Abstract
Efforts to increase the potency of transcriptional activators are generally unsuccessful because poor expression of activators in mammalian cells limits their delivery to target promoters. Here we report that the effectiveness of chimeric activators can be dramatically improved by expressing them as noncovalent tetrameric bundles. Bundled activation domains are much more effective at activating a reporter gene than simple monomeric activators, presumably because, at similar expression levels, up to 4 times as many the activation domains are delivered to the target promoter. These bundled activation domains are also more effective than proteins in which activation domains are tandemly reiterated in the same polypeptide chain, because such proteins are very poorly expressed and therefore not delivered effectively. These observations suggest that there is a threshold number of activation domains that must be bound to a promoter for activation, above which promoter activity is simply a function of the number of activators bound. We show that bundling can be exploited practically to enhance the sensitivity of mammalian two-hybrid assays, enabling detection of weak interactions or those between poorly expressed proteins. Bundling also dramatically improves the performance of a small-molecule-regulated gene expression system when the expression level of regulatory protein is limiting, a situation that may be encountered in gene therapy applications.
Many emerging technologies in gene therapy and biological research utilize chimeric transcriptional activators to control the expression of a target gene (1–6). Transcriptional activators bind to specific DNA sequences in the promoter region of a gene and, through their activation domains, interact with and recruit components of the transcription machinery (7–9). Typically, chimeric activator proteins are composed of a DNA-binding domain that is foreign to the host cell and an activation domain derived from proteins such as the herpes simplex virus regulator VP16 or the p65 subunit of human transcription factor NF-κB (10–13). Both of these activation domains have been shown to function as potent inducers of transcription when recruited to a wide range of promoters (15–17). Furthermore, they differ from many other activation domains in their ability to activate stably integrated promoters (18, 19). Consistent with their shared ability to function as potent inducers of transcription of both integrated and episomal promoters, these activation domains share many structural properties (20–22) and interact with many of the same components of transcription machinery (23–25).
Most practical uses of chimeric activators would benefit from activation domains of increased potency. One strategy commonly used for increasing the potency of transcriptional activators is simple tandem reiteration of the activation domain in the context of a single polypeptide chain (22, 26–29). In theory, this strategy generates higher local concentrations of the activation domain in the vicinity of the promoter, in turn enhancing the efficiency with which the transcriptional apparatus is recruited. Several studies have shown that multimerization of short amino acid motifs from well-characterized activators can lead to potent and synergistic activation of gene expression (26–30). For example, tandemly reiterated amino acid motifs derived from the activation domains of OCT-1 and VP16 strongly stimulate transcription (26, 27). However, reiteration of such motifs generally does not lead to activators significantly more potent than the intact parental domain. One possible explanation for the unexpectedly poor performance of tandemly reiterated activation domains is low protein expression. Indeed, proteins containing either the VP16 or the p65 activation domain are poorly expressed in eukaryotic cells, perhaps because of cytotoxic effects (30–33). Proteins with multiple activation domains may therefore be expressed at levels so low that they are not effectively recruited to target promoters.
In this report, we show that chimeric activators containing multiple copies of the VP16 or p65 activation domains are indeed expressed at very low levels in mammalian cells and that they are ineffective inducers of transcription. We present a strategy for enhancing the delivery of multiple activators to a target promoter. We show that activation domain fusion proteins expressed as noncovalent tetrameric “bundles” are expressed at higher levels than tandemly reiterated multimers and that they are significantly more potent than simple monomeric activators at similar levels of expression. Bundled activation domains greatly enhance the level of target gene expression in engineered cells and enhance the sensitivity of two-hybrid assays.
Materials and Methods
Plasmid Constructions.
Transcription factor fusion proteins were expressed from pCGNN (34). Individual components of the transcription factors were synthesized by PCR as fragments containing an XbaI site immediately upstream of the first codon and an SpeI site, an in-frame stop codon, and a BamHI site immediately downstream of the last codon. Chimeric proteins comprising multiple components were assembled by stepwise insertion of XbaI–BamHI fragments into SpeI/BamHI-opened vectors as described by Rivera et al. (5). The individual components used and their abbreviations are as follows: G, yeast GAL4 DNA-binding domain, amino acids 1–94; F, human FKBP12, amino acids 1–107; R, FKBP12-rapamycin-binding (FRB) domain of human FKBP12-rapamycin-associated protein, amino acids 2025–2113; S, activation domain from the p65 subunit of human NF-κB, amino acids 361–551; V, activation domain from herpesvirus VP16, amino acids 410–490; and L, Escherichia coli lactose repressor, amino acids 46–360. For example, pCGNN-GF2 was made by insertion of the GAL4 DNA-binding domain into pCGNN to generate pCGNN-G, followed by the sequential insertion of two FKBP domains.
Stable Cell Lines.
