Abstract
Evolution modulates the quantitative characteristics of protein interactions and often uses combinations of weak interactions to achieve a particular specificity. We addressed how quantitative optimization might be used in the design of multidomain proteins, using a chimera containing epidermal growth factor (EGF) as a cell targeting element and interferon-α-2a (IFNα-2a) to initiate signal transduction. We first connected EGF and IFNα-2a via a linker that allows both ligands to bind to their receptors on a cell surface and then incorporated a series of mutations into the IFNα-2a portion that progressively decrease both the on rate and the dissociation constant of the IFNα-2a-IFNα receptor 2 (IFNAR2) interaction. Using this strategy, we designed chimeric proteins in which the activation of the IFNα receptor in HeLa, A431, and engineered Daudi cells depends on the presence of EGF receptor on the same cell. The mutant chimeric proteins also inhibited proliferation of IFNα-sensitive cells in an EGF receptor-dependent manner. These results provide insights into the quantitative requirements for specific binding to multisubunit receptors and illustrate the value of a quantitative approach in the design of synthetic-biological constructs.
Biological recognition events are often mediated by modular protein and nucleic acid segments that can be arbitrarily linked to give functional combinations. In the course of evolution, multidomain proteins have been repeatedly generated and constitute a large fraction of the proteins encoded by metazoan genomes. In the evolutionary improvement of such chimeras after a genetic rearrangement, an important but under studied process is the quantitative optimization of the individual modules.
For many years, researchers have also constructed chimeric proteins with properties that derive from the parental modules. For example, one therapeutic approach has sought to use cell surface proteins as addresses to direct the delivery of specific molecules, such as toxins to tumor cells. Pastan's group (1) described a chimeric protein consisting of Pseudomonas exotoxin and interleukin-2, in which the interleukin-2 moiety directed the toxin to cells bearing interleukin-2 receptor; they later described a P. exotoxin-tumor growth factor-α chimeric protein that binds to EGFR3 (2–4), a hallmark of many tumors (5). Similar strategies have been adapted by many groups (6, 7). A universal problem with this kind of approach is that when any targeted agent is administered to a patient, unwanted effects will occur as the drug travels through the body before reaching its target.
We therefore sought a different strategy based on quantitative modulation of the signaling part of a targeted molecule. Our strategy builds on the ideas of Adam and Delbrück (8), who proposed that in biological systems, reaction rates are often enhanced by reduction of the dimension of a space in which diffusion occurs. Because a cell surface is effectively two-dimensional, we reasoned that an initial rapid binding reaction to one cell surface protein could drive a second, weak interaction on the same cell surface. We further reasoned that if we could tune the binding affinities of both the targeting agent and the activating ligand appropriately, we could develop a mutant chimeric protein that would show negligible activation on cells expressing just one of the relevant receptors. We set out to test this strategy using EGF as the targeting agent and IFNα-2a as the toxin.
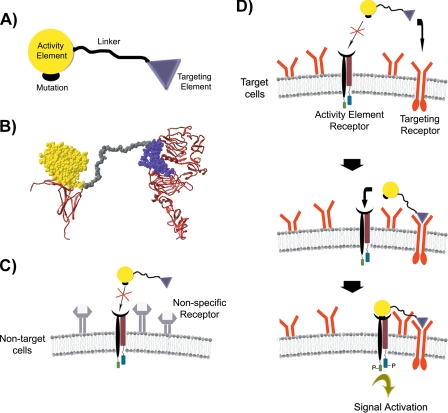
By combining quantitative information about protein binding kinetics and affinities with structural information about protein ligands and their cell-surface receptors, we developed a new class of artificial proteins that simultaneously bind to distinct cell-surface receptors to create ligands with new cell type specificities. These proteins, termed chimeric activators, have the following general structure: an activity element containing at least one mutation, a linker, and a targeting element (see Fig. 1A). This design is analogous to natural designs that use multivalent interactions between ligands and cell surfaces to ensure precise delivery of biological activities.
FIGURE 1.
