Abstract
Bacteria communicate with each other to coordinate expression of specific genes in a cell density-dependent fashion, a phenomenon called quorum sensing and response. Although we know that quorum sensing via acyl-homoserine lactone (HSL) signals controls expression of several virulence genes in the human pathogen Pseudomonas aeruginosa, the number and types of genes controlled by quorum sensing have not been studied systematically. We have constructed a library of random insertions in the chromosome of a P. aeruginosa acyl-HSL synthesis mutant by using a transposon containing a promoterless lacZ. This library was screened for acyl-HSL induction of lacZ. Thirty-nine quorum sensing-regulated genes were identified. The genes were organized into classes depending on the pattern of regulation. About half of the genes appear to be in seven operons, some seem organized in large patches on the genome. Many of the quorum sensing-regulated genes code for putative virulence factors or production of secondary metabolites. Many of the genes identified showed a high level of induction by acyl-HSL signaling.
Many host-associated bacteria use chemical signals to monitor their own species population density and control expression of specific genes in response to population density. This type of gene regulation is termed quorum sensing (1). Several Gram-negative bacteria use acylated homoserine lactone (HSL) signals in quorum sensing. Quorum sensing in Pseudomonas aeruginosa, an opportunistic human pathogen responsible for persistent and often incurable infections in immunocompromised people and individuals with cystic fibrosis, has been well studied. The sequence of the P. aeruginosa genome is available publicly. Expression of a number of extracellular virulence factors produced by P. aeruginosa is controlled by quorum sensing (for recent reviews see refs. 2 and 3).
Two quorum sensing systems, the las and rhl systems, have been identified in P. aeruginosa. In the las quorum sensing system, the lasI gene product directs the formation of the diffusible extracellular signal, N-(3-oxododecanoyl)-l-HSL (3OC12-HSL) (4), which interacts with LasR (5, 6) to activate a number of virulence genes, including lasB, lasA, apr, toxA, and lasI itself (6–10). Synthesis of the siderophore pyoverdine also is activated by the las system (11). P. aeruginosa strains lacking a functional LasR are avirulent in animal models (12). Although 3OC12-HSL is diffusible, it appears to partition into cell membranes, and P. aeruginosa efflux pumps aid in the movement of this signal to the external environment (13, 14).
The rhlI product catalyzes the synthesis of N-butyryl-l-HSL (C4-HSL) (15, 16). This diffusible signal (14) in conjunction with RhlR activates expression of the rhlAB rhamnolipid synthesis genes, rhlI, and to some extent lasB (17–20). Other virulence factors and secondary metabolites, including pyocyanin, cyanide, and chitinase, are positively regulated by the rhl system (18, 21), although direct transcriptional regulation of the genes involved in synthesis of these compounds has not been shown. A quorum sensing hierarchy exists with the las system controlling expression of the transcriptional activator RhlR (20, 22). Therefore genes controlled by the rhl system require a functional las system for full activation.
Recently, genes not directly involved in virulence, including the stationary phase sigma factor rpoS (20), and genes coding for components of the general secretory pathway (xcp) (23), have been reported to be controlled by quorum sensing. Furthermore, the las system is required for maturation of P. aeruginosa biofilms (24). Thus it seems that quorum sensing represents a global gene regulation system in P. aeruginosa. However, the current view of quorum sensing circuits in P. aeruginosa derives from an assortment of different types of studies, some with recombinant Escherichia coli (9, 20, 23), some with reporters or regulatory genes on multicopy plasmids in P. aeruginosa (10, 20, 23), and some involving nonquantitative Northern analysis techniques (8, 10). Furthermore, with few exceptions, lasB and rhlAB for example, levels of activation are low, 2- to 3-fold (10, 20, 23).
To begin a systematic investigation of global gene regulation by quorum sensing in P. aeruginosa we have generated random lacZ transcriptional fusions in the chromosome of a lasI-rhlI double mutant. Insertions in quorum sensing-regulated genes were identified by monitoring β-galactosidase expression in the presence and absence of 3OC12-HSL and C4-HSL, and the insertion mutants were characterized.
Materials and Methods
Bacterial Strains, Plasmids, and Media.
The P. aeruginosa strains were PAO1 (41), PDO100 a rhlI∷Tn501 derivative of PAO1 (42), MW1, and MW10, which are described below. The E. coli strains were DH5α (25), HB101 (25), SY327 λpir (43), and S17-1 (44). The plasmids used were pJPP4 (ori6K, mobRP4, ΔlasI, tetracycline and ampicillin resistance) (17), pTL61T (lacZ transcriptional fusion vector, ampicillin resistance) (45), pGMΩ1, (aacC1 flanked by transcriptional stops) (46), pTL61T-GmΩ1 (pTL61T with aacC1 gene from pGMΩ1 upstream of lacZ), pMW100 [pJPP4 with tetA(B) in place of tetA(C)], pRK2013 (ColE1, tra+, RK2, kanamycin resistance) (47), pSUP102 (48), pSUP102-lasB (pSUP102 with lasB on a 3.1-kb P. aeruginosa DNA fragment, chloramphenicol and, tetracycline resistance), pMW300 (pSUP102-lasB with lacZ-aacC1 from pTL61T-GmΩ1), and pTN5-B22 (28).