The HT1080B stable cell line used in this study has been described previously (19). This cell line contains a stably integrated copy of the secreted alkaline phosphatase (SEAP) reporter gene placed under the control of five GAL4-binding sites. pCGNN-RLN/Z1F3/Neo and pCGNN-RN/Z1F3/Neo tricistronic vectors were stably transfected into HT1080L cells (5) to generate HT34 and HT35 cells. Over 100 G418-resistant colonies were pooled. Addition of drugs and SEAP assays were as described by Rivera et al. (5). Background SEAP activity, obtained from untransfected HT1080B cells, was subtracted from each value.
Results
Multimerization of Activation Domains Failed to Increase Their Potency in Vivo.
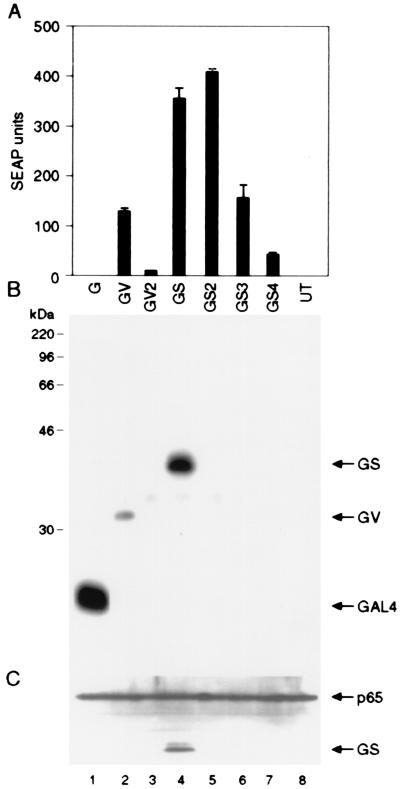
To determine whether simple multimerization of activation domains increases their activity, we generated a series of chimeric activators containing two or more tandemly reiterated copies of the p65 (S) or VP16 (V) activation domains fused to the GAL4 DNA-binding domain (G). We analyzed the function of these activators in HT1080B cells, a human fibrosarcoma line carrying a stably integrated SEAP gene driven by a minimal IL-2 promoter flanked by five GAL4-binding sites. We have shown previously that the stably integrated SEAP reporter gene is expressed in HT1080B cells (19). As expected, transfection into these cells of an expression plasmid producing a GAL4-p65 fusion protein (GS) resulted in significant activation of the SEAP reporter gene (Fig. 1A). A similar activator containing two tandemly reiterated p65 activation domains (GS2) induced reporter gene expression to about the same level. In contrast, GAL4 fusion proteins containing three or four copies of p65 activation domain, GS3 or GS4, respectively, were considerably less effective. Thus, tandem reiteration of the p65 activation domain failed to increase reporter gene expression and, at higher levels of reiteration, actually resulted in sharply lower reporter gene expression. For GAL4-VP16, the inhibitory effect of activation domain reiteration was even more acute. A fusion protein carrying just two reiterated VP16 activation domains (GV2) elicited significantly lower levels of gene expression than a fusion carrying a single domain (GV) (Fig. 1A).
Figure 1.
Multimerization of potent activation domains increases their specific activity but decreases their intracellular concentration. (A) Indicated GAL4 activator plasmids (100 ng each) were introduced into HT1080B cells that carry a stably integrated SEAP reporter gene placed under the control five GAL4-binding sites. Recombinant proteins GV and GV2 contain GAL4 DNA-binding domains fused with one and two copies of VP16 activation domains. GS, GS2, GS3, and GS4 contain GAL4 DNA-binding domains fused with one, two, three, and four copies of p65 activation domains, respectively. G denotes the GAL4 DNA-binding domain only. Mean values of the SEAP activity secreted into the medium are shown (±SD). (B and C) Western blot analysis of the total cellular extracts from transfected cells. The membrane was probed first with anti-hemagglutinin to detect the transfected proteins (B) and then with anti-p65 antibodies to confirm that roughly equal amounts of protein are present in each lane (C).
To examine the levels of expression of the various fusion proteins, we performed immunoblot analysis of extracts of transfected cells (Fig. 1B). This analysis showed that GAL4 fusion proteins containing two or more copies of the p65 or VP16 activation domains were expressed very poorly compared with fusion proteins containing a single activation domain. A control blot probed with an antibody that detects endogenous p65 confirmed that similar amounts of protein were loaded in each lane (Fig. 1C). Taken together, these observations suggest two conclusions. First, tandem reiteration of activation domains leads to the generation of activator proteins with increased specific activity. This is most readily apparent by comparing GS and GS2, which elicit similar levels of reporter gene activity despite a significant difference in protein levels. Second, tandem reiteration of activation domains leads to low protein expression levels, and this reduction in protein concentration can quantitatively exceed the increase in specific activity. Consequently, despite their greater inherent potency, these proteins are not delivered to the target promoter in sufficient numbers to achieve a net increase in gene expression.
Noncovalent “Bundling” of Activation Domains.
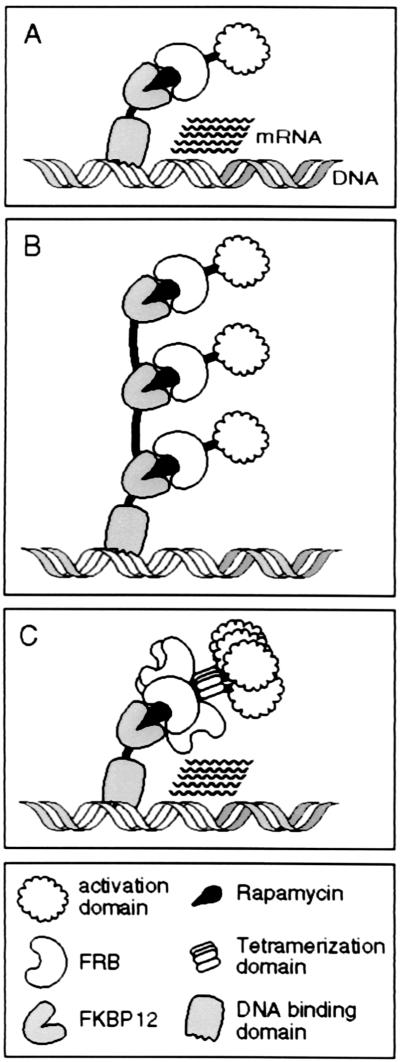
Our observations suggest that higher levels of transcription of the reporter gene could be achieved either by enhancing the expression of highly potent activators or, in the face of low activator expression in the cell, by enhancing the efficiency with which the activators are delivered to the promoter. With this in mind, we modified our previously described system for small-molecule-regulated gene expression (5). Our goals were to achieve higher levels of target gene expression and to enhance the efficiency of target gene expression in applications, such as gene therapy, in which expression of the regulatory transcription factors may be low. The basic system for small-molecule-regulated gene expression (Fig. 2A) is composed of a GAL4 DNA-binding domain fused to a single copy of FKBP12 (GF) and an activation domain from the p65 subunit of NF-κB fused to the FRB domain of FKBP12-rapamycin-associated protein (RS) (5, 35). In the presence of the natural-product compound rapamycin, which binds simultaneously to FKBP and FRB, the FRB-p65 fusion protein is recruited to the GAL4-FKBP fusion protein, resulting in activation of a target gene. This arrangement results in the delivery of a maximum of one p65 activation domain per GAL4 monomer (Fig. 2A). In this system, the number of activation domains recruited to the promoter can be increased in two ways. First, multiple FKBP domains can be linked to GAL4, enabling each GAL4 monomer to recruit multiple activators (Fig. 2B). Alternatively, multiple activation domains can be linked to FRB, which enables a single FKBP domain to recruit multiple activators.
Figure 2.
Diagrammatic description of strategies used to increase the number of activation domains delivered to the promoter. (A) In the basic method, two fusion proteins, one containing a GAL4 DNA-binding domain fused to FKBP12 and the other containing a p65 activation domain fused to FRB, are expressed in cells. Addition of rapamycin leads to the recruitment of a single activation domain to each DNA-binding domain monomer. (B) Fusion of multiple FKBPs to the DNA-binding domain allows rapamycin to recruit multiple activation domains to each DNA-binding domain monomer. (C) Addition of the lactose repressor tetramerization domain to the FRB-activation domain fusion protein, producing RLS, allows rapamycin to recruit four activation domains to each FKBP fused to the DNA-binding domain. The number of activation domains recruited to the promoter can be increased by attaching more FKBP moieties to the DNA-binding domain and/or multiplying the number of binding sites for the activator. In theory, as many as 160 activation domains can be delivered to a promoter containing five GAL4-binding sites by treating the cells expressing GF4 and RLS fusion proteins with rapamycin. The actual number of activation domains delivered to the promoter of the reporter gene in vivo by using the bundling strategy has not been determined.
Bundling Increases the Potency of Activation Domains.
Consistent with what was shown above, increasing the number of activation domains in a chimeric activator by tandem reiteration is not an effective strategy for delivering more activation domains to a promoter with rapamycin (data not shown). Instead, we constructed a chimeric activator that carried a single activation domain plus a domain that promotes noncovalent protein multimerization, such that each delivery event brings a “bundle” of activation domains to the promoter. This protein, RLS, contained a portion of lactose repressor (36) containing its tetramerization domain (L) placed between the FRB (R) and p65 (S) domains. The resulting protein should exist in cells as a noncovalent tetramer, carrying four FRB domains and four activation domains. In the presence of the dimerizer rapamycin, a single molecule of rapamycin (Fig. 2C) could recruit the entire tetrameric bundle. By combining tandem reiteration of the FKBP moiety on the GAL4 DNA-binding domain with the bundling strategy, it should be possible to recruit up to 16 p65 activation domains to a single GAL4 monomer.