A, general structure of chimeric activators, showing a targeting element connected by a peptide linker to an activity element with a mutation that reduces binding to the receptor for the activity element. B, molecular model of the IFNα-2a-EGF chimeric activator (space-filling structure), showing how the IFNα-2a and EGF components can simultaneously interact with their receptors (ribbons). Models for EGF·EGFR complex (12, 13), and the IFNα-2a·IFNAR2 complex (14, 15), are shown with the C termini of the receptor extracellular domains at the bottom; in each case, these C termini are followed by the membrane-spanning segment of the receptor. C and D, mechanism of specific binding of chimeric activators to target cells. C, the chimeric activator binds poorly to non-target cells because the intrinsic binding affinity of the mutant activity element to its receptor is low. D, in contrast, the targeting element binds to receptors on a target cell at a high rate. After the targeting element complexes with its receptor, the activity element is in a high local concentration relative to its receptor so that the activity element can then bind and stimulate signal transduction.
EXPERIMENTAL PROCEDURES
Cell Lines and Culture Conditions—Daudi, 293-T, and HeLa cells were obtained from the American Type Culture Collection (Manassas, VA). A431 cells were a gift from Thomas M. Roberts (Dana Farber Cancer Institute). A431, 293-T, and HeLa cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 μgml-1 penicillin, and 100 μgml-1 streptomycin. Daudi cells were maintained in RPMI 1640 modified medium supplemented with 10% fetal bovine serum, 100 μgml-1 penicillin, and 100 μgml-1 streptomycin. Stable Daudi-pLPCX-EGFR (Daudi-EGFR) and Daudi-pLPCX cell lines were generated by retroviral infection of Daudi cells.4 Infected Daudi cells were first selected with puromycin (1.5 μg/ml), and then cells were isolated with Dynabeads Pan mouse IgG from Invitrogen precoated with anti-EGFR mAb following the manufacturer's instructions. The constructs pLPCX and pLPCX-EGFR were kindly provided by Joan S. Brugge, Harvard Medical School (9).
Antibodies—Anti-phospho-STAT1 (Tyr701) rabbit pAb and anti-EGFR rabbit pAb were purchased from Cell Signaling Technology (Beverly, MA). PhosohoDetect™ anti-phospho-EGFR (Tyr1068) rabbit pAb, anti-EGFR mouse mAb (EGFR.1), anti-EGFR mouse mAb (528), and anti-EGF rabbit pAb were purchased from Calbiochem. Anti-actin mouse mAb was purchased from Chemicon International (Temecula, CA). Anti-human IFNα/β R1-phycoerythrin mouse mAb was purchased from R&D systems (Minneapolis, MN). Anti-EGFR R-phycoerythrin-conjugated mouse mAb was purchased from BD Biosciences.
Gene Synthesis, Protein Expression, and Purification—The coding sequence for the “wild-type” chimeric activator, INFα-2a-(Gly4-Ser)7-EGF, consisting of the 165 amino acids of mature INFα-2a (GI:2781226), the 35-amino-acid linker and the 53 amino acids of mature EGF (GI:24987355), was synthesized by Top Gene Technologies (Quebec, Canada). This sequence was codon-optimized for expression in Pichia pastoris. The sequence was subcloned (with XhoI and XbaI restriction sites) into the pPICZα A vector (Invitrogen), which includes the alcohol oxidase promoter and the α-factor leader sequence, a c-Myc epitope tag, and a His6 tag for purification. The final sequence of the constructs was confirmed by DNA sequencing. Approximately 20 μg of the DNA construct were linearized with PmeI prior to transformation of P. pastoris X33 (Mut+) and KM71H (MutS) cells. The electroporation method of the EasySelect™ Pichia expression kit (version H; Invitrogen) was used for transformation, and the transformants were plated on minimal dextrose and minimal methanol agar plates to screen for the methanol utilization (Mut) phenotype. Several Mut+ and MutS clones were put on plates with high zeocin concentrations (0.5–1 mg/ml) to select for clones with multiple integration events. A MutS clone was selected for the protein expression. Transformants were grown and induced with methanol according to the instructions from Invitrogen. The INFα-2a-(Gly4-Ser)7-EGF chimeric activator was secreted into the medium and was purified with the ProBond™ purification system (for purification of polyhistidine-containing recombinant proteins, version K, Invitrogen). Purity was checked by Coomassie Blue stain and by immunoblotting against EGF. The final yield was ∼1 mg/ml of cell culture.
We constructed variants of INFα-2a-(Gly4-Ser)7-EGF containing the IFNα-2a mutations K133A, R144A, and R149A by standard recombinant DNA techniques using the QuikChange™ site-directed mutagenesis kit from Stratagene. To produce control proteins, a set of Pichia expression vectors encoding IFNα-2a wild-type and mutant proteins lacking the linker and EGF but containing the c-Myc and His6 tags was also constructed by analogous techniques. The correctness of the constructs was confirmed by DNA sequencing. The transformation, expression, and purification of the different constructs were performed as described above.
