Abstract
The rate of spontaneous mutation is a key parameter in modeling the genetic structure and evolution of populations. The impact of the accumulated load of mutations and the consequences of increasing the mutation rate are important in assessing the genetic health of populations. Mutation frequencies are among the more directly measurable population parameters, although the information needed to convert them into mutation rates is often lacking. A previous analysis of mutation rates in RNA viruses (specifically in riboviruses rather than retroviruses) was constrained by the quality and quantity of available measurements and by the lack of a specific theoretical framework for converting mutation frequencies into mutation rates in this group of organisms. Here, we describe a simple relation between ribovirus mutation frequencies and mutation rates, apply it to the best (albeit far from satisfactory) available data, and observe a central value for the mutation rate per genome per replication of μg ≈ 0.76. (The rate per round of cell infection is twice this value or about 1.5.) This value is so large, and ribovirus genomes are so informationally dense, that even a modest increase extinguishes the population.
Rates of spontaneous mutation are critical to understanding the genetic structure of populations over time and thus to understanding the course of evolution. Mutation provides the prime variation on which selection, recombination, and genetic drift operate. Recombination and its enabling partner, sex, probably persist primarily because of the deleterious consequences of mutation (1). In a species capable of contemplating its own genetic health, the products of mutation are viewed as major threats to public health, and the consequences of increasing the mutation rate are viewed with alarm (2).
When rates of spontaneous mutation are expressed per genome per genome replication, different broad groups of organisms display characteristic values (3): roughly 0.2 for retroelements; close to 0.0034 for DNA-based microbes (including both viral and cellular organisms); and roughly 0.01 for higher eukaryotes. Riboviruses (RNA viruses exclusive of retroviruses) tend to display very high mutation rates (4); however, quantifying those rates has proved difficult, because the mutational targets have been too small to sample the genome reliably and because it was uncertain how to combine mutation frequencies and population history to calculate mutation rates (5).
Here, we describe a robust relationship among the mutation frequency f, the easily determined growth parameter c, and the mutation rate μ. Applying this formulation to the available data provides the best estimate to date of the rate of spontaneous mutation in riboviruses. In addition, the formulation also suggests ways to measure hitherto opaque parameters of viral replication.
Theory
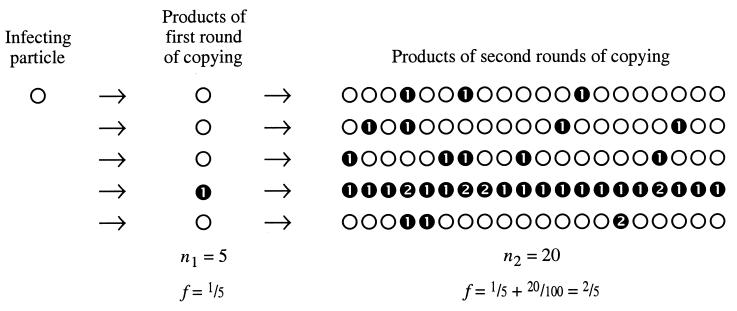
The replication of riboviruses whose chromosomes are composed of single-stranded RNA follows a simple scheme (Fig. 1). A cell is infected with one (or more) virus particles. Each infecting genome is copied iteratively such that complementary strands accumulate. Subsequently, the complementary strands are themselves copied iteratively, producing final strands of the same polarity as the infecting strand, and these final strands are packaged and released. We make the reasonable assumption that “final” strands rarely or never reenter the beginning of the cycle within a single infection. The key attribute of iterative replication is that the mutation frequency f equals the mutation rate μ per copying event: if n complementary strands are copied from a template and if μ is the mutation rate per copying event, then the number of mutations will be nμ, and f = nμ/n = μ.
Figure 1.
The accumulation of mutations during a single round of infection. The diagram shows the consequences of an arbitrary μg = 0.2 at both the first and second rounds, and the scheme is simplified by setting n2 = 20 for all second rounds of copying. The burst size is n1n2 = 100. ○, unmutated genomes; ➊, genomes with a single mutation; ➋, genomes with two mutations, which may arise in sequential replications (as in the fourth line) or during a single replication (as in the bottom line).
When a ribovirus infects a cell, the population of complementary strands accumulating from the first round of repeated copying has f1 = μ1. Ignoring rare precise back mutations and in the absence of selection, this mutant subpopulation will persist at the frequency f1. In the second round of repeated copying, a new fraction of mutations will be contributed, amounting to f2 = μ2. Thus, in one cycle of cell infection, f = f1 + f2 = μ1 + μ2. It is presently unknown whether μ1 and μ2 differ, and we must combine them into f = 2μ. This process of linear accumulation will continue as a virus stock grows through c cycles of cell infection; c will generally be a small (but not necessarily an integer) number, from one for a single round of infection (high multiplicity of infection), to perhaps four or five for a large population of host cells and a small inoculum. For stocks grown to a size sufficient to accumulate numerous mutations, the general formulation is f = 2cμ. Note that this expression holds only in the absence of selection, which is likely to be weak or absent during the first cycle of infection but may become strong in subsequent cycles. The mutational targets used in the studies we cite are believed to be largely free of selection. Given specific information about selection coefficients, it is possible to describe their effects on mutation accumulation over multiple cycles.
An alternative approach, used frequently with DNA-based microbes and occasionally with riboviruses, is the null-class method (6). In this method, numerous parallel cultures are each seeded with a small amount of virus and grown to a population size such that only roughly half of the cultures have accumulated any mutants. Each culture is then screened simply for the presence or absence of mutants, and the average number N of virus particles per culture is also determined. On the assumption that mutational events are randomly distributed among replication events and because the number of replication events per culture is close to N, the proportion of cultures with no mutants P(0) = e−Nμ.
Calculations
General Method.
By using either the accumulation method or the null-class method described above, a published mutation frequency f for a particular trait can be converted into a mutation rate μ. A mutation rate μb per base can be obtained by dividing μ by the mutational target size T (the number of bases at which the event can occur) and multiplying by a correction factor for mutations other than base substitutions. For mutations studied in riboviruses, T is usually very small; for instance, if the mutations consist exclusively of a single base substitution (such as G → A) at a single site, then T = 1/3. (Strictly speaking, T is a third of the total number of different base substitutions that can be monitored.) In these riboviral systems, only base-pair substitutions are scored. Thus, it is necessary to correct for all other kinds of mutations. Because riboviral mutational spectra are unavailable, we must instead fall back on the correction factor [(all mutations)/(base pair substitutions)] of 1.462 determined in several DNA-based microbial systems (5, 7). Multiplying by the genome size G then yields the mutation rate per genome per replication μg. When these calculations are strung together, the mutation accumulation μg = 1.462fG/2cT, and the null-class μg = −1.462[lnP(0)]G/NT.
Previous Values Accepted or Updated.
Several mutation frequencies and references to supplementary information were considered previously (5) and either are used unchanged or are recalculated with the new equation for mutation rate. (i) In two experiments measuring C → T mutations at poliovirus base 5,310 (8), f = 3.05 × 10−5 and 2.28 × 10−5; G = 7,433; c ≈ 2.8 and 2.8; T = 1/3; and μg = 0.177 and 0.132, respectively. (ii) Three measurements were made of mutation to guanidine resistance in poliovirus (9). The first was transformed by an arcane method that does not require updating and for which μg = 0.758 (5). In the second and third, f = 1.11 × 10−4 and 5.73 × 10−4; c ≈ 2.5 and 2.5; T = 4/3; and μg = 0.182 and 0.876, respectively. (iii) In two measurements with vesicular stomatitis virus (VSV; ref. 10), f = 1.75 × 10−4 and 2.35 × 10−4; G = 11,162; c ≈ 2 and 2.5; T = 2/3; and μg = 1.07 and 1.15, respectively.
New Values.
(i) Frequencies of revertants of drug-dependent mutants of human rhinovirus 16 to drug independence were measured somewhat incidentally (11). This virus has a genome of 7,124 bases (12). The mean revertant frequency (reciprocal of the mean relative plaquing efficiency) of the V1210A mutant was 1/6,457 = 1.55 × 10−4. Assuming that reversion was exclusively to the wild type, T = 1/3; c ≈ 3.6, including the growth of plaques into stocks. Thus, μg = (1.462 × 1.55 × 10−4 × 7,124)/(2 × 3.6 × 1/3) = 0.672. (ii) Mutation rates to resistance to monoclonal antibodies were screened in measles virus by using the null-class method (13). When the authors’ calculations are extended to take into account the effects of the inoculum size on the number of total replications and the content of preexisting mutants in the inocula, the mean mutation rate to drug independence is μ = 1.08 × 10−4. The number of sites at which mutation could occur was estimated from the observation that five sequenced mutations fell into four sites; assuming a Poisson distribution of mutations among sites, the most probable number of sites is 7.5. The size of the measles genome is 15,894 bases (14). Thus, μg = (1.462 × 1.08 × 10−4 × 15,894 × 3)/7.5 = 1.00.