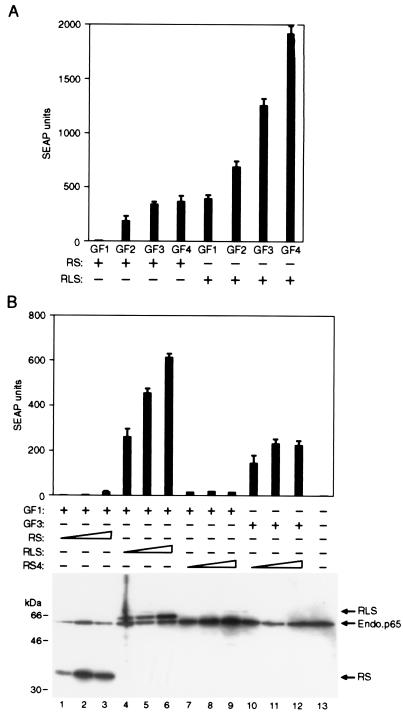
To examine how bundled activation domain fusion proteins function in this system, we transfected HT1080B cells with plasmids expressing various combinations of transcription factor fusion proteins and treated the cells with 10 nM rapamycin to deliver the activation domains to the promoter. We observed that delivering a single copy of the FRB-p65 fusion protein, RS, to each GAL4 monomer induced the reporter gene only marginally above background (GF1 + RS, Fig. 3A). In contrast, delivering a single RLS bundle to each GAL4 monomer (GF1 + RLS) induced reporter gene transcription strongly. Immunoblot analysis indicated that the RS and RLS proteins were expressed at similar levels in the transfected cells (see Fig. 3B). Thus, increasing the number of activation domains delivered to a target promoter leads to significant increases in gene expression.
Figure 3.
The level of expression of a stably integrated gene correlates with the number and strength of the activation domains bound to its promoter. (A) The indicated DNA-binding domain and activation domain fusion proteins (see Materials and Methods for abbreviations used) were transfected into HT1080B cells that carry a stably integrated SEAP reporter gene placed under the control of five GAL4-binding sites. In all cases, SEAP expression values are plotted for cultures receiving 100 ng of activation domain expression plasmid, which gives peak expression values in transiently transfected cells and slightly below peak levels in the stably transfected cell line. The background SEAP activity (24 SEAP units) was subtracted from each value before plotting. In this experiment, expressing the GF1 + RS combination of fusion proteins produced four SEAP units above background levels in the presence of rapamycin. (B) The DNA-binding domain and activation domain expression plasmids were transfected into HT1080B cells. In all cases, mean values of SEAP activity secreted into the medium after the addition of 10 nM rapamycin are shown (±SD). Western blot analysis of the total cell lysates with anti-hemagglutinin antibody allowed an assessment of transcription factor component levels in cells.
Indeed, by testing various combinations of fusion proteins, it was possible to systematically vary the number of activation domains delivered to each DNA-binding domain from 1 to 16 (Fig. 3A). Under these conditions, there was an excellent correlation between the number of activation domains delivered to the promoter and induced reporter gene activity. This observation suggests that, in general, expression of an engineered gene may be limited solely by the number and potency of activators that can be effectively delivered to the promoter of a stably integrated gene.
Activation Domain Bundles Are More Potent and Expressed at Higher Levels than Multimerized Activation Domains.
Next we compared the transcriptional activity of RLS bundles with the RS4 fusion protein, which is capable of delivering the same number of p65 activation domains to the promoter upon each rapamycin-induced delivery event. For this purpose, we expressed the DNA-binding domain fusion protein, GF1, with either RS4 or RLS, and delivered the activators to the promoter with 10 nM rapamycin. We again observed that the recruitment of RLS bundles to the promoter strongly induced reporter gene expression (Fig. 3B). In contrast, rapamycin failed to induce reporter gene expression in cells expressing GF1 and RS4 fusion proteins. Because each delivery event should bring the same number of activation domains to the promoter, it is likely that this difference in activity is caused by reduced delivery of RS4 relative to RLS. Consistent with this interpretation, RS4, like the GS4 protein, is expressed at very low levels in the transfected cells (Fig. 3B). Its low concentration would lead to reduced complex formation in the presence of rapamycin. Further support for this idea comes from the observation that rapamycin-induced RS4 activity is indeed detectable in conjunction with a GAL4 DNA-binding protein containing three tandemly reiterated FKBP domains (Fig. 3B), suggesting that the low concentration of RS4 can be partially compensated for by increasing the concentration of its receptor.