Bacteria were routinely grown in LB broth or LB agar (25) with antimicrobial agents when necessary. The antimicrobial agents were used at the following concentrations: HgCl2, 15 μg/ml in agar and 7.5 μg/ml in broth; 20 μg/ml nalidixic acid; 300 μg/ml carbenicillin; tetracycline, 50 μg/ml for P. aeruginosa and 20 μg/ml for E. coli; and gentamicin, 100 μg/ml for P. aeruginosa and 15 μg/ml for E. coli. Synthetic acyl-HSL concentrations were 2 μM for 3OC12-HSL and 5 μM for C4-HSL, and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) was used at 50 μg/ml.
DNA Manipulations and Plasmid Constructions.
DNA manipulations followed standard methods (26). Plasmid isolation was performed by using QIAprep spin miniprep kits (Qiagen, Chatsworth, CA), and DNA fragments were excised and purified from agarose gels by using GeneClean spin kits (Bio 101). DNA was sequenced at the University of Iowa DNA core facility by using standard automated sequencing technology.
To construct pMW100 we replaced the pBR322 tetA(C) gene-containing ClaI–NotI DNA fragment in pJPP4 with a tetA(B)-containing BstB1–NotI fragment from Tn10. It was necessary to use tetA(B) rather than tetA(C) to inactivate lasI because the tetA(C) gene from pBR322 was a hot spot for Tn5-B22 mutagenesis (ref. 27 and data not shown).
To construct pMW300 a 1.6-kb SmaI fragment from pGMΩ1 that contained the aacC1 gene (gentamicin acetyltransferase-3–1) was cloned into EagI-digested pTL61T, which had been polished with T4 polymerase. The resulting plasmid pTL61T-GMΩ1 was digested with SmaI and MscI to release a 6.5-kb lacZ-aacC1 fragment. A 3.1-kb P. aeruginosa chromosomal DNA fragment containing the lasB gene was amplified by PCR using the Expand Long Template PCR System (Boehringer Mannheim). This fragment was cloned into BamHI-digested pSUP102. The resulting plasmid, pSUP102-lasB, was digested with NotI, polished with T4 polymerase, and ligated with the 6.5-kb lacZ-aacC1 fragment from pTL61T-GMΩ1 to generate pMW300. The promoterless lacZ gene in pMW300 is 549 nt from the start of the lasB ORF, is flanked by 1.5 kb upstream and 1.6 kb downstream P. aeruginosa DNA, and contains the p15A ori, which does not function in P. aeruginosa.
Construction of P. aeruginosa Mutants.
A lasI, rhlI mutant of P. aeruginosa PAO-MW1 was made by insertional mutagenesis of lasI in the rhlI deletion mutant, PDO100. For mutagenesis, the lasI∷tetA(B) plasmid, pMW100 was mobilized from E. coli SY327 λpir into PDO100 by triparental mating with E. coli HB101 containing pRK2013. Because pMW100 has a λpir-dependent ori, it cannot replicate in P. aeruginosa. We selected a tetracycline-resistant, carbenicillin-sensitive exconjugant, which was shown by a Southern blot analysis (see below) to contain lasI∷tetA but not lasI or pMW100. To confirm inactivation of lasI in this strain, PAO-MW1, the amount of 3OC12-HSL in the fluid from a stationary phase culture was measured (4). As expected, we found no detectable 3-OC12-HSL (<5 nM).
A mutant, P. aeruginosa PAO-MW10, which contains a lacZ reporter in the chromosomal lasB gene, was constructed by introduction of pMW300 into PAO-MW1 by triparental mating as described above. Exconjugants resistant to gentamicin and sensitive to chloramphenicol were selected as potential recombinants. Southern blotting of chromosomal DNA with lasB and lacZ probes indicated that the pMW300 lasB-lacZ insertion had replaced the wild-type lasB gene.
Southern Blotting.
Approximately 2 μg of chromosomal DNA was digested with restriction endonucleases, separated on a 0.7% agarose gel, and transferred to a nylon membrane (26). DNA probes and probing were by standard techniques (Boehringer Mannheim).
Tn5 Mutagenesis.
We used Tn5-B22, which carries a promoterless lacZ gene (28), to mutagenize PAO-MW1. Equal volumes of a late logarithmic phase culture of E. coli S17–1 carrying pTn5-B22 grown at 37°C with shaking and a late logarithmic phase culture of PAO-MW1 grown at 42°C without shaking were mixed. The mixture was centrifuged at 6,000 × g for 10 min at room temperature, suspended in LB (5% of the original volume), and spread onto LB plates (100 μl per plate). After 16–24 h at 30°C, the cells on each plate were suspended in 500 μl of LB and 100-μl volumes were spread onto LB agar plates containing HgCl2, gentamicin, tetracycline, and nalidixic acid. After 48–72 h at 37°C, 20 colonies were selected from each mating and grown on LB selection agar plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). Ten of the 20 were picked for further study. The colonies picked showed a range in the intensity of the blue color on the X-Gal plates. A Southern blot using a probe to lacZ performed on 20 randomly chosen transconjugants indicated that the Tn5 insertion in each was in a unique location (data not shown).
The Screen for Quorum Sensing Controlled (qsc) Fusions.
We used a microtiter dish assay to identify mutants showing acyl-HSL-dependent β-galactosidase expression (qsc mutants). Each transconjugant was grown in four wells containing LB broth without added autoinducer, with added 3OC12-HSL, C4-HSL, or both 3OC12-HSL and C4-HSL for 12–16 h at 37°C. Inocula were 10 μl of an overnight culture, and final culture volumes were 70 μl. The β-galactosidase activity of cells in each microtiter dish well was measured with a luminescence assay (Tropix, Bedford, MA). Luminescence was measured with a Lucy I microtiter dish luminometer (Anthos, Salzburg, Austria). This semiautomated method for measuring β-galactosidase activity allowed us to perform the 28,000 assays required to complete our screen (see Results).