Data Not Used.
In several instances, either recently appearing or previously analyzed data (5) are not well suited to the approach employed here. The latter include pioneering bacteriophage studies (15, 16), studies of VSV and poliovirus genomic RNA based on limit ribonuclease digestions rather than on genetic approaches (17, 18), and studies based on sequencing clonal copies of poliovirus and influenza virus genomes or involving sampling procedures that may have perturbed mutant frequencies (19, 20).
Mutation Rates Tabulated.
The independently measured μg values are arranged by increasing magnitude in Table 1. For a set of nine values varying by about 9-fold, the median is likely to be a better estimator than the mean. The 96% confidence interval around the median is 0.18–1.07 (21).
Table 1.
Genomic mutation rates in riboviruses
| Virus | μg |
|---|---|
| P | 0.13 |
| P | 0.18 |
| P | 0.18 |
| R | 0.67 |
| P | 0.76 |
| P | 0.88 |
| M | 1.00 |
| V | 1.07 |
| V | 1.15 |
| Median | 0.76 |
| Mean | 0.67 |
M, measles virus; P, poliovirus; R, rhinovirus; V, VSV.
Discussion
The mutation process in riboviruses can be described by a simple linear equation that reflects the repeated copying of templates that is characteristic of these organisms. The mutational consequences of iterative replication have not been explored much in the several decades since the primordial analysis of a “stamping machine” model (22). An exception is the single-stranded DNA bacteriophage φX174. This phage replicates in a complicated but fundamentally linear manner (23, 24), such that the mutation equations (25, 26) are similar to the equation used here for riboviruses, although the observed rates are far lower than those of riboviruses and are instead characteristic of DNA-based microbes (3). However, uncertainty persists concerning the extent to which second-generation strands reenter the beginning of the cycle within a single φX174 infection (24).
Applying mainly mutation-accumulation or null-class analyses to published data yields a set of nine rates of mutation per genome per replication that vary fairly smoothly over about an order of magnitude (Table 1). Just such variation is expected from the tiny mutational target sizes characteristic of the cited measurements: 1/3 base for seven entries, 2/3 base for two entries, and roughly 7.5 bases for the measles-virus entry. Most mutational spectra display a wide range of site-specific mutabilities, and even a specific substitution such as G:C → A:T in DNA can vary by over 2,000-fold depending on the neighboring sequences (27). The smooth spread of values in Table 1 and the similarity of mean and median lead us to propose the median (0.76) as the best current value for the rate of spontaneous mutation in riboviruses generally. A similar value, 1.2, was recently obtained by applying the fundamentally different Bateman–Mukai analysis to data obtained with VSV (28). These values are very high in comparison to values in other organisms but were anticipated in classical studies of mutation in an RNA phage (15, 16) and have long been known to be generally high (29).
Bacteriophage φ6 has a segmented, double-stranded RNA genome and thus differs profoundly from the viruses in Table 1. Nevertheless, the φ6 mutation rate must be much higher than that of DNA-based microbes, because stocks accumulate about 0.5% temperature-sensitive mutations (30). Nonsense mutations in φ6 display revertant frequencies ≥10−4 in stocks where c ≈ 5 (L. Chao, personal communication). For φ6, G = 13,379 (31). Assuming negligible selection and T = 1.5, μg ≥ 0.13, a value compatible with the values shown in Table 1. Thus, the high ribovirus mutation rate seems to encompass both animal viruses and phages.
