Abstract
BRCA2 mutations predispose carriers mainly to breast cancer. The vast majority of BRCA2 mutations are predicted to result in a truncated protein product. The smallest known cancer-associated deletion removes from the C terminus only 224 of the 3,418 residues constituting BRCA2, suggesting that these terminal amino acids are crucial for BRCA2 function. A series of green fluorescent protein (GFP)-tagged BRCA2 deletion mutants revealed that nuclear localization depends on two nuclear localization signals that reside within the final 156 residues of BRCA2. Consistent with this observation, an endogenous truncated BRCA2 mutant (6174delT) was found to be cytoplasmic. Together, these studies provide a simple explanation for why the vast majority of BRCA2 mutants are nonfunctional: they do not translocate into the nucleus.
Germ-line mutations in the breast cancer susceptibility genes BRCA1 and BRCA2 predispose carriers mostly to breast cancer, but also to other cancers (reviewed in ref. 1). BRCA1 and BRCA2 account for 50% and 30%, respectively (2), of the inherited cases of breast cancer, which account for about 5–10% of all cases of breast cancer (3). These genes are classified as tumor suppressors because most BRCA1- and BRCA2-linked tumors have undergone loss of heterozygosity (LOH) at these loci. They have further been classified as “caretakers” on the basis of their proposed genome integrity maintenance functions as well as the infrequency of linked sporadic tumors (4).
Although the precise biochemical functions of the BRCA1 and BRCA2 gene products have yet to be determined, there is substantial evidence linking both of them to transcriptional control and DNA repair (reviewed in refs. 5 and 6), activities consistent with their nuclear localization (7–17). For BRCA2, we present evidence that its inability to be translocated to the nucleus may explain why the vast majority of BRCA2 mutations are nonfunctional. Since nearly all BRCA2 mutations are predicted to encode truncated proteins (18), we sought to determine the biochemical consequence of the C terminus. The smallest known cancer-associated deletion of BRCA2 is predicted to remove 224 amino acids (7% of the coding sequence) from its C terminus (19). These C-terminal amino acids appear to be critical because the nuclear localization signals (NLSs) of BRCA2 reside within this region. Using a series of green fluorescent protein (GFP)-tagged BRCA2 deletion mutants, we found that nuclear localization depends upon two NLSs that reside within the final 156 residues of BRCA2. Consistent with this observation, an endogenous cancer-associated truncated BRCA2 mutant (6174delT) was found to be cytoplasmic. Consequently, truncation mutant forms of BRCA2 are predicted to encode nucleus-excluded gene products, providing a simple explanation for why these mutants are nonfunctional: they do not translocate into the nucleus.
Materials and Methods
Cell Culture and Transfection.
293T and MCF-7 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum. Capan-1 cells were cultured in Iscove’s modified Dulbecco’s medium (IMDM) supplemented with 20% fetal calf serum. Transfection of 293T cells was performed by the calcium phosphate method adapted from ref. 20.
Plasmid Constructions.
Full-length human BRCA2 was modified to contain NotI and SalI restriction sites that permitted directional cloning into mammalian expression vector pRK5 (21), thereby creating pRK5-B2. To make the BRCA2 minigene expression vector MG-B2, a PCR amplicon containing the first exon and intron of BRCA2 was cloned into pRK5-B2. This amplicon and all other amplicons were sequenced to verify that no mutations had been introduced. To permit introduction of tags at the C terminus, this construct was engineered by PCR to include in-frame BglII and RsrII restriction sites just before the termination codon. To make MG-B2GFP, a PCR amplicon of GFP (pEGFP-N1 from CLONTECH was used as template) with flanking in-frame BglII and RsrII sites was introduced into MG-B2. MG-B2GFPΔ1529–3418 was made by deleting sequences between the NheI and BglII sites, using an in-frame bridging oligonucleotide. pRK5-B2GFPΔ1–1526 was made by replacing sequences from the 5′ end of the cDNA through the NheI site with an in-frame oligonucleotide that introduces a start ATG and sequences optimal for translation initiation (22). MG-B2GFPΔ2889–3418 was made by digesting MG-B2GFP with PmlI and BglII, treating with the Klenow fragment of DNA polymerase, and then religating. MG-B2GFPΔ1–2886 was made with a PCR amplicon extending from the PmlI site to codon 3418 of BRCA2. MG-B2GFPΔ3263–3418, MG-B2GFPΔ3270–3418, and MG-B2GFPΔ3270–3418/K3266T were made with PCR amplicons extending from the PmlI site to codons 3262, 3269, and 3269, respectively. For MG-B2GFPΔ3263–3418, an in-frame NotI site was introduced before the termination codon so that additional sequences could be added (see below). For MG-B2GFPΔ3270–3418/K3266T, the PCR amplicon included the K3266T mutation (codon 3266 5′-AAA-3′ was mutated to 5′-ACA-3′) which was introduced by the reverse PCR primer. These amplicons were used to replace the PmlI/BglII fragment of MG-B2GFP. MG-B2GFPΔ3263–3270 was made by adding a PCR amplicon containing codons 3270 through 3418 into MG-B2GFPΔ3263–3418. MG-B2GFPΔ3263–70/Δ3381–85 was made in two steps. First, a PCR amplicon from codons 3271 through 3380 was added to MG-B2GFPΔ3263–3418. This amplicon contained an in-frame ApaI restriction site before the termination codon. This site was used to introduce a PCR amplicon containing codons 3386 through 3418. MG-B2GFPΔ3263–3380 and MG-B2GFPΔ3263–3385 were made with PCR amplicons extending from codons 3381 or 3386 through codon 3418, respectively. MG-B2GFP-6174delT was constructed as follows. Using Capan-1 genomic DNA as template, we PCR amplified the sequences between the NheI site and the premature 2003ter codon such that the naturally occurring missense mutations that occur because of the frameshift mutation were maintained; also, a BglII site was introduced at the 3′ end. After NheI and BglII digestion, the PCR product was cloned into MG-B2GFPΔ1529–3418. Immunoblotting using anti-GFP Ab (polyclonal Ab from CLONTECH) was done to confirm expression of the GFP fusion products.