Patterns of Acyl-HSL Induction of β-Galactosidase Activity in qsc Mutants.
We analyzed the patterns of β-galactosidase expression in response to acyl-HSLs in the qsc mutants identified in our screen (see Results). Each mutant was grown in 1 ml of Mops (50 mM, pH 7.0) buffered LB broth containing one, the other, both, or neither acyl-HSL signal in an 18-mm culture tube at 37°C with shaking. A midlogarithmic phase culture was used as an inoculum, and initial ODs at 600 nm were 0.1. Growth was monitored as OD at 600 nm, and we measured β-galactosidase activity in 0.1-ml samples taken at 0, 2, 5, and 9 h after inoculation.
DNA Sequencing and Sequence Analysis.
To identify DNA sequences flanking Tn5-B22 insertions, arbitrary PCR (29) was performed with primers and conditions as described (30). Sequences that could not be identified by using arbitrary PCR were cloned. For cloning, chromosomal DNA was digested with EcoRI and ligated with EcoRI-digested, phosphatase-treated pBR322. E. coli DH5α was transformed by electroporation, and plasmids from gentamicin-resistant colonies were used for sequencing Tn5-flanking DNA.
DNA sequences flanking Tn5-B22 insertions were located on the P. aeruginosa PAO1 chromosome by searching the chromosomal database at the P. aeruginosa Genome Project web site (www.pseudomonas.com). The ORFs containing the insertions are those described at the web site. Functional coupling (ref. 31, http://wit.mcs.anl.gov/WIT2), sequence analysis, and expression patterns of the qsc mutants were used to identify potential operons (see Results).
Results
Identification of P. aeruginosa qsc Genes.
We screened 7,000 Tn5-B22 mutants of P. aeruginosa PAO-MW1. Tn5-B22 contains a promoterless lacZ, and P. aeruginosa PAO-MW1 is a lasI, rhlI mutant that does not make acyl-HSL signals. Thus transcription of the Tn5-B22 lacZ in a qsc gene should respond to an acyl-HSL signal. The screen involved growth of each mutant in a complex medium in a microtiter dish well with no added acyl-HSL, 3OC12-HSL, C4-HSL, or both 3OC12-HSL and C4-HSL. After 12–16 h, β-galactosidase activity was measured. Two hundred-seventy mutants showed >2-fold stimulation of β-galactosidase activity in response to either or both acyl-HSLs. Of these, 70 showed a >5-fold stimulation of β-galactosidase activity and were studied further. Each mutant was grown with shaking in culture tubes and 47 showed a reproducible >5-fold stimulation of β-galactosidase activity in response to either or both of the acyl-HSL signals. These were considered to have Tn5-B22 insertions in qsc genes. Southern blotting with a lacZ probe indicated each mutant contained a single Tn5-B22 insertion.
We do not believe this collection of 47 mutants represents the entire set of qsc genes. Our threshold of >5-fold induction may be too stringent. We have not performed a saturation mutagenesis, and there may be conditions other than those we used that reveal other genes not detected in our screen. Nevertheless, we have identified a set of 47 insertions in genes that show significant activation in response to acyl-HSL.
Responses of qsc Mutants to Acyl-HSL Signals.
For cultures of each of the 47 qsc mutants we measured β-galactosidase at different times after addition of signals. The basal levels of β-galactosidase varied depending on the mutant. The responses to the acyl-HSLs could be grouped into four general classes based on which of the two signals was required for activation of lacZ, and whether the response to the signal(s) occurred immediately or was delayed until stationary phase. A response was considered immediate if there was a 5-fold or greater response within 2 h of acyl-HSL addition (the ODs of the cultures ranged from 0.5 to 0.7 at 2 h). A response was considered delayed or late if there was <5-fold induction at 2 h but >5-fold induction of β-galactosidase at 5 h or later (ODs of 2 or greater). In some strains activation of lacZ required 3OC12-HSL, others required both 3OC12-HSL and C4-HSL for full activation of lacZ. A number of strains responded to either signal alone but showed a much greater response with both 3OC12-HSL and C4-HSL. None of the mutants responded well to C4-HSL alone (Table 1). This result was expected because expression of RhlR, which is required for a response to C4-HSL depends on 3OC12-HSL (22). Therefore at least some of the insertions exhibiting a response to both acyl-HSLs may be responding to the rhl system, which requires activation by the las system.
Table 1.
qsc genes in P. aeruginosa
| Classification | Identity* | Signal
response†
|
||
|---|---|---|---|---|
| 30C12-HSL | C4-HSL | Both | ||
| Class I | ||||
| qsc100 | Peptide synthetase (2,535,711) | 65 | 3 | 69 |
| qsc101 | No match (2,065,297) | 145 | 1 | 184 |
| qsc102 | No match (2,067,716) | 350 | 1 | 400 |
| qsc103 | No match (4,375,793) | 85 | 1 | 95 |
| qsc104 | Polyamine binding protein (2,935,208) | 7 | 2 | 8 |
| qsc105 | FAD-binding protein (2,927,668) | 40 | 1 | 42 |
| qsc106A&B | No match (5,467,402 for 106A) | 9 | 1 | 10 |
| qsc107 | No match (2,538,070) | 44 | 2 | 50 |
| Class II | ||||
| qsc108 | ORF 5 (2,720,329) | 13 | 1 | 7 |
| qsc109 | Bacitracin synthetase 3 (2,678,258) | 13 | 1 | 8 |
| qsc110A&B | Pyoverdine synthetase D (2,676,014 for 100A) | 10 | 1 | 7 |
| qsc111 | Pyoverdine synthetase D (2,671,429) | 11 | 1 | 7 |
| qsc112A&B | Aculeacin A acylase (2,636,707 for 112A) | 15 | 1 | 12 |
| qsc113 | Transmembrane protein (4,566,558) | 5 | 1 | 5 |
| qsc114‡ | No match (3,128,663) | 9 | 1 | 7 |
| qsc115§ | No match (131,753) | 4 | 1 | 5 |
| qsc116 | No match (934,322) | 5 | 1 | 5 |
| Class III | ||||
| qsc117§ | ACP-like protein (2,031,833) | 22 | 22 | 186 |
| qsc118 | RhII (3,889,744) | 38 | 14 | 70 |
| qsc119 | RhlAB (3,890,793) | 9 | 7 | 100 |
| qsc120 | Chloramphenicol resistance (3,745,609) | 3 | 7 | 24 |
| qsc121 | 3-Oxoacyl ACP synthase (3,742,723) | 13 | 27 | 105 |
| qsc122A&B | Cytochrome p450 (3,742,173 for 122A) | 2 | 10 | 90 |
| qsc123 | 9-Cis retinol dehydrogenase (3,740,171) | 14 | 28 | 96 |
| qsc124A&B | Pyoverdine synthetase D (3,739,430 for 124A) | 35 | 50 | 148 |
| qsc125 | Zeaxanthin synthesis (3,737,612) | 20 | 65 | 140 |
| qsc126 | Pristanimycin I synthase 3 & 4 (3,734,193) | 3 | 5 | 24 |
| qsc127‡ | No match (3,728,924) | 5 | 2 | 15 |
| qsc128 | Hydrogen cyanide synthase HcnB (2,412,909) | 19 | 12 | 42 |
| qsc129A&B | Cellulose binding protein p40 (931,539) | 15 | 1 | 100 |
| qsc130 | glc operon transcription activator (6,023,975) | 5 | 1 | 14 |
| qsc131 | PhzC (4,715,256) | 50 | 168 | 742 |
| Class IV | ||||
| qsc132A&B | Unknown (B. pertusis) (4,721,118 for 132A) | 1 | 1 | 40 |
| qsc133A&B | AcrB (4,709,375 for 133A) | 1 | 1 | 9 |
| qsc134 | Saframycin Mx1 synthetase A (4,556,461) | 6 | 1 | 28 |
| qsc135 | Cytochrome C precursor (3,395,532) | 3 | 1 | 6 |
| qsc136‡ | No match (1,221,771) | 7 | 3 | 10 |
| qsc137 | Asparagine synthetase (66,507) | 1 | 1 | 10 |
| qsc138 | No match (58,785,410) | 3 | 5 | 32 |
The bold letters indicate matches were to known P. aeruginosa genes. The numbers in parenthesis are the chromosomal locations of the insertions (OriC is defined as base number 1).
† The signal response is given as β-galactosidase activity in cells grown in the presence of the indicated signal(s) divided by the β-galactosidase activity of cells grown in the absence of added signals. Maximum responses are indicated.
‡ The lacZ insertions in these strains are in the opposite orientation of the ORFs described in the P. aeruginoas Genome Project web site. We have indicated that the insertions are in locations with no reported identity.
§ Insertions do not lie in but are near the putative ORFs indicated. In qsc117 the insertion is 129 bp downstream of the ACP ORF and interrupts a potential rho-independent transcription terminator. The qsc115 insertion is 60 bp upstream of the ORF.
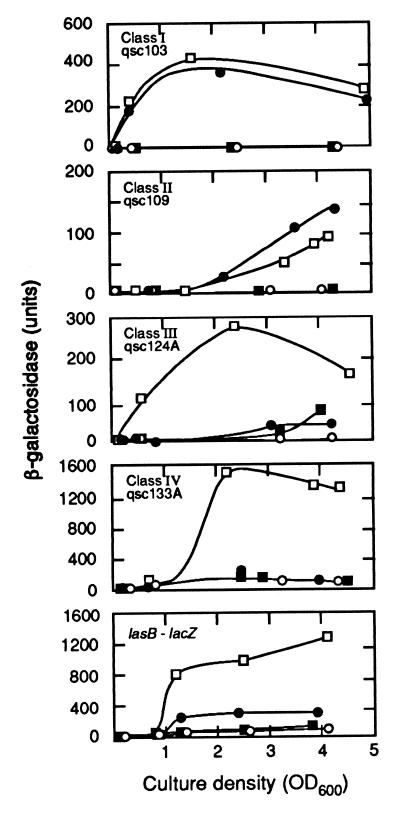
Class I mutants responded to 3OC12-HSL immediately, class II responded to 3OC12-HSL late, class III responded best to both signals early, and class IV responded to both signals late. Fig. 1 shows responses of representative members of each class to acyl-HSLs. Most early genes (class I and III genes) showed a much greater induction than most late genes (class II and IV). Many of the class III mutants showed some response to either signal alone but showed a greater response in the presence of both signals (Table 1 and Fig. 1).