Cell Stimulation, Protein Extraction, Immunoblotting, and Immunoprecipitation—A431 and HeLa cells were seeded in 60-mm plates, and Daudi and Daudi-EGFR were seeded in 50-ml bottles. Once they reached confluence, the growth medium was replaced with fresh Dulbecco's modified Eagle's medium without fetal bovine serum or antibiotics for 3 h (to reduce background signal from the serum).
EGFR Stimulation—Cells were stimulated with human recombinant EGF from Escherichia coli (Cell Signaling Technology), the chimeric activators, and interferon-α A (Calbiochem) or vehicle (as negative controls) for 5 min at 37 °C and 5% CO2. Stimulations were terminated by washing the cells once with ice-cold phosphate-buffered saline, and cells were lysed in Nonidet P-40 lysis buffer (20 mm Tris-HCl at pH 7.5, 150 mm NaCl, 1% Nonidet P-40, 25 mm NaF, 10 mm β-glycerophosphate, 250 μm Na3VO4 supplemented with protease inhibitor mixture tablets (Roche Applied Science)). Cells were solubilized for 15 min at 4 °C in lysis buffer follow by centrifugation at 15,000 × g for 15 min at 4 °C, and the detergent extracts (supernatant) were then subjected to immunoprecipitation. Lysates were incubated with 1 μg of anti-EGFR (EGFR.1) mouse monoclonal antibody overnight at 4 °C. EGFR was immunoprecipitated with a mixture of protein A and G-Sepharose beads (Amersham Biosciences) for 1 h. at 4 °C. Beads were washed with Nonidet P-40 lysis buffer and boiled in SDS sample buffer. Proteins were resolved on 8% SDS-polyacrylamide, transferred to nitrocellulose, and detected by immunoblotting using the anti-phospho-EGFR(Tyr1068) antiserum.
STAT1 Stimulation—As described above, cells were stimulated with the chimeric activators, the IFNα-only counterparts, interferon-α A, or EGF or vehicle (as negative controls) for 30 min at 37 °C and 5% CO2. Stimulations were terminated by washing the cells once with ice-cold phosphate-buffered saline and lysed in radioimmune precipitation buffer (150 mm NaCl, 1% deoxycholate, 10 mm Tris-HCl at pH 7.2, 0.1% SDS, 1.0% Triton X-100, 5 mm EDTA, 25 mm NaF, 10 mm β-glycerophosphate, 250 μm Na3VO4 supplemented with protease inhibitor mixture tablets (Roche Applied Science)). Proteins were resolved on 10% SDS-polyacrylamide, transferred to nitrocellulose, and detected by immunoblotting with anti-phospho-STAT1(pTyr701) antiserum.
Neutralization of EGF Receptor and STAT1 Stimulation—HeLa cells were seeded as described previously. Once they reached confluence, the medium was changed for fresh Dulbecco's modified Eagle's medium, and cells were incubated with anti-EGFR mouse mAb 528. After 2 h of treatment, cells were tested for STAT1 stimulation as described above.
Anti-proliferative Assay—The anti-proliferative activity of IFNα-2a-linker-EGF chimeras, corresponding IFNα-2a proteins, and commercial IFNα A on Daudi and Daudi-EGFR cell lines was assayed as follows. Proteins that had been previously filtered thorough a 0.2-μm polyvinylidene difluoride filter unit (Millipore) and then quantitated were serially diluted. Twenty serial dilutions were prepared in flat-bottomed 96-well plates for each tested protein. Daudi and Daudi-EGFR cells grown in RPMI-1680 medium were added (3 × 104 cells in 100 μl) to each well and were grown for an additional 60 h in the presence of the different proteins. The number of living cells was then determined using a cell staining kit (Cell proliferation reagent WST-1, Roche Applied Science) based on the colorimetric detection of the cleavage of the tetrazolium salt WST-1 into formazan. The WST-1 reaction solution was added according to the manufacturer's recommendation for a period of 4 h, after which the absorbance at 450 and 650 nm (reference wavelength) was recorded in a Victor3V multilabel reader (Perkin Elmer Life Sciences).