Subcellular Fractionation.
Cytoplasmic and nuclear fractions were prepared in buffers A and C, respectively (23), with the cytoplasmic fractions additionally clarified by centrifugation at 35,000 rpm for 30 min in an SW-60 swinging-bucket rotor (Beckman) (24). Whole-cell extracts were prepared and Western blot analyses were performed as reported (11) except that 3–8% gradient gels were used (Novex, San Diego, CA). For detection of BRCA2, anti-BRCA2A Ab was used (8). For detection of BRCA2–6174delT-GFP, anti-GFP Ab was used (mouse mAb from CLONTECH). The anti-p100 Ab was from Santa Cruz Biotechnology.
Microscopy.
All images were acquired with a Bio-Rad MRC1000 confocal microscope. 293T cells were cultured on poly(l-lysine)-coated slides, transfected with BRCA2-GFP fusions, then fixed, stained with 4′,6-diamidino-2-phenylindole (DAPI), and imaged 40 hr after transfection. For indirect immunofluorescence, cells were fixed, permeabilized, and stained essentially as described (8) except for the following modifications: 5% normal donkey serum was added to the permeabilization and primary antibody solutions, secondary antibodies were FITC-conjugated donkey anti-rabbit IgG (Jackson ImmunoResearch), and 5 μg/ml DAPI was used to stain nuclei. Primary antibodies were used at a concentration of 2 μg/ml. Rabbit IgG (Jackson ImmunoResearch) was used as a primary antibody negative control; these samples were negative (data not shown).
Results
BRCA2 NLSs Reside in the Extreme C Terminus.
Approximately 100 cancer-associated mutations have been reported for BRCA2, and about 95% of these are predicted to result in a truncated protein product (18). The most C-terminal of these cancer-associated mutations results in a termination signal at codon 3195 (Thr3195ter), thereby eliminating only 7% (224 residues) of BRCA2 (19). This observation suggests that the most C-terminal portion of BRCA2 is critical. Demarcating the C-terminal end of this critical region is the Lys3326ter polymorphism, which removes 92 residues from the C terminus, apparently without consequence (25). In the Breast Cancer Information Core database (18), a variant has been reported that is even further downstream (Glu3342ter). However, it was not identified as part of an affected breast cancer family and, because it occurs downstream of the Lys3326ter polymorphism, it is unlikely to be cancer associated (T. Frank, personal communication). Therefore, the most C-terminal cancer-associated truncation mutant, Thr3195ter, and the Lys3326ter polymorphism define a stretch of 131 residues that appears to be essential to BRCA2 (Fig. 1).
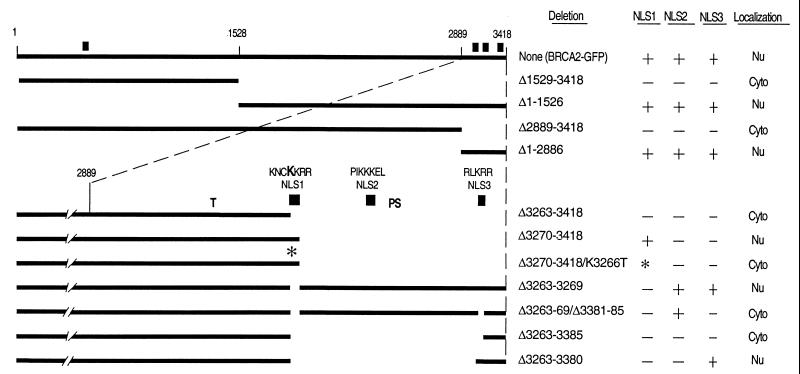
Figure 1.
