Abstract
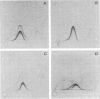
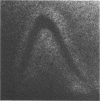
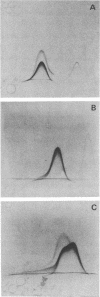
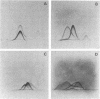
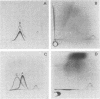
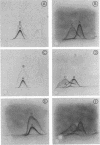
The cell-wall-associated proteolytic systems of several Streptococcus cremoris strains were analyzed by crossed immunoelectrophoresis. At least four immunologically different components of the proteolytic system's were found. One of these proteins was produced by all strains tested. The proteolytic activity of this enzyme was demonstrated with a zymogram staining technique which is based on the degradation of Coomassie-brilliant-blue-stainable casein. The crossed-immunoelectrophoresis patterns of the proteolytic systems of different S. cremoris strains indicated that each strain produces a characteristic combination of proteins. On the basis of these combinations, the different S. cremoris strains were classified into four groups.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K., Favre M. Maturation of the head of bacteriophage T4. I. DNA packaging events. J Mol Biol. 1973 Nov 15;80(4):575–599. doi: 10.1016/0022-2836(73)90198-8. [DOI] [PubMed] [Google Scholar]
- Law B. A., Kolstad J. Proteolytic systems in lactic acid bacteria. Antonie Van Leeuwenhoek. 1983 Sep;49(3):225–245. doi: 10.1007/BF00399500. [DOI] [PubMed] [Google Scholar]
- Otto R., Sonnenberg A. S., Veldkamp H., Konings W. N. Generation of an electrochemical proton gradient in Streptococcus cremoris by lactate efflux. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5502–5506. doi: 10.1073/pnas.77.9.5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto R., de Vos W. M., Gavrieli J. Plasmid DNA in Streptococcus cremoris Wg2: Influence of pH on Selection in Chemostats of a Variant Lacking a Protease Plasmid. Appl Environ Microbiol. 1982 Jun;43(6):1272–1277. doi: 10.1128/aem.43.6.1272-1277.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R. H., Morris H. A., Castberg H. B., McKay L. L. Hydrolysis of milk proteins by bacteria used in cheese making. J Agric Food Chem. 1976 Nov-Dec;24(6):1106–1113. doi: 10.1021/jf60208a026. [DOI] [PubMed] [Google Scholar]
- Sogawa K., Takahashi K. Use of fluorescamine-labeled casein as a substrate for assay of proteinases. J Biochem. 1978 Jun;83(6):1783–1787. doi: 10.1093/oxfordjournals.jbchem.a132094. [DOI] [PubMed] [Google Scholar]
- van der Plas J., Hellingwerf K. J., Seijen H. G., Guest J. R., Weiner J. H., Konings W. N. Identification and localization of enzymes of the fumarate reductase and nitrate respiration systems of escherichia coli by crossed immunoelectrophoresis. J Bacteriol. 1983 Feb;153(2):1027–1037. doi: 10.1128/jb.153.2.1027-1037.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]