RESULTS
Quantitative Rationale for Design of Chimeric Activators—The design of chimeric activators with a desired cell specificity requires an understanding of the on rates, off rates, and equilibrium constants of each of the receptor-binding elements. An important aspect of ligand-receptor interactions is that kon is primarily limited by diffusion. The off rate for ligand-receptor interactions is often slower than the process of receptor-mediated endocytosis, and in such cases, koff is not relevant to signaling. As a result, the binding of a chimeric protein composed of a targeting element and an activity element might not be significantly influenced by the supposed targeting element. For example, EGF and IFNα-2a both have similar on rates (Table 1) (10, 11) so that fusion of EGF to IFNα-2a may have very little effect on the binding of the latter to its receptor. We imagined that for a chimeric activator to have a cell specificity driven by the targeting element, one or both of the following conditions should be met; the on rate of the activity element should be lower than that of the targeting element, or if the off rates are faster than the internalization rate, then the equilibrium constant (kon/koff) of the targeting element should be higher than that of the activity element. Once such a chimeric activator binds to a cell surface via the receptor for the targeting element, binding of the activity element to its receptor should in principle be driven by its high local concentration relative to its receptor (Fig. 1, C and D).
TABLE 1.
On-rates, off-rates, and dissociation constants of EGF and wild-type and mutant IFNα2
| Kon | Koff | Kd | Chimeric activators (CA) | |
|---|---|---|---|---|
| 1/m·seg | 1/s | m | ||
| EGF | 1 × 106 | 2.6 × 10–3 | 2.6 × 10–9 | |
| IFNα-2a (wild-type) | 3.7 × 106 | 1 × 10–2 | 3 × 10–9 | IFNα-2a(wild-type)-(Gly4Ser)7-EGF (CA-WT) |
| IFNα-2a (K133A) | 0.7 × 106 | 1.8 × 10–2 | 26 × 10–9 | IFNα-2a(K133A)-(Gly4Ser)7-EGF (CA-K133A) |
| IFNα-2a (R144A) | 0.36 × 106 | 4 × 10–2 | 120 × 10–9 | IFNα-2a(R144A)-(Gly4Ser)7-EGF (CA-R144A) |
| IFNα-2a (R149A) | Too low to measure | 538 × 10–9 | IFNα-2a(R149A)-(Gly4Ser)7-EGF (CA-R149A) |
A rationally designed set of chimeric activators was constructed consisting of wild-type and mutant forms of IFNα-2a as the activating element and EGF as the targeting element (Fig. 1A). Our choice of these elements was based on the following considerations: 1) the three-dimensional structures of the EGF·EGFR and the IFNα-2a·IFNAR2 complexes have been solved or modeled (12–14), allowing us to choose appropriate positions for linking and to design a linker of the right length; 2) a precise characterization of the on rates, off rates, and KD values of a series of IFNα alanine-scanning mutants was previously reported (11) so that we could choose a series of IFNα-2a mutants with stepwise reductions in on rates and affinity (Table 1); and 3) the on rates and binding constants of wild-type EGF and IFNα are similar so that the effect of mutations should be significant. The on rates of both proteins are thought to be faster than diffusion-limited because they are driven by charge complementarity between these ligands and their receptors (11, 16). Another consideration was that the molecules described here could serve as a proof-of-concept for a protein drug and that targeting might improve the therapeutic index of a toxic molecule only by a limited amount. Thus, the fact that IFNα already has a preferential activity against abnormal cells such as cancer cells and virus-infected cells made it an attractive candidate (17).
For the chimeric activator concept to work, the targeting element and activation element must be able to bind simultaneously to their respective receptors. The published structure of EGFR with a ligand and a model of the IFNα-2a structure indicate that in these receptor-ligand complexes, the EGF is about 90 Å from the cell surface, and the IFNα-2a is about 50 Å from the cell surface, so that a linker of at least 40 Å is needed to bridge the two elements. We therefore selected a standard glycine-serine linker of 35 amino acids ([Gly4Ser]7, with a length of roughly 120 Å; Fig. 1B) (18). Four chimeric activators were constructed (Table 1). These consist of wild-type or mutant IFNα-2a at the N terminus followed by the 35-amino acid linker, EGF, a Myc epitope tag, and a His6 purification tag at the C terminus. The IFNα-2a mutations used were K133A, R144A, and R149A, which allow the protein to fold correctly but cause stepwise reductions in the on rate and equilibrium binding of IFNα-2a for its receptor (Table 1) (11). The chimeric activators carrying wild-type INFα-2a and these mutant forms were termed CA-WT, CA-K133A, CA-R144A, and CA-R149A, respectively.