Deletion analysis to identify the functional NLSs of BRCA2. GFP was fused to the C terminus of the depicted BRCA2 regions to facilitate analysis by fluorescence microscopy. Noted at the top are the positions of the first and last BRCA2 residues and the approximate location of the residues corresponding to the NheI (1528) and PmlI (2889) restriction sites used to construct many of these vectors (see Materials and Methods). Thick lines indicate the presence of BRCA2 regions, whereas no line indicates deletions. The composition and relative position of NLS1 (residues 3263–3269), NLS2 (residues 3311–3317), and NLS3 (residues 3381–3385) are given for the constructs depicted in the lower portion of the figure. Note that all C-terminal deletion constructs maintain the entire N-terminal region of BRCA2. ■ marks the approximate positions of the NLS candidates. ∗ indicates the presence of the K3266T mutation introduced into NLS1. The bold K in the NLS1 sequence indicates the residue mutated in this construct. Indicated in the columns to the right are the specific residues deleted, the status of the three C-terminal NLS candidates (+, present; −, absent; and *, mutated) and where the fusion products appear to be localized. “Nu” indicates that the green fluorescence appeared to be predominantly nuclear and “Cyto” indicates predominantly cytoplasmic. “T” marks codon 3195, where the most C-terminal cancer-associated truncation is predicted to terminate (19). “PS” marks the polymorphic stop that has been identified at codon 3326 (25).
Sequence analysis of BRCA2 revealed the presence of three possible NLSs in the C terminus and a fourth candidate in the N terminus (26) (Fig. 1). Each of these is an NLS like that of simian virus 40 (reviewed in ref. 27). To test the functionality of these NLSs, a series of GFP-tagged BRCA2 deletion mutants was transfected into 293T cells. The parent construct MG-B2GFP, containing full-length BRCA2 fused to the GFP gene, resulted in a nuclear BRCA2-GFP fusion product as judged by direct fluorescence (Figs. 1 and 2 a–c). Results from N-terminal and C-terminal expression constructs (Δ1529–3418 and Δ1–1526, respectively) indicated that the potential N-terminal NLS (amino acid residues 433–436, KRKK) is not functional and that the functional NLS(s) reside in the C terminus (Fig. 1). Results from expression constructs MG-B2GFPΔ2889–3418 and MG-B2GFPΔ1–2886 supported this conclusion and delimited the NLS-containing region to 530 amino acids (2889–3418) containing the critical residues defined by Thr3195ter and Lys3326ter (Fig. 1).
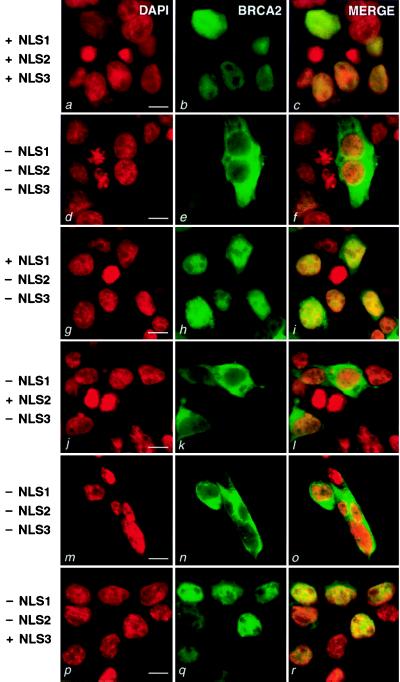
Figure 2.
Fluorescent confocal microscopy of cells expressing GFP-tagged full-length BRCA2 and deleted forms of BRCA2. 293T cells were transiently transfected with the following constructs: MG-B2GFP (a–c), MG-B2GFPΔ3263–3418 (d–f), MG-B2GFPΔ3270–3418 (g–i), MG-B2GFPΔ3263–70/Δ3381–85 (j–l), MG-B2GFPΔ3263–3385 (m–o) and MG-B2GFPΔ3263–3380 (p–r). (Left) DAPI was used to stain nuclei (red, a, d, g, j, m, p). (Center) Green fluorescence from the GFP fusions (green, b, e, h, k, n, q). (Right) The merged images are presented (c, f, i, l, o, r). To the left, the status of NLS1, NLS2, and NLS3 is indicated (+, present; −, absent). (Bar = 10 μm.)
We termed the clustered C-terminal NLS candidates NLS1 (amino acids 3263–3269, KNCKKRR), NLS2 (amino acids 3311–3317, PIKKKEL), and NLS3 (amino acids 3381–3385, RLKRR). NLS1 is well conserved among human, rat, and mouse BRCA2/Brca2 sequences, whereas NLS2 and NLS3 are not (28). Since NLS3 is positioned downstream of the Lys3326ter polymorphism, it was expected to be nonessential, but possibly functional. Testing of the individual NLS candidates revealed that NLS1 and NLS3 are functional, whereas NLS2 is not (Figs. 1 and 2). Deletion of the 156 amino acid region containing NLS1, NLS2, and NLS3 (Δ3263–3418 in Fig. 1) resulted in a nucleus-excluded GFP fusion product (Fig. 2 d–f). When NLS1 was included in a similar expression construct (Δ3270–3418 in Fig. 1), the GFP signal was observed to be nuclear (Fig. 2 g–i). Introducing a mutation (K3266T) in NLS1 similar to one known to block nuclear localization of simian virus 40 large T antigen (29) resulted in a nucleus-excluded GFP fusion product (Δ3270–3418/K3266T in Fig. 1). These data indicate that NLS1 is functional. However, deletion of only NLS1 (Δ3263–3269 in Fig. 1) still gave rise to a nuclear protein, revealing that NLS1 is not necessary in the context of an otherwise complete C terminus. Further deletion studies suggested that NLS2 does not function as an NLS, but that NLS3 does. Specifically, inclusion of NLS2 and deletion of NLS1 and NLS3 (Δ3263–69/Δ3381–85 in Fig. 1) resulted in a fusion product that was predominantly cytoplasmic (Fig. 2 j–l). Finally, an expression construct containing only NLS3 (Δ3263–3380 in Fig. 1) encoded a nuclear product (Fig. 2 p–r), whereas a similar construct without NLS3 (Δ3263–3385 in Fig. 1) led to a nucleus-excluded product (Fig. 2 m–o). In summary, within the final 156 amino acids of BRCA2 reside NLS1 and NLS3, the two functional NLSs of BRCA2. Each of these NLSs is downstream of all known cancer-associated truncating mutations, thereby giving rise to the hypothesis that these mutants cannot be functional because their products do not reach the nucleus.
Endogenous BRCA2 Is Nuclear.
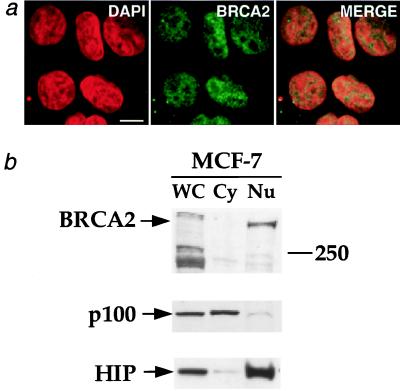
Using polyclonal antisera developed by Chen et al. (8), we confirmed that endogenous BRCA2 is nuclear in MCF-7 cells by indirect immunofluorescence (Fig. 3a). We further demonstrated that BRCA2 is nuclear by biochemical fractionation of MCF-7 cells (Fig. 3b).
Figure 3.
Endogenous BRCA2 is nuclear. (a) Indirect immunofluorescence of human breast cancer cells (MCF-7) expressing full-length BRCA2. Cells were prepared as described in Materials and Methods, probed with anti-BRCA2A Ab (8) (BRCA2), stained with DAPI, and then imaged by confocal microscopy. The merged images are presented on the right. (Bar = 10 μm.) (b) Biochemical fractionation of MCF-7 cells. MCF-7 cells were separated into cytoplasmic (Cy) and nuclear (Nu) fractions. WC, whole-cell extract. Equal numbers of cells were used to prepare the whole-cell and fractionated lysates. (Top) Anti-BRCA2A Ab was used to probe for BRCA2. The 250-kDa protein marker band is marked on the right. To validate the fractionation procedure, separate blots of protein from the subcellular fractions were probed with antibodies against the cytoplasmic NF-κB2 p100 precursor protein (38) (Middle) and the nuclear HIP protein (39) (Bottom). These proteins fractionated as expected.
Endogenous Cancer-Associated BRCA2 Truncation Mutant Is Cytoplasmic.
We sought to determine the subcellular localization of a cancer-associated endogenous truncated BRCA2 mutant. There is only one cell line, Capan-1, currently known to express a truncated form of BRCA2. Capan-1 cells express only the 6174delT mutant form of BRCA2 (8, 17, 30). This mutation eliminates ≈40% of the BRCA2 coding sequence, including NLS1 and NLS3. Many other cancer-associated truncation mutants have been identified that eliminate even less of the BRCA2 coding sequences (Table 1) (18).
Table 1.
C-terminal BRCA2 truncation mutations
| Nucleotide change* | Termination codon† | Ref(s).‡ |
|---|---|---|
| 6174delT | 2003 | 40–42 |
| 6299delA | 2039 | 40 |
| 6503delTT | 2098 | 42–45 |
| 6601delA | 2136 | 19 |
| 6630delTAACT | 2137 | 44 |
| 6635delTAAAT | 2137 | 19 |
| 6690delTC | 2174 | 44 |
| 6696delTC§ | 2174 | 46 |
| 6819delTG | 2201 | 44, 45 |
| 7057delCTTAT | 2290 | 44 |
| 7231delTTTCG | 2337 | 45 |
| 7253delAA | 2358 | 47 |
| 7786C→T | 2520 | 19 |
| 7989delC | 2647 | 40 |
| 8138delCCTTT | 2638 | 44 |
| 8162delG | 2645 | 44 |
| 8525delC | 2776 | 41 |
| 8535delAG | 2780 | 48 |
| 8664insA | 2819 | 40 |
| 8702delC | 2862 | 19 |
| 9179C→G | 2984 | 44 |
| 9208ins4 | 2994 or 3018¶ | 44 |
| 9254delATCAT | 3015 | 41, 42 |
| 9326insA | 3043 | 42, 43 |
| 9433delGT | 3170 | 48 |
| 9636delT | 3162 | 19, 43 |
| 9654delTT | 3148 | 49 |
| 9808delCC | 3195 | 19 |
The nucleotide positions are based on the GenBank entry of BRCA2 (accession no. U43746).