Figure 1.
Patterns of β-galactosidase expression in representative P. aeruginosa MW1 qsc mutants and in a strain with a lasB∷lacZ chromosomal fusion generated by site-specific mutation. Units of β-galactosidase are given as a function of culture density for cells grown without added signal molecules (○), with added 3OC12-HSL (●), with added C4-HSL (■), or with both signals added (□). The background levels of β-galactosidase without added signals were 4–8 units for qsc103, 6–11 units for qsc109, 1–2 units for qsc124, 190 units for qsc133A, and 90 units for lasB∷lacZ. Stationary phase begins at a culture density of about 1.5.
Identity and Analysis of qsc Genes.
The Tn5-B22-marked qsc genes were identified by sequencing flanking DNA. The sequences were located in the P. aeruginosa PAO1 chromosome by searching the P. aeruginosa Genome Project web site. To confirm the locations of the Tn5-B22 insertions in each qsc mutant we performed a Southern blot analysis with Tn5-B22 as a probe. The sizes of Tn5-B22 restriction fragments agreed with those predicted based on the P. aeruginosa genomic DNA sequence (data not shown). The 47 qsc mutations mapped in or adjacent to 39 different ORFs.
We identified only two genes already known to be controlled by quorum sensing, rhlI and rhlB. Several other genes potentially involved in processes known to be regulated by quorum sensing also were identified, including phzC (phenazine synthesis), a putative cyanide synthesis gene (related to the Pseudomonas fluorescens hcnB), and ORF 5 (pyoverdine synthesis) (21, 32). Of note we did not identify lasB in our search, yet the LasI-LasR quorum sensing system originally was described as regulating lasB (5). We constructed a lasB-lacZ chromosomal fusion in PAO-MW1 so that we could compare regulation of lasB by quorum sensing to the other genes we identified. The lasB-lacZ fusion responded only slightly to 3OC12-HSL (3-fold stimulation). The full response (12- to 13-fold over background) required both C4-HSL and 3OC12-HSL and was late (Fig. 1). Thus lasB shows the characteristics of a class IV gene.
Some of the qsc mutants had obvious phenotypes. Unlike the parent, on LB agar, colonies of the class II mutants qsc108, qsc109, qsc110A, qsc110B, and qsc111 were not fluorescent. Because pyoverdine is a fluorescent pigment, and because the qsc110 and qsc111 mutations were in genes coding for pyoverdine synthetase-like proteins, we suspect that these mutations define a region involved in pyoverdine synthesis. The insertion in qsc131 is in phzC, which is required for pyocyanin synthesis. Although the parent strain produced a blue pigment in LB broth, qsc131 did not. The two qsc132 mutants also did not produce detectable levels of pyocyanin but did produce a water-soluble red pigment.
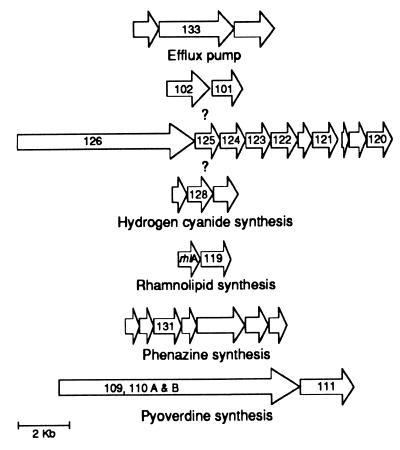
We used functional coupling (31) and sequence analysis to identify seven putative qsc operons, one of which is the previously described rhlAB operon (Fig. 2). The functional coupling algorithm identifies gene clusters as possible operons when genes are clustered in other bacteria and code for functionally related polypeptides. Functional coupling will not organize genes encoding polypeptides without known relatives into operons, and we disallowed organization of genes in an operon when there was >250 bp between two adjacent ORFs. The qsc101 and qsc102 genes are an example of a putative operon that was not identified by functional coupling (Fig. 2). These two ORFs do not show significant similarities with other polypeptides. Nevertheless, they are transcribed in the same direction, closely juxtaposed, both class I genes, and there is a las- box-like element upstream of these ORFs. The las box is a palindromic sequence found upstream of and involved in LasR-dependent activation of lasB (33).
Figure 2.
Diagrams of seven putative qsc operons. ORFs are indicated by the arrows. ORFs discovered in the qsc screen are indicated by their qsc numbers.
The qsc133A and qsc133B insertions are in a putative three-gene operon with similarity to acrAB-tolC from E. coli and the mex-opr family of efflux pump operons in P. aeruginosa (34–36). The qsc133 mutations are within a gene encoding a MexF homolog. The qsc133 mutants show class IV regulation (Table 1 and Fig. 1). We could not identify any las-box-like sequences upstream of this suspected efflux pump operon.
A third possible operon identified by functional coupling is about 8 kb and contains 10 genes. We have eight strains with insertions in six of the 10 genes, all of which are class III mutants (Table 1). A las box-like sequence was identified upstream of the first gene of this operon. The function of these 10 genes is unknown but the similarities shown in Table 1 suggest that they may encode functions for synthesis and resistance to an antibiotic-like compound.
The qsc128 mutation is within a gene coding for a polypeptide that shows similarity to the P. fluorescens hcnB product and appears to be in a three-gene operon (Table 1, Fig. 2). By analogy to the P. fluorescens hcn operon, this operon is likely required for the production of hydrogen cyanide. Previous investigations have shown that hydrogen cyanide production is reduced in P. aeruginosa rhl quorum sensing mutants. Consistent with this finding, qsc128 is a class III mutant (Table 1). A las-box-like sequence was identified in the region upstream of the translational start codon of the first gene in this operon.