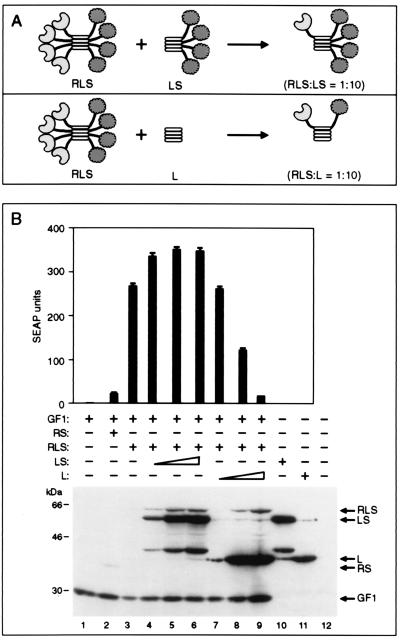
An alternative explanation for the increased activity of the bundled activator RLS in the presence of rapamycin is that bundling of the FRB domains creates an avidity effect for the rapamycin-mediated interaction of the fusion proteins. That is, RLS bundles function as highly potent inducers of transcription not because of their ability to deliver more activation domains to the promoter but because of the cooperative interaction between multiple FRB domains with the FKBP moieties linked to the DNA-binding domain. To rule out this hypothesis, we performed the experiment diagrammed at the top of Fig. 4A. We expressed a limiting quantity of RLS in the presence of increasing quantities of a truncated protein (LS) that lacked the FRB domain. We expected the LS protein to dilute FRB out of the bundled complexes, thereby reducing any potential avidity effect. Fig. 4B shows that, even in the presence of a large excess of LS over RLS, the level of reporter gene expression in the presence of 10 nM rapamycin was not reduced. This observation suggests that avidity is not responsible for the increased potency of bundled activation domains. Indeed, at the highest concentrations of LS, it is likely that most bundles contained no more than one FRB domain, suggesting that recruitment of four activation domains through a single FRB is sufficient to achieve efficient promoter activation.
Figure 4.
The transcriptional activity of RLS bundles depends on the presence of multiple activation but not with the FRB domains in the bundle. (A) Diagrammatic description of the composition of the activation domain-containing protein bundle when RLS and increasing concentrations of LS or L region are coexpressed in the cell. (B) (Upper) GF1-encoding plasmid (20 ng) was cotransfected with 100 ng of RLS alone or with indicated concentrations of LS or L regions. The cells were stimulated with 10 nM rapamycin, and the SEAP activity in the medium was measured 18 h after transfection. Mean values of SEAP activity secreted into the medium after the addition of rapamycin are shown (±SD). (Lower) Western blot analysis of lysates probed with 12CA5 antibody.
To test the hypothesis that it is indeed the number of activation domains per recruitment event that is critical to achieving efficient promoter activation, we performed the experiment diagrammed at the lower part of Fig. 4A. Here we expressed just the tetramerization domain (L), thus diluting out both FRB and the p65 activation domain from the bundles. In this case, there was a concentration-dependent reduction in reporter gene expression. At the highest concentration of L tested, in which L is at very large excess to RLS such that most bundles would contain at most a single activation domain, reporter gene expression was equivalent to that achieved with the conventional RS fusion protein. Thus, it appears that the enhanced potency of bundled activation domains derives from their ability to deliver multiple activation domains to the promoter with each molecular delivery event.
Bundling Increases Transcription at Very Low Activator Expression Levels.
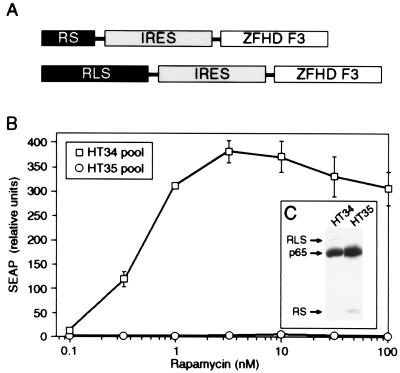
The increased potency of bundled activators has several potential practical applications. In scenarios in which activator expression is too low to support target gene activation, such as may often be encountered in gene therapy or in cell lines recalcitrant to transfection, bundled activators may work. To recreate such a scenario, we generated stable reporter cell lines in which expression of the chimeric transcription factors was deliberately limited by placing the activator expression under the control of the relatively weak Rous sarcoma virus promoter instead of the strong cytomegalovirus promoter (Fig. 5A). One pool of stable cell lines (HT34) expressed the bundled activator RLS, whereas the other pool (HT35) expressed the conventional activator RS. The two pools differed dramatically in their responsiveness to rapamycin; HT34 responded robustly, whereas HT35 did not respond at all (Fig. 5B). In contrast, the expression levels of RLS and RS were similar in the two pools (Fig. 5C). Similar results were obtained with isolated clones from the two pools (data not shown). Thus, at equal levels of protein expression, the bundled activator can successfully activate a target gene under conditions in which a conventional activator cannot.
Figure 5.
The threshold amount of activators required to stimulate gene expression can be reduced by improving the efficiency of delivering activation domains to the promoter. (A) Diagram showing the transcriptional activator coding sequences stably integrated into HT1080 cells. The activation domain and DNA-binding domain fusion proteins were deliberately expressed at lower levels by placing their corresponding coding sequences under the control of the relatively weak Rous sarcoma virus promoter. The internal ribosome entry sequence (IRES) is derived from encephalomyocarditis virus. The composite DNA-binding domain fusion protein ZFHD F3 has been described previously (5). (B) Pools of HT34 and HT35 cells were treated with indicated concentrations of rapamycin in the medium for 24 h and the SEAP activity secreted into the medium during this period was measured. In all cases, mean values of SEAP activity secreted into the medium 24 h after the addition of rapamycin are shown (±SD). (C) Western blot analysis of the lysates from pools of HT34 and HT35 cells probed with anti-p65 antibody. RLS and RS proteins are expressed at very low levels and appear as very faint bands.
Bundling Enhances the Sensitivity of Two-Hybrid Assays.
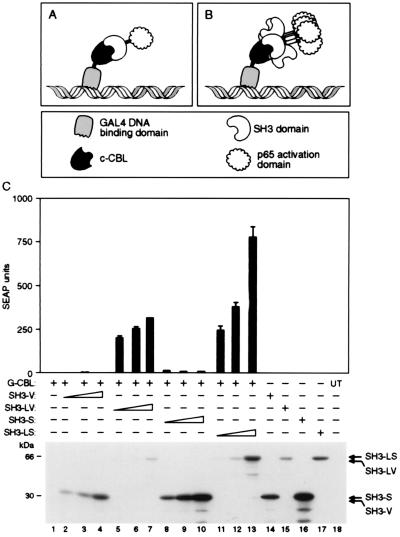
A related scenario may occur when two-hybrid assays are used to detect protein–protein interactions (37–39). The output of this assay depends on the interaction of two fusion proteins, one containing a DNA-binding domain and the other an activation domain, resulting in the activation of a reporter gene. Because a positive signal in this assay requires the assembly of a two-component transcription factor complex, both fusion proteins must be expressed at sufficient levels relative to their affinity for one another for enough of these complexes to form. In many cases, however, these fusion proteins are poorly expressed, or their affinity is below the detection threshold. To test whether bundling of the activation domain partner might overcome such limitations, we examined the interaction of the c-Src SH3 domain and its partner c-Cbl (40, 41). Although the interaction of these proteins can be detected in a yeast-based two-hybrid assay, no interaction could be detected between GAL4-CBL and SH3-p65 in a mammalian system. Immunoblot analysis of transfected mammalian cells suggested that this failure was caused by poor expression of the GAL4-CBL fusion protein (data not shown). We asked whether this limitation could be overcome by bundling the SH3-p65 fusion protein (Fig. 6B). Indeed, as shown in Fig. 6C, the bundled protein SH3-LS led to an extremely strong signal from the reporter gene, whereas the signal obtained with the conventional SH3-S fusion was barely detectable. Similar results were obtained with SH3-VP16 fusion proteins. Thus, bundling of the activation domain partner in a two-hybrid assay permits the measurement of interactions that escape detection in a conventional system.
Figure 6.
Noncovalent bundling of activation domain fusion protein enhances the detection of protein–protein interactions in mammalian cells. (A) Diagrammatic representation of two-hybrid assays with bundled fusion protein containing the target and activation domains. GAL4 DNA-binding domain fused to c-Cbl (G-CBL in C) is shown interacting with its target protein SH3 fused to p65 activation domain (SH3-S or V in C) lactose repressor tetramerization domain p65 activation domain sequences (SH3-LS in C). (C) (Upper) HT1080B cells carrying SEAP reporter genes placed under the control of five GAL4-binding sites were transfected with 100 ng of indicated expression plasmids. Mean values of SEAP activity secreted into the medium 24 h after transfection are shown (±SD). (Lower) Western blot analysis of extracts prepared from transiently transfected cells probed with anti-hemagglutinin antibody.
Discussion
Our data show that activators with reiterated activation domains are expressed at very low levels in mammalian cells and perhaps for this reason fail to function as potent inducers of transcription in vivo. Therefore, by using conventional methods, it is not possible to improve the potency of transcriptional activators to achieve higher levels of activator-directed gene expression in engineered cells. The strategy of bundling appears to be effective at circumventing the problem of low expression levels because noncovalently bundled activators have the potency of covalently multimerized proteins but the apparent toxicity profile of simpler activators. Consequently, bundling permits the delivery of multiple activation domains to a target promoter with high efficiency.
The efficiency of delivery of an activator to a target promoter—whether mediated by a small-molecule dimerizer, a DNA-protein interaction, or a protein–protein interaction—is determined simply by the intrinsic affinity constant for the interaction and the concentration of the reactants. Covalently multimerized activators fail to activate target genes effectively because their low concentration keeps them from reaching their target. Low concentration of activators can also account for poor target gene expression after experimental gene transfer and, as we have shown, for the failure of two-hybrid assays. Bundled activators are therefore more effective than covalently reiterated activators presumably because their higher intracellular concentration permits them to achieve higher target occupancy.