†Natural termination occurs at codon 3419.
‡ For space reasons, this reference list is not comprehensive. Most of the mutations in this table are also listed in the Breast Cancer Information Core (BIC) database (18). The BIC database also contains other mutations that are not published elsewhere and are not listed above.
§Mutation reported as 6697delTC, but TC sequence begins at position 6696.
¶ Termination codon depends upon inserted bases, the sequence of which was not given.
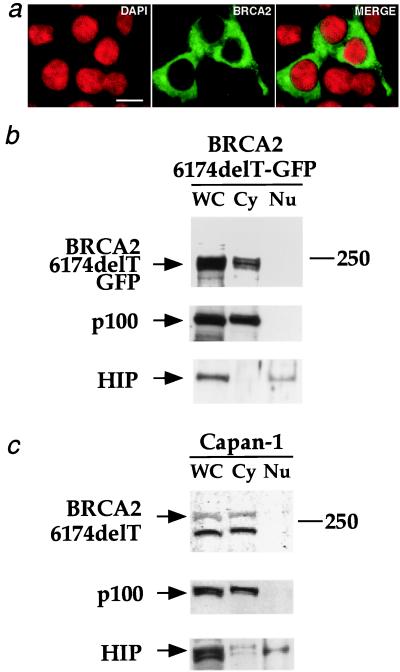
To determine the subcellular localization of the cancer-associated 6174delT truncated form of BRCA2, three experiments were conducted. First, a BRCA2–6174delT-GFP expression construct was transiently transfected into 293T cells to facilitate direct examination of the truncated product’s localization. The GFP signal emanating from BRCA2–6174delT-GFP was clearly excluded from the nucleus (Fig. 4a). Second, 293T cells transiently transfected with the BRCA2–6174delT-GFP expression construct were subjected to biochemical fractionation. The data from this fractionation confirmed that transiently expressed BRCA2–6174delT-GFP is excluded from the nucleus (Fig. 4b).
Figure 4.
Cancer-associated BRCA2–6174delT is cytoplasmic. (a) Direct observation of BRCA2–6174delT-GFP. 293T cells were transiently transfected with MG-B2–6174delT-GFP. (Left) DAPI was used to stain nuclei. (Center) Green fluorescence from the BRCA2–6174delT-GFP fusion. (Right) The merged image. (Bar = 10 μm.) (b) Biochemical fractionation of cells transiently expressing BRCA2–6174delT-GFP. 293T cells transiently transfected with MG-B2–6174delT-GFP were separated into cytoplasmic (Cy) and nuclear (Nu) fractions. WC, whole-cell extract. The fractionation was done as described for Fig. 3b. Anti-GFP Ab (mouse mAb from CLONTECH) was used to probe the blot for BRCA2–6174delT-GFP; the control blots were probed as described for Fig. 3b. (c) Biochemical fractionation of Capan-1 cells. Human pancreatic cancer cells (Capan-1) expressing only the 6174delT truncation mutant form of BRCA2 were separated into cytoplasmic (Cy) and nuclear (Nu) fractions. WC, whole-cell extract. Fractionation and immunoblotting were done as described for Fig. 3b.
Since the above experiments used transiently expressed BRCA2–6174delT-GFP, it was critical to evaluate the subcellular localization of endogenously expressed BRCA2–6174delT. For the third experiment, Capan-1 cells expressing BRCA2–6174delT were subjected to biochemical fractionation. The endogenous 6174delT truncated form of BRCA2 was observed to be cytoplasmic (Fig. 4c). These data support the hypothesis that expressed truncated forms of BRCA2 behave as mutants because of improper localization.
Discussion
The subcellular localization of a cancer-associated BRCA2 truncation mutant was shown to be cytoplasmic. This localization was observed for both endogenous and transiently expressed protein. A panel of GFP-tagged BRCA2 deletion mutants revealed the presence of two NLSs within the final 156 amino acids of BRCA2. Taken together, these data indicate that all of the known disease-linked truncations of BRCA2 are likely to be nonfunctional because they do not translocate to the nucleus.
The next stage of testing for our hypothesis will involve evaluating primary tumor samples and their matched controls. In normal tissue from carriers of BRCA2 truncation mutants, we expect to find distinct cytoplasmic and nuclear pools of BRCA2; in tumor tissue that has undergone loss of heterozygosity, we expect to see only the cytoplasmic form. If this is the case, then individuals who may carry any truncating mutation that results in an expressed product could be screened by immunostaining methods. Such testing might be particularly useful in cases where prior family history is unavailable. It will also be worthwhile to ascertain whether sporadic breast tumors exhibit changes in BRCA2 localization.