The phz operon, required for phenazine biosynthesis, has been described in other pseudomonads (37, 38) and the insertion in strain qsc131 is located in a gene encoding a phzC homolog. Analysis of the sequence around this phzC homolog revealed an entire phenazine biosynthesis operon (phzA-G). As discussed above, qsc131 does not produce the blue phenazine pigment pyocyanin. The phz operon in P. aeruginosa also contains a las-box-like sequence upstream of the first gene of the operon.
The final putative operon consists of two or three genes, qsc109–111, which appear to be involved in pyoverdine synthesis (see above). These ORFs were not identified in the P. aeruginosa Genome Project web site but were identified and shown to be functionally coupled with the Argonne National Laboratory web site.
For three of the insertions, lacZ was in an orientation opposite to the ORF described in the Genome Project web site (qsc114, qsc127, and qsc136). Further studies will be required to determine whether the annotation of the genome sequence needs to be amended or if there is something more interesting about these insertions.
Locations of qsc Genes on the P. aeruginosa Chromosome.
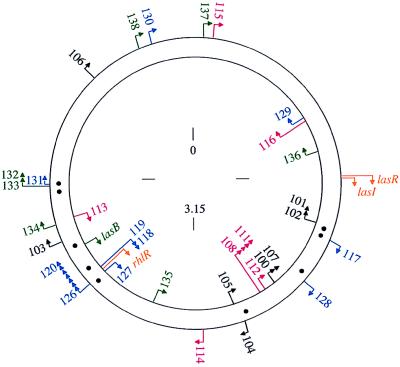
The qsc genes were mapped to sites on the P. aeruginosa chromosome (Fig. 3). In additionlasB, lasR, lasI, and rhlR were placed on this map. The distribution of the genes we have identified is patchy. For example, a 400-kb region flanked by qsc101 and qsc105 contained nine of the 17 class I and II 3OC12-HSL-dependent genes. Another cluster of 15 genes representing all of the classes and including lasB and rhlR was localized to a region of about 800 kb.
Figure 3.
Map of the qsc insertions on the P. aeruginosa chromosome. Arrowheads indicate the direction of lacZ transcription. Class I insertions are black, class II red, class III blue, and class IV green. In addition to the qsc mutants we identified, we have mapped lasR, lasI, and rhlR (gold) and lasB (green). The locations of las-box-like elements are shown as black dots between the two DNA strands. The numbers indicate distance in Mb on the approximately 6.3-Mb chromosome. Numbering starts at oriC.
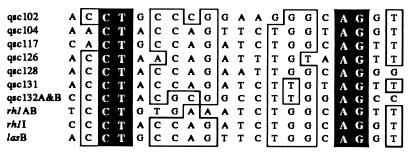
Identification of las-Box-Like Sequences that Could Be Involved in qsc Gene Control.
As discussed above the las box is found upstream of and involved in LasR-dependent activation of lasB (33). The las box shows similarity to the lux box, which is the promoter element required for quorum control of the Vibrio fischeri luminescence genes (39). Elements similar to a las box were identified upstream of qsc ORFs. Our search was for sequences with at least 50% identity to the las box centered 42 bp upstream of the lasB transcription start site (33). In all we identified las-box-like sequences that we suspect are involved in the regulation of 14 of the 39 qsc genes listed in Table 1 (Fig. 4). Because there is little information on the transcription starts of most of the genes identified in our screen, we may have missed relevant las boxes and some of the sequences we have identified may not be in relevant positions.
Figure 4.
las-Box-like sequences in upstream DNA regions of qsc mutants. Bases highlighted in black are conserved in all sequences and those in boxes are identical in eight of the 10 sequences.
Discussion
By screening a library of lacZ promoter probes in P. aeruginosa we have identified 39 qsc genes. Most of these genes were not identified as qsc. We did not find all of the genes that have been described as qsc. We did not find mutations in every gene in putative qsc operons (Fig. 2). We did not study mutants that showed only a small degree of acyl-HSL-dependent lacZ induction in the screen. Thus we presume that we have not identified all qsc genes. A conservative estimate is that about 1% of the genes in P. aeruginosa are controlled by quorum sensing (we confirmed 39 of about 5,000–6,000 genes in the P. aeruginosa chromosome without saturating the mutagenesis). A more liberal estimation of 3–4% can be drawn from our finding of 270 mutants showing at least a 2-fold induction in response to one or both of the acyl-HSL signals in the initial screen of 7,000 mutants. It seems clear that the percent of qsc genes in P. aeruginosa is somewhere between or around these values.
Several mutants, for example qsc101 and qsc102, showed an immediate and relatively large response to 3OC12-HSL (class I mutants, Table 1). Several mutants showed a relatively large and immediate response when both signals were supplied in the growth medium. Examples are qsc119 (pblAB), qsc121–125, qsc129A, and qsc129B. The qsc mutant showing the largest response was qsc131 (Table 1). Many of the mutants that responded best to both signals early (class III mutants) showed a small response when exposed to one or the other signal. The reasons for the small response to either signal are unclear. These genes may be subject to signal cross talk, or they may show a response to either LasR or RhlR. One reason they may respond to both signals better than they respond to C4-HSL alone is that 3OC12-HSL and LasR are required to activate RhlR, the transcription factor required for a response to C4-HSL (20, 22).
There were two mutant classes that showed a delayed response to the signals: class II mutants, which required only 3OC12-HSL, and class IV mutants, which required both signals for full induction. There are a number of possible explanations for a delayed response. Some of these genes may be stationary phase genes. Some may be iron repressed. For example, we know that the synthesis of pyoverdine is regulated by iron (32, 40) and the qsc109–111 mutations are in genes involved in pyoverdine synthesis. It is also possible that some of these genes are not regulated by quorum sensing, directly. The acyl-HSLs might control other factors that influence expression of any of the genes we have identified and this possibility seems most likely with the late genes. However, we do not believe that indirect regulation will be the rule for late genes, because the lasB-lacZ chromosomal insertion generated by site-specific mutation was late, and we know from other investigations that lasB responds to LasR and 3OC12-HSL, directly (6, 33). It would not be surprising to find that the late qsc genes consist of several subclasses.
At least some qsc genes appear to be organized in patches or islands on the chromosome (Fig. 3). The rhlI-rhlR quorum sensing modulon occurs on one of the qsc islands, but none of the qsc genes are tightly linked to the lasR-lasI modulon. Genes representing each of the four classes occur over the length of the chromosome and on both DNA strands. This finding is consistent with the view that quorum sensing is a global regulatory system in P. aeruginosa. Of interest there is a third LuxR family member in P. aeruginosa. This gene is adjacent to qsc131.
We know that quorum sensing is critical for virulence of P. aeruginosa and for the development of mature biofilms. We would like to know what qsc genes are responsible for these phenotypes. The mutants we have identified and the strategy for identification of qsc genes provide a manageable group of genes to test for function in virulence and biofilm development. Furthermore, the availability of the P. aeruginosa genome sequence likely will lead to the development of a gene expression array for this organism. The studies described here provide a set of genes that respond to specific treatments in a predictable way (Table 1) and will provide a means to validate P. aeruginosa microarray technology.
Although this investigation has revealed some interesting features of the global control of gene expression by quorum sensing in P. aeruginosa, it clearly raises more questions than it answers. How many more qsc genes are there? Are there genes showing repression by quorum sensing? Which qsc genes are required for biofilm formation or virulence? Why is there a delay in the response of some genes to quorum sensing signals? What defines the regulatory DNA involved in the qsc response? With this beginning of our understanding of quorum sensing in P. aeruginosa at the genomic level it is now feasible to address these questions.
Acknowledgments
We thank Q. Pham and M. Olson for providing genomic sequencing information before its posting on the Pseudomonas Genome Sequencing web site. We thank C. Manoil and R. Kolter for sharing insights about transposon mutagenesis of P. aeruginosa and R. Overbeek for help with functional coupling. This research was supported by grants from the National Science Foundation (MCB 9808308) and the National Institutes of Health (GM59026). M.W. is supported by a National Science Foundation Research Training Grant (DBI9602247).
Abbreviations
- HSL
homoserine lactone
- 3OC12-HSL
N-(3-oxododecanoyl)-l-HSL
- C4-HSL
N-butyryl-l-HSL
- qsc
quorum sensing controlled
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Fuqua W C, Winans S C, Greenberg E P. J Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fuqua C, Greenberg E P. Curr Opin Microbiol. 1998;1:183–189. doi: 10.1016/s1369-5274(98)80009-x. [DOI] [PubMed] [Google Scholar]
- 3.Pesci E C, Iglewski B H. Trends Microbiol. 1997;5:132–135. doi: 10.1016/S0966-842X(97)01008-1. [DOI] [PubMed] [Google Scholar]
- 4.Pearson J P, Gray K M, Passador L, Tucker K D, Eberhard A, Iglewski B H, Greenberg E P. Proc Natl Acad Sci USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gambello M J, Iglewski B H. J Bacteriol. 1991;173:3000–3009. doi: 10.1128/jb.173.9.3000-3009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Passador L, Cook J M, Gambello M J, Rust L, Iglewski B H. Science. 1993;260:1127–1130. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- 7.Toder D S, Ferrell S J, Nezezon J L, Rust L, Iglewski B H. Infect Immun. 1994;62:1320–1327. doi: 10.1128/iai.62.4.1320-1327.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toder D S, Gambello M J, Iglewski B H. Mol Microbiol. 1991;5:2003–2010. doi: 10.1111/j.1365-2958.1991.tb00822.x. [DOI] [PubMed] [Google Scholar]
- 9.Seed P C, Passador L, Iglewski B H. J Bacteriol. 1995;177:654–659. doi: 10.1128/jb.177.3.654-659.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gambello M J, Kaye S, Iglewski B H. Infect Immun. 1993;61:1180–1184. doi: 10.1128/iai.61.4.1180-1184.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stintzi A, Evans K, Meyer J, Poole K. FEMS Microbiol Lett. 1998;166:341–345. doi: 10.1111/j.1574-6968.1998.tb13910.x. [DOI] [PubMed] [Google Scholar]
- 12.Tang H B, DiMango E, Bryan R, Gambello M, Iglewski B H, Goldberg J B, Prince A. Infect Immun. 1996;64:37–43. doi: 10.1128/iai.64.1.37-43.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans K, Passador L, Srikumar R, Tsang E, Nezezon J, Poole K. J Bacteriol. 1998;180:5443–5447. doi: 10.1128/jb.180.20.5443-5447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pearson J P, Van Delden C, Iglewski B H. J Bacteriol. 1999;181:1203–1210. doi: 10.1128/jb.181.4.1203-1210.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pearson J P, Passador L, Iglewski B H, Greenberg E P. Proc Natl Acad Sci USA. 1995;92:1490–1494. doi: 10.1073/pnas.92.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winson M K, Camara M, Latifi A, Foglino M, Chhabra S R, Daykin M, Bally M, Chapon V, Salmond G P C, Bycroft B W, et al. Proc Natl Acad Sci USA. 1995;92:9427–9431. doi: 10.1073/pnas.92.20.9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pearson J P, Pesci E C, Iglewski B H. J Bacteriol. 1997;179:5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ochsner U A, Reiser J. Proc Natl Acad Sci USA. 1995;92:6424–6428. doi: 10.1073/pnas.92.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ochsner U A, Koch A K, Fiechter A, Reiser J. J Bacteriol. 1994;176:2044–2054. doi: 10.1128/jb.176.7.2044-2054.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Latifi A, Foglino M, Tanaka K, Williams P, Lazdunski A. Mol Microbiol. 1996;21:1137–1146. doi: 10.1046/j.1365-2958.1996.00063.x. [DOI] [PubMed] [Google Scholar]
- 21.Latifi A, Winson K M, Foglino M, Bycroft B W, Stewart G S A B, Lazdunski A, Williams P. Mol Microbiol. 1995;17:333–344. doi: 10.1111/j.1365-2958.1995.mmi_17020333.x. [DOI] [PubMed] [Google Scholar]
- 22.Pesci E C, Pearson J P, Seed P C, Iglewski B H. J Bacteriol. 1997;179:3127–3132. doi: 10.1128/jb.179.10.3127-3132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chapon-Herve V, Akrim M, Latifi A, Williams P, Lazdunski A, Bally M. Mol Microbiol. 1997;24:1169–1178. doi: 10.1046/j.1365-2958.1997.4271794.x. [DOI] [PubMed] [Google Scholar]
- 24.Davies D G, Parsek M R, Pearson J P, Iglewski B H, Costerton J W, Greenberg E P. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 26.Ausubel F, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Short Protocols in Molecular Biology. New York: Wiley; 1997. [Google Scholar]
- 27.Berg D E, Schmandt M A, Lowe J B. Genetics. 1983;105:813–828. doi: 10.1093/genetics/105.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simon R, Quandt J, Klipp W. Gene. 1989;80:161–169. doi: 10.1016/0378-1119(89)90262-x. [DOI] [PubMed] [Google Scholar]
- 29.Caetano-Annoles G. PCR Methods Appl. 1993;3:85–92. [Google Scholar]
- 30.O’Toole G A, Kolter R. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 31.Overbeek R, Fonstein M, D’Souza M, Pusch G D, Maltsev N. Proc Natl Acad Sci USA. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cunliffe H E, Merriman T R, Lamont I L. J Bacteriol. 1995;177:2744–2750. doi: 10.1128/jb.177.10.2744-2750.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rust L, Pesci E C, Iglewski B H. J Bacteriol. 1996;178:1134–1140. doi: 10.1128/jb.178.4.1134-1140.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kohler T, Michea-Hamzehpour M, Henze U, Gotoh N, Curty L K, Pechere J C. Mol Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 35.Poole K, Krebes K, McNally C, Neshat S. J Bacteriol. 1993;175:7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poole K, Gotoh N, Tsujimoto H, Zhao Q, Wada A, Yamasaki T, Neshat S, Yamagishi J-I, Li X-Z, Nishino T. Mol Microbiol. 1996;21:713–724. doi: 10.1046/j.1365-2958.1996.281397.x. [DOI] [PubMed] [Google Scholar]
- 37.Georgakopoulos D G, Hendson M, Panopoulos N J, Schroth M N. Appl Environ Microbiol. 1994;60:2931–2938. doi: 10.1128/aem.60.8.2931-2938.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mavrodi D V, Ksenzenko V N, Bonsall R F, Cook R J, Boronin A M, Thomashow L S. J Bacteriol. 1998;180:2541–2548. doi: 10.1128/jb.180.9.2541-2548.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Devine J H, Shadel G S, Baldwin T O. Proc Natl Acad Sci USA. 1989;86:5688–5692. doi: 10.1073/pnas.86.15.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rombel I T, McMorran B J, Lamont I L. Mol Gen Genet. 1995;246:519–528. doi: 10.1007/BF00290456. [DOI] [PubMed] [Google Scholar]
- 41.Holloway B W, Krishnapillai V, Morgan A F. Microbiol Rev. 1979;43:73–102. doi: 10.1128/mr.43.1.73-102.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brint J M, Ohman D E. J Bacteriol. 1995;177:7155–7163. doi: 10.1128/jb.177.24.7155-7163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller V L, Mekalonos J J. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simon R, Priefer U, Puhler A. Bio/Technology. 1983;1:37–45. [Google Scholar]
- 45.Linn T, St Pierre R. J Bacteriol. 1990;172:1077–1084. doi: 10.1128/jb.172.2.1077-1084.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schweizer H P. Biotechniques. 1993;15:831–833. [PubMed] [Google Scholar]
- 47.Figurski D H, Helinski D R. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simon R, O’Connell M, Labes M, Puhler A. Methods Enzymol. 1986;118:640–659. doi: 10.1016/0076-6879(86)18106-7. [DOI] [PubMed] [Google Scholar]