibitor, HCV-769, in combination with pegylated interferon alpha-2b in treatment-naive patients with chronic HCV. J. Hepatol. 46(Suppl. 1):S24. [Google Scholar]
- 55.Vrolijk, J. M., A. Kaul, B. E. Hansen, V. Lohmann, B. L. Haagmans, S. W. Schalm, and R. Bartenschlager. 2003. A replicon-based bioassay for the measurement of interferons in patients with chronic hepatitis C. J. Virol. Methods 110:201-209. [DOI] [PubMed] [Google Scholar]
- 56.Wasley, A., and M. J. Alter. 2000. Epidemiology of hepatitis C: geographic differences and temporal trends. Semin. Liver Dis. 20:1-16. [DOI] [PubMed] [Google Scholar]
- 57.Westby, M., J. Mori, C. Smith-Burchnell, M. Lewis, M. Mosley, F. Perruccio, R. Mansfield, P. Dorr, and M. Perros. 2005. Maraviroc (UK-427,857)-resistant HIV-1 variants, selected by serial passage, are sensitive to CCR5 antagonists and T-20. Antivir. Ther. 10:S72. [Google Scholar]
- 58.Yan, Y., Y. Li, S. Munshi, V. Sardana, J. L. Cole, M. Sardana, C. Steinkuehler, L. Tomei, R. De Francesco, L. C. Kuo, and Z. Chen. 1998. Complex of NS3 protease and NS4A peptide of BK strain hepatitis C virus: a 2.2 Å resolution structure in a hexagonal crystal form. Protein Sci. 7:837-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yao, N., P. Reichert, S. S. Taremi, W. W. Prosise, and P. C. Weber. 1999. Molecular views of viral polyprotein processing revealed by the crystal structure of the hepatitis C virus bifunctional protease-helicase. Structure Fold Des. 7:1353-1363. [DOI] [PubMed] [Google Scholar]