In light of the cytoplasmic localization of human BRCA2 truncation mutants, it would be of interest to determine the subcellular localization of the truncated mutant forms of murine Brca2 that are putatively expressed in the viable deletion mutant strains (31, 32). Since these truncation mutants delete the conserved NLS (NLS1) yet still permit survival, unlike more severe Brca2 truncations, which are always embryonic lethal (33–35), it may be that the positioning of the NLSs in murine Brca2 is different from that in human BRCA2. Indeed, this is the case at least for human NLS3, which is not present in murine Brca2. Ultimately, it will be interesting to determine whether there is any functional significance to change(s) in NLS positioning that may have occurred during the course of evolution.
Other issues raised by our findings involve the interactions of BRCA2 with BRCA1 and RAD51. It has been observed that BRCA1 and RAD51 interact with the 6174delT truncated form of BRCA2 present in Capan-1 cells (8), a result that would not have been predicted on the basis of the cytoplasmic localization of truncated BRCA2 and the typically predominant nuclear localization of BRCA1 and RAD51. Plausible explanations for this apparent discrepancy include that the observed complex(es) may form in the cytoplasm or after lysis of the cells.
Another issue involves a possible RAD51 interaction domain identified in the C terminus of murine Brca2 immediately downstream of the conserved NLS (NLS1) (35, 36). This region in human BRCA2 would be eliminated by all known cancer-associated truncations, raising the possibility that RAD51 interaction is also affected by the truncation mutants. However, there is some controversy regarding this candidate RAD51-binding region because another group using similar methods did not observe RAD51 binding to the C terminus of human BRCA2 (17). RAD51 binding has also been found to be independent of this C-terminal region (8), consistent with the observation that RAD51 interacts with the BRC repeats in the central portion of human BRCA2 (17, 37). In summary, there are many known cancer-associated truncation mutants for human BRCA2 that eliminate the NLSs and the candidate C-terminal RAD51 interaction region, but should retain the capacity to bind RAD51 by means of the BRC repeats (Table 1). However, on the basis of our results, these truncation mutants should not be localized to the nucleus. Resolution of these issues awaits development of biological assays for BRCA2 that correlate well with cancer predisposition.
Acknowledgments
We thank Junjie Chen and David Livingston for providing anti-BRCA2A Ab, Kathy Jones for providing anti-HIP116 antibody, Sean Tavtigian (Myriad Genetics) for providing a full-length BRCA2 cDNA clone, Matthew Weitzman for providing mammalian genomic DNA, Cindy Wilson for valuable advice and assistance, Heinz Ruffner for excellent technical advice, and Tom Frank (Myriad Genetics) and members of the Verma laboratory for valuable discussions. C.J.L. was supported by a fellowship from the Cancer Research Institute, B.H.S. was supported by a fellowship from the American Cancer Society, and L.S.S. was supported by a fellowship from the Paralyzed Veterans of America Spinal Cord Research Foundation. F.H.G.’s laboratory is supported by a grant from the National Institute of Aging. I.M.V.’s laboratory is supported by grants from the National Institutes of Health and the American Cancer Society. We also acknowledge support from the Wayne and Gladys Valley Foundation and the H. N. and Frances C. Berger Foundation. I.M.V. is an American Cancer Society Professor of Molecular Biology.
Abbreviations
- NLS
nuclear localization signal
- GFP
green fluorescent protein
- DAPI
4′,6-diamidino-2-phenylindole
References
- 1.Rahman N, Stratton M R. Annu Rev Genet. 1998;32:95–121. doi: 10.1146/annurev.genet.32.1.95. [DOI] [PubMed] [Google Scholar]
- 2.Ford D, Easton D F, Stratton M, Narod S, Goldgar D, Devilee P, Bishop D T, Weber B, Lenoir G, Chang-Claude J, et al. Am J Hum Genet. 1998;62:676–689. doi: 10.1086/301749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szabo C I, King M C. Am J Hum Genet. 1997;60:1013–1020. [PMC free article] [PubMed] [Google Scholar]
- 4.Kinzler K W, Vogelstein B. Nature (London) 1997;386:761–763. doi: 10.1038/386761a0. [DOI] [PubMed] [Google Scholar]
- 5.Bertwistle D, Ashworth A. Curr Opin Genet Dev. 1998;8:14–20. doi: 10.1016/s0959-437x(98)80056-7. [DOI] [PubMed] [Google Scholar]
- 6.Zhang H, Tombline G, Weber B L. Cell. 1998;92:433–436. doi: 10.1016/s0092-8674(00)80936-8. [DOI] [PubMed] [Google Scholar]
- 7.Marmorstein L Y, Ouchi T, Aaronson S A. Proc Natl Acad Sci USA. 1998;95:13869–13874. doi: 10.1073/pnas.95.23.13869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J, Silver D P, Walpita D, Cantor S B, Gazdar A F, Tomlinson G, Couch F J, Weber B L, Ashley T, Livingston D M, Scully R. Mol Cell. 1998;2:317–328. doi: 10.1016/s1097-2765(00)80276-2. [DOI] [PubMed] [Google Scholar]
- 9.Wilson C A, Ramos L, Villasenor M R, Anders K H, Press M F, Clarke K, Karlan B, Chen J J, Scully R, Livingston D, et al. Nat Genet. 1999;21:236–240. doi: 10.1038/6029. [DOI] [PubMed] [Google Scholar]
- 10.Bertwistle D, Swift S, Marston N J, Jackson L E, Crossland S, Crompton M R, Marshall C J, Ashworth A. Cancer Res. 1997;57:5485–5488. [PubMed] [Google Scholar]
- 11.Ruffner H, Verma I M. Proc Natl Acad Sci USA. 1997;94:7138–7143. doi: 10.1073/pnas.94.14.7138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas J E, Smith M, Rubinfeld B, Gutowski M, Beckmann R P, Polakis P. J Biol Chem. 1996;271:28630–28635. doi: 10.1074/jbc.271.45.28630. [DOI] [PubMed] [Google Scholar]
- 13.Thakur S, Zhang H B, Peng Y, Le H, Carroll B, Ward T, Yao J, Farid L M, Couch F J, Wilson R B, Weber B L. Mol Cell Biol. 1997;17:444–452. doi: 10.1128/mcb.17.1.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y, Chen C F, Riley D J, Allred D C, Chen P L, Von Hoff D, Osborne C K, Lee W H. Science. 1995;270:789–791. doi: 10.1126/science.270.5237.789. [DOI] [PubMed] [Google Scholar]
- 15.Wilson C A, Payton M N, Elliott G S, Buaas F W, Cajulis E E, Grosshans D, Ramos L, Reese D M, Slamon D J, Calzone F J. Oncogene. 1997;14:1–16. doi: 10.1038/sj.onc.1200924. [DOI] [PubMed] [Google Scholar]
- 16.Chen Y, Farmer A A, Chen C F, Jones D C, Chen P L, Lee W H. Cancer Res. 1996;56:3168–3172. [PubMed] [Google Scholar]
- 17.Chen P-L, Chen C-F, Chen Y, Xiao J, Sharp Z D, Lee W-H. Proc Natl Acad Sci USA. 1998;95:5287–5292. doi: 10.1073/pnas.95.9.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friend S, Borresen A L, Brody L, Casey G, Devilee P, Gayther S, Goldgar D, Murphy P, Weber B L, Wiseman R. Nat Genet. 1995;11:238–239. doi: 10.1038/ng1195-238. [DOI] [PubMed] [Google Scholar]
- 19.Hakansson S, Johannsson O, Johansson U, Sellberg G, Loman N, Gerdes A M, Holmberg E, Dahl N, Pandis N, Kristoffersson U, et al. Am J Hum Genet. 1997;60:1068–1078. [PMC free article] [PubMed] [Google Scholar]
- 20.Wigler M, Pellicer A, Silverstein S, Axel R, Urlaub G, Chasin L. Proc Natl Acad Sci USA. 1979;76:1373–1376. doi: 10.1073/pnas.76.3.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schall T J, Lewis M, Koller K J, Lee A, Rice G C, Wong G H, Gatanaga T, Granger G A, Lentz R, Raab H, et al. Cell. 1990;61:361–370. doi: 10.1016/0092-8674(90)90816-w. [DOI] [PubMed] [Google Scholar]
- 22.Kozak M. Nucleic Acids Res. 1984;12:857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andrews N C, Faller D V. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abrams H D, Rohrschneider L R, Eisenman R N. Cell. 1982;29:427–439. doi: 10.1016/0092-8674(82)90159-3. [DOI] [PubMed] [Google Scholar]
- 25.Mazoyer S, Dunning A M, Serova O, Dearden J, Puget N, Healey C S, Gayther S A, Mangion J, Stratton M R, Lynch H T, et al. Nat Genet. 1996;14:253–254. doi: 10.1038/ng1196-253. [DOI] [PubMed] [Google Scholar]
- 26.Nakai K, Kanehisa M. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hicks G R, Raikhel N V. Annu Rev Cell Dev Biol. 1995;11:155–188. doi: 10.1146/annurev.cb.11.110195.001103. [DOI] [PubMed] [Google Scholar]