Bundled activators are probably more effective than monomers because each delivery event brings multiple activation domains to the target promoter. Although this idea by itself is quite simple, it is surprising that the effect is so quantitatively dramatic. In almost all of the experiments shown here, bundled activators generated robust gene expression under conditions in which conventional activators were almost completely ineffective. The simplest explanation for why bundled activators are so much more effective is that there is a threshold requirement for the number of activators that must be delivered to a promoter before transcription can be initiated.
If, for example, two bound activation domains are required for transcriptional initiation, simple activators would require two independent delivery events, whereas bundles would require only one. If the frequency of delivery—as determined by the concentration of activator and its affinity for its receptor site—was one event per 10 promoters, then only 1 in 100 promoters would ever reach the threshold number if each delivery event brought a single activation domain to the promoter. In contrast, each delivery of a bundled activator would take the promoter over the threshold. Consequently, at equal protein concentrations, bundled activators would be ≥10-fold more effective. If the threshold value is greater than two activators, or the frequency of delivery lower, then the quantitative advantage of bundled activators could be enormous.
The bundling strategy has practical applications. We demonstrated here that activation domain bundling substantially improves the sensitivity of mammalian two-hybrid assays, allowing interactions between poorly expressed proteins to be detected. Bundled activation domains will likely also allow the detection of very weak interactions and potentially of transient interactions that do not lead to a signal in conventional assays.
Chimeric activators also form the basis of systems that place gene transcription under small-molecule control (6), for regulating the expression of transgenes in animal models and in gene therapy applications. We showed here that bundling allows the rapamycin-regulated system to function when the regulatory proteins are expressed at very low levels—a likely situation in many gene therapy procedures. Interestingly, we also observed that bundling shifts the dose-response of rapamycin activation approximately 10-fold, with maximal activation at 1 nM rapamycin (Fig. 5B) compared with the usual 10 nM (5). Thus, incorporating bundling domains may improve the practicality of regulated gene therapies by substantially decreasing the doses of drug required. For such clinical applications, we have developed an alternative tetramerization domain of human origin that should be minimally immunogenic (unpublished data).
Ultimately, chimeric activators might be used to activate otherwise untranscribed endogenous genes, by expressing them as fusions to natural or designed DNA-binding domains targeting the promoter of interest. The ability of bundled activators to drive high levels of activation without apparent cellular toxicity may be critical to the success of such approaches.
Acknowledgments
We thank members of the ARIAD our gene therapy department for their support. We thank Tim Clackson, Roy Pollock, Dorre Grueneberg, and Karen Zoller for their helpful comments on the manuscript.
Abbreviations
- SEAP
secreted alkaline phosphatase
- FRB
FKBP12-rapamycin binding
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Gossen M, Bujard H. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Y, DeMayo F J, Tsai S Y, O’Malley B W. Nat Biotechnol. 1997;15:239–243. doi: 10.1038/nbt0397-239. [DOI] [PubMed] [Google Scholar]
- 3.No D, Yao T-P, Evans R M. Proc Natl Acad Sci USA. 1996;93:3346–3351. doi: 10.1073/pnas.93.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belshaw P J, Ho S N, Crabtree G R, Schreiber S L. Proc Natl Acad Sci USA. 1996;93:4604–4607. doi: 10.1073/pnas.93.10.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rivera V M, Clackson T, Natesan S, Pollock R, Amara J F, Keenan T, Magari S R, Phillips T, Courage N L, Cerasoli F, Jr, et al. Nat Med. 1996;2:1028–1032. doi: 10.1038/nm0996-1028. [DOI] [PubMed] [Google Scholar]
- 6.Clackson T. Curr Opin Chem Biol. 1997;1:210–218. doi: 10.1016/s1367-5931(97)80012-9. [DOI] [PubMed] [Google Scholar]
- 7.Ptashne M, Gann A. Nature (London) 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 8.Koh S S, Ansari A Z, Ptashne M, Young R A. Mol Cell. 1998;1:895–904. doi: 10.1016/s1097-2765(00)80088-x. [DOI] [PubMed] [Google Scholar]