The viral particles emerging from individual infected cells contain occasional mutant clones descended from new mutations. Mutation in riboviruses should produce mutant clone size distributions very different from those characteristic of exponentially replicating chromosomes. In riboviruses, fewer mutations will occur in the first round of genome copying than in the second round, because fewer copying events occur in the first round. Therefore, a few clones will contain several mutants, whereas most clones will contain only one mutant. This prediction has already been verified for the single-stranded DNA phage φX174, where roughly 5 large clones and about 309 single mutants (clones of size 1) were observed; the uncertainties reflect probable coincidences of clones of size 1 and a few preexisting mutations producing large clones (23). In riboviruses, the mutant clone size distribution can be described in terms of the mutation rates characteristic of the first and second rounds of copying (μ1 and μ2, respectively) and the mean numbers of genomes produced per template in the first and second rounds (n1 and n2, respectively). Specifically, the first round contributes μ1n1 mutant clones of mean size n2; the second round contributes μn1n2 mutant clones of size 1; and the total virus progeny per cell is n1n2. These are experimentally resolvable parameters.
Because a single cycle of infection produces a mutant frequency twice that of the rate per genome replication, few progeny viruses escape mutation. In general, for 1.5c mutations distributed randomly among genomes, the mutation-free fraction of a population would be e−1.5c (0.22 for c = 1; 0.05 for c = 2; etc.). However, many or most mutations in riboviruses are deleterious, and selection against deleterious mutations will reduce both mutant frequency and virus yield even within a single cycle of infection. Their high mutation rate renders the riboviruses particularly vulnerable to the consequences of rate increases, and both poliovirus and VSV populations are extinguished by chemical mutagenesis sufficient to increase the rate by a mere 2.5-fold (32). Merely tripling the viral mutation rate, if this increase could be achieved without harming the host, could cure ribovirus infections. Conversely, the normal mutation rate does not seem to be limiting for adaptation (29, 33).
Another consequence of this high rate of mutation is that all but the most mild mutator mutations will be lethal. Mutation pressure alone is likely to generate many mutator mutations, and an analysis of the VSV polymerase gene, encompassing almost 0.6 of the genome, suggested considerable polymerase compositional variation within a population (34). Influenza virus populations seem to accumulate a high frequency (≈12%) of mutators, but these increase mutation rates only by 2- to 4-fold (35); stronger mutators probably die out rapidly. The strength of a VSV mutator (36) cannot be quantitated.
Because recombination can regenerate unmutated genomes, it can reduce the danger of a very high mutation rate. Crossing over is frequent among many riboviruses, and independent assortment of segmented genomes also occurs (30). Perhaps most importantly, however, riboviruses have high fecundity, with yields of 100–10,000 per infected cell, such that the product of yield times the proportion of mutation-free progeny tends to exceed unity.
Acknowledgments
We thank Lin Chao, Jim Crow, Dave Denhardt, Esteban Domingo, Dmitry Gordenin, Mike Resnick, Roel Schaaper, and Charlie Steinberg for their critical comments on evolving stages of this article.
Abbreviation
- VSV
vesicular stomatitis virus
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Barton N H, Charlesworth B. Science. 1998;281:1986–1990. [PubMed] [Google Scholar]
- 2.Muller H J. Am J Hum Genet. 1950;2:111–176. [PMC free article] [PubMed] [Google Scholar]
- 3.Drake J W, Charlesworth B, Charlesworth D, Crow J F. Genetics. 1998;148:1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holland J, Spindler K, Horodyski F, Grabau E, Nichol S, VandePol S. Science. 1982;215:1577–1585. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- 5.Drake J W. Proc Natl Acad Sci USA. 1993;90:4171–4175. doi: 10.1073/pnas.90.9.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luria S E, Delbrück M. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drake J W. Proc Natl Acad Sci USA. 1991;88:7160–7164. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de la Torre J C, Giachetti C, Semler B L, Holland J J. Proc Natl Acad Sci USA. 1992;89:2531–2535. doi: 10.1073/pnas.89.7.2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de la Torre J C, Wimmer E, Holland J J. J Virol. 1990;64:664–671. doi: 10.1128/jvi.64.2.664-671.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holland J J, de la Torre J C, Steinhauer D A, Clarke D, Duarte E, Domingo E. J Virol. 1989;63:5030–5036. doi: 10.1128/jvi.63.12.5030-5036.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang W, Lee W-M, Mosser A G, Rueckert R R. J Virol. 1998;72:1210–1218. doi: 10.1128/jvi.72.2.1210-1218.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee W-M, Wang W, Rueckert R R. Virus Genes. 1995;9:177–181. doi: 10.1007/BF01702661. [DOI] [PubMed] [Google Scholar]
- 13.Schrag S J, Rota P A, Bellini W J. J Virol. 1999;73:51–54. doi: 10.1128/jvi.73.1.51-54.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffin D E, Bellini W J. In: Field’s Virology. 3rd Ed. Field B N, Knipe D M, Howley P M, editors. Philadelphia: Lippincott–Raven; 1996. pp. 1267–1312. [Google Scholar]
- 15.Domingo E, Flavell R A, Weissmann C. Gene. 1976;1:3–25. doi: 10.1016/0378-1119(76)90003-2. [DOI] [PubMed] [Google Scholar]
- 16.Batschelet E, Domingo E, Weissmann C. Gene. 1976;1:27–32. doi: 10.1016/0378-1119(76)90004-4. [DOI] [PubMed] [Google Scholar]
- 17.Steinhauer D A, de la Torre J C, Holland J J. J Virol. 1989;63:2063–2071. doi: 10.1128/jvi.63.5.2063-2071.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ward C D, Flanegan J B. J Virol. 1992;66:3784–3793. doi: 10.1128/jvi.66.6.3784-3793.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parvin J D, Moscona A, Pan W T, Leider J M, Palese P. J Virol. 1986;59:377–383. doi: 10.1128/jvi.59.2.377-383.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sedivy J M, Capone J P, RajBhandary U L, Sharp P A. Cell. 1987;50:379–389. doi: 10.1016/0092-8674(87)90492-2. [DOI] [PubMed] [Google Scholar]
- 21.Dixon W J, Massey F J., Jr . Introduction to Statistical Analysis. New York: McGraw–Hill; 1969. [Google Scholar]
- 22.Luria S E. Cold Spring Harbor Symp Quant Biol. 1951;16:463–470. doi: 10.1101/sqb.1951.016.01.033. [DOI] [PubMed] [Google Scholar]
- 23.Denhardt D T, Silver R B. Virology. 1966;30:10–19. doi: 10.1016/s0042-6822(66)81004-8. [DOI] [PubMed] [Google Scholar]
- 24.Denhardt D T. In: Encyclopedia of Virology. 2nd Ed. Granoff A, Webster R G, editors. London: Academic; 1999. pp. 274–281. [Google Scholar]
- 25.Loeb L A, Kunkel T A, Schaaper R M. ICN-UCLA Symp Mol Cell Biol. 1980;19:735–751. [Google Scholar]
- 26.Fersht A R, Knill-Jones J W. J Mol Biol. 1983;165:633–654. doi: 10.1016/s0022-2836(83)80271-x. [DOI] [PubMed] [Google Scholar]
- 27.Ronen A, Rahat A. Mutat Res. 1976;34:21–34. doi: 10.1016/0027-5107(76)90258-x. [DOI] [PubMed] [Google Scholar]
- 28.Elena S F, Moya A. J Evol Biol. 1999;12:1078–1088. [Google Scholar]
- 29.Domingo E, Holland J J. Annu Rev Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 30.Chao L, Tran T T, Tran T T. Genetics. 1997;147:953–959. doi: 10.1093/genetics/147.3.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gottlieb P, Metzger S, Romantschuk M, Carton J, Strassman J, Bamford D H, Kalkkinen N, Mindich L. Virology. 1988;163:183–190. doi: 10.1016/0042-6822(88)90245-0. [DOI] [PubMed] [Google Scholar]
- 32.Holland J J, Domingo E, de la Torre J C, Steinhauer D A. J Virol. 1990;64:3960–3962. doi: 10.1128/jvi.64.8.3960-3962.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee C H, Gilbertson D L, Novella I S, Huerta R, Domingo E, Holland J J. J Virol. 1997;71:3636–3640. doi: 10.1128/jvi.71.5.3636-3640.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schubert M, Harmison G, Meier E. J Virol. 1984;51:505–514. doi: 10.1128/jvi.51.2.505-514.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suárez P, Valcárcel J, Ortín J. J Virol. 1992;66:2491–2949. doi: 10.1128/jvi.66.4.2491-2494.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pringle C R, Devine V, Wilkie M, Preston C M, Dolan A, McGeoch D J. J Virol. 1981;39:377–389. doi: 10.1128/jvi.39.2.377-389.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]