- 28.McAllister K A, Haugen-Strano A, Hagevik S, Brownlee H A, Collins N K, Futreal P A, Bennett L M, Wiseman R W. Cancer Res. 1997;57:3121–3125. [PubMed] [Google Scholar]
- 29.Kalderon D, Richardson W D, Markham A F, Smith A E. Nature (London) 1984;311:33–38. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- 30.Su L K, Wang S C, Qi Y, Luo W, Hung M C, Lin S H. Oncogene. 1998;17:2377–2381. doi: 10.1038/sj.onc.1202162. [DOI] [PubMed] [Google Scholar]
- 31.Friedman L S, Thistlethwaite F C, Patel K J, Yu V P, Lee H, Venkitaraman A R, Abel K J, Carlton M B, Hunter S M, Colledge W H, et al. Cancer Res. 1998;58:1338–1343. [PubMed] [Google Scholar]
- 32.Connor F, Bertwistle D, Mee P J, Ross G M, Swift S, Grigorieva E, Tybulewicz V L, Ashworth A. Nat Genet. 1997;17:423–430. doi: 10.1038/ng1297-423. [DOI] [PubMed] [Google Scholar]
- 33.Ludwig T, Chapman D L, Papaioannou V E, Efstratiadis A. Genes Dev. 1997;11:1226–1241. doi: 10.1101/gad.11.10.1226. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki A, de la Pompa J L, Hakem R, Elia A, Yoshida R, Mo R, Nishina H, Chuang T, Wakeham A, Itie A, et al. Genes Dev. 1997;11:1242–1252. doi: 10.1101/gad.11.10.1242. [DOI] [PubMed] [Google Scholar]
- 35.Sharan S K, Morimatsu M, Albrecht U, Lim D S, Regel E, Dinh C, Sands A, Eichele G, Hasty P, Bradley A. Nature (London) 1997;386:804–810. doi: 10.1038/386804a0. [DOI] [PubMed] [Google Scholar]
- 36.Mizuta R, LaSalle J M, Cheng H L, Shinohara A, Ogawa H, Copeland N, Jenkins N A, Lalande M, Alt F W. Proc Natl Acad Sci USA. 1997;94:6927–6932. doi: 10.1073/pnas.94.13.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong A K C, Pero R, Ormonde P A, Tavtigian S V, Bartel P L. J Biol Chem. 1997;272:31941–31944. doi: 10.1074/jbc.272.51.31941. [DOI] [PubMed] [Google Scholar]
- 38.Mercurio F, Didonato J, Rosette C, Karin M. DNA Cell Biol. 1992;11:523–537. doi: 10.1089/dna.1992.11.523. [DOI] [PubMed] [Google Scholar]
- 39.Sheridan P L, Schorpp M, Voz M L, Jones K A. J Biol Chem. 1995;270:4575–4587. doi: 10.1074/jbc.270.9.4575. [DOI] [PubMed] [Google Scholar]
- 40.Couch F J, Farid L M, DeShano M L, Tavtigian S V, Calzone K, Campeau L, Peng Y, Bogden B, Chen Q, Neuhausen S, et al. Nat Genet. 1996;13:123–125. doi: 10.1038/ng0596-123. [DOI] [PubMed] [Google Scholar]
- 41.Tavtigian S V, Simard J, Rommens J, Couch F, Shattuck-Eidens D, Neuhausen S, Merajver S, Thorlacius S, Offit K, Stoppa-Lyonnet D, et al. Nat Genet. 1996;12:333–337. doi: 10.1038/ng0396-333. [DOI] [PubMed] [Google Scholar]
- 42.Neuhausen S L, Godwin A K, Gershoni-Baruch R, Schubert E, Garber J, Stoppa-Lyonnet D, Olah E, Csokay B, Serova O, Lalloo F, et al. Am J Hum Genet. 1998;62:1381–1388. doi: 10.1086/301885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haraldsson K, Loman N, Zhang Q X, Johannsson O, Olsson H, Borg A. Cancer Res. 1998;58:1367–1371. [PubMed] [Google Scholar]
- 44.Gayther S A, Mangion J, Russell P, Seal S, Barfoot R, Ponder B A, Stratton M R, Easton D. Nat Genet. 1997;15:103–105. doi: 10.1038/ng0197-103. [DOI] [PubMed] [Google Scholar]
- 45.Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G. Nature (London) 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 46.Krainer M, Silva-Arrieta S, FitzGerald M G, Shimada A, Ishioka C, Kanamaru R, MacDonald D J, Unsal H, Finkelstein D M, Bowcock A, et al. N Engl J Med. 1997;336:1416–1421. doi: 10.1056/NEJM199705153362003. [DOI] [PubMed] [Google Scholar]
- 47.Friedman L S, Gayther S A, Kurosaki T, Gordon D, Noble B, Casey G, Ponder B A, Anton-Culver H. Am J Hum Genet. 1997;60:313–319. [PMC free article] [PubMed] [Google Scholar]
- 48.Phelan C M, Lancaster J M, Tonin P, Gumbs C, Cochran C, Carter R, Ghadirian P, Perret C, Moslehi R, Dion F, et al. Nat Genet. 1996;13:120–122. doi: 10.1038/ng0596-120. [DOI] [PubMed] [Google Scholar]
- 49.Serova O M, Mazoyer S, Puget N, Dubois V, Tonin P, Shugart Y Y, Goldgar D, Narod S A, Lynch H T, Lenoir G M. Am J Hum Genet. 1997;60:486–495. [PMC free article] [PubMed] [Google Scholar]