- 9.Keaveney M, Struhl K. Mol Cell. 1998;1:917–924. doi: 10.1016/s1097-2765(00)80091-x. [DOI] [PubMed] [Google Scholar]
- 10.Gaudreau L, Adam M, Ptashne M. Mol Cell. 1998;1:913–916. doi: 10.1016/s1097-2765(00)80090-8. [DOI] [PubMed] [Google Scholar]
- 11.Sadowski I, Ma J, Triezenberg S, Ptashne M. Nature (London) 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 12.Cress W D, Triezenberg S J. Science. 1990;251:87–90. doi: 10.1126/science.1846049. [DOI] [PubMed] [Google Scholar]
- 13.Moore P A, Ruben S M, Rosen C A. Mol Cell Biol. 1993;13:1666–1674. doi: 10.1128/mcb.13.3.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmitz M L, Baeuerle P A. EMBO J. 1991;10:3805–3817. doi: 10.1002/j.1460-2075.1991.tb04950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blau J, Xiao H, McCracken S, O’Hare P, Greenblat J, Bentley D. Mol Cell Biol. 1996;16:2044–2055. doi: 10.1128/mcb.16.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das G, Hinkley C S, Herr W. Nature (London) 1995;374:657–659. doi: 10.1038/374657a0. [DOI] [PubMed] [Google Scholar]
- 17.Emami K H, Navarre W W, Smale S T. Mol Cell Biol. 1995;15:5906–5916. doi: 10.1128/mcb.15.11.5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bunker C A, Kingston R E. Proc Natl Acad Sci USA. 1996;93:10820–10825. doi: 10.1073/pnas.93.20.10820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Natesan S, Rivera V M, Molinari E, Gilman M Z. Nature (London) 1997;390:349–350. doi: 10.1038/37019. [DOI] [PubMed] [Google Scholar]
- 20.Triezenberg S J. Curr Biol. 1995;5:190–196. doi: 10.1016/0959-437x(95)80007-7. [DOI] [PubMed] [Google Scholar]
- 21.Uesugi M, Nyanguile O, Lu H, Levine A J, Verdine G L. Science. 1997;277:1310–1313. doi: 10.1126/science.277.5330.1310. [DOI] [PubMed] [Google Scholar]
- 22.Blair W S, Bogerd H P, Madore S J, Cullen B R. Mol Cell Biol. 1994;14:7226–7234. doi: 10.1128/mcb.14.11.7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giniger E, Ptashne M. Nature (London) 1987;330:670. doi: 10.1038/330670a0. [DOI] [PubMed] [Google Scholar]
- 24.Chi T, Carey M. Mol Cell Biol. 1993;13:7045–7055. doi: 10.1128/mcb.13.11.7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmitz M L, dos Santos Silva M A, Altmann H, Czisch M, Holak T A, Baeuerle P A. J Biol Chem. 1994;269:25613–25620. [PubMed] [Google Scholar]
- 26.Tanaka M, Clouston W M, Herr W. Mol Cell Biol. 1994;14:6046–6055. doi: 10.1128/mcb.14.9.6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanaka M. Proc Natl Acad Sci USA. 1996;93:4311–4315. doi: 10.1073/pnas.93.9.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emami K H, Carey M. EMBO J. 1992;11:5005–5012. doi: 10.1002/j.1460-2075.1992.tb05607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohashi Y, Brickman J M, Furman E, Middleton B, Carey M. Mol Cell Biol. 1994;14:2731–2739. doi: 10.1128/mcb.14.4.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerber H-P, Seipel K, Georgiev O, Hofferer M, Hug M, Rusconi S, Schaffner W. Science. 1994;263:808–811. doi: 10.1126/science.8303297. [DOI] [PubMed] [Google Scholar]
- 31.Gill G, Ptashne M. Nature (London) 1988;344:721–724. doi: 10.1038/334721a0. [DOI] [PubMed] [Google Scholar]
- 32.Berger S L, Pina B, Silverman N, Marcus G A, Agapite J, Regier J L, Triezenberg S J, Guarente L. Cell. 1992;70:251–265. doi: 10.1016/0092-8674(92)90100-q. [DOI] [PubMed] [Google Scholar]
- 33.Baron U, Gossen M, Bujard H. Nucleic Acids Res. 1997;25:2723–2729. doi: 10.1093/nar/25.14.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Attar R M, Gilman M Z. Mol Cell Biol. 1992;12:2432–2443. doi: 10.1128/mcb.12.5.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sabatini D M, Erdjument-Bromage H, Lui M, Tempst P, Snyder S H. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 36.Friedman A M, Fischmann T O, Steitz T A. Science. 1995;268:1721–1727. doi: 10.1126/science.7792597. [DOI] [PubMed] [Google Scholar]
- 37.Fields S, Song O-K. Nature (London) 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 38.Fields S, Sternglanz R. Trends Genet. 1994;10:286–292. doi: 10.1016/0168-9525(90)90012-u. [DOI] [PubMed] [Google Scholar]
- 39.Allen J B, Walberg M W, Edwards M C, Elledge S J. Trends Biochem Sci. 1995;20:511–516. doi: 10.1016/s0968-0004(00)89119-7. [DOI] [PubMed] [Google Scholar]
- 40.Ribon V, Printen J A, Hoffman N G, Kay B K, Saltiel A R. Mol Cell Biol. 1998;18:872–879. doi: 10.1128/mcb.18.2.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robertson H, Langdon W Y, Thien C B, Bowtell D D. Biochem Biophys Res Commun. 1997;240:46–50. doi: 10.1006/bbrc.1997.7608. [DOI] [PubMed] [Google Scholar]