We expressed these chimeric activators and the corresponding individual IFNα-2a and IFNα-2a mutants in the yeast P. pastoris according to published methods and purified them from culture supernatant (19, 20). All proteins were epitope-tagged and His6-tagged for further detection and purification. The proteins were expressed at high levels and recovered at high purity (see “Experimental Procedures”).
Functionality of Parts within Chimeric Activators—The biological activity of the targeting element, EGF, was verified in the chimeric activators by testing its ability to stimulate phosphorylation of tyrosine 1068 in EGFR. Activation of EGFR was evaluated in HeLa cells by treating them with the four chimeric proteins, a commercial recombinant EGF as a positive control, and IFNα-2a tagged with c-Myc and His6 as a negative control. Fig. 2 illustrates the induction of phosphorylation of the EGFR by the different chimeric proteins as well for the recombinant epidermal growth factor control (lanes 2–7). Cells treated only with phosphate-buffered saline (vehicle) or with tagged IFNα do not show detectable activity (lanes 1 and 8). To test the activity of the IFNα portion of the chimeric activators, we examined by Western blot the phosphorylation of STAT1 tyrosine 701 (a consequence of IFN-α receptor activation; Fig. 3 and supplemental Fig. 1). These experiments demonstrated that the EGF and IFNα-2a components of the chimeric proteins were both active and that the mutations K133A, R144A, and R149A reduced the activity of the tagged IFNα-2a to a degree similar to the reductions previously reported (11).
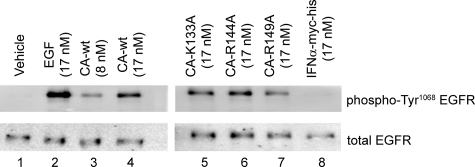
FIGURE 2.
EGFR activation upon treatment with IFNα-2a-EGF chimeric activators. HeLa cells were stimulated for 5 min with vehicle (phosphate-buffered saline; lane 1), EGF (lane 2), chimeric activators containing wild-type EGF linked to wild-type or mutant IFNα-2a (lanes 3–7), or wild-type IFNα-2a protein expressed from P. pastoris in the same manner as the chimeric activators (lane 8). EGF receptor immunoprecipitates from stimulated HeLa cell lysates were separated by SDS-PAGE and then immunoblotted for Tyr1068-phosphorylated EGF receptor (top) or total EGF receptor (bottom).
FIGURE 3.
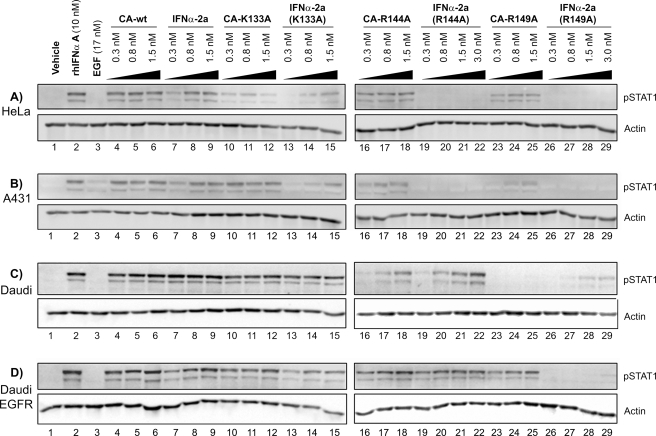
STAT1 activation upon treatment with IFNα-2a-EGF chimeric activators. Starved HeLa (A), A431 (B), Daudi (C), or Daudi-EGFR (D) cells were incubated for 30 min with phosphate-buffered saline (vehicle), commercial IFNα A, EGF, and IFNα·EGF chimeric activator proteins containing wild-type or mutant IFNα-2a (CA-WT CA-K133A, CA-R144A, or CA-R149A) or the corresponding wild-type or mutant IFNα-2a proteins produced from Pichia (see “Experimental Procedures”). Lysates were prepared, and immunoblots were performed as described under “Experimental Procedures” probing with an anti-STAT1(pTyr701) antibody and with an anti-actin antibody as a loading control. (See also supplemental Fig. 1, which shows similar immunoblots in which lanes were scanned with a densitometer and the phospho-STAT1 signal was normalized to the actin signal.) HeLa, A431, and Daudi-EGFR cells express both EGFR and IFNAR, whereas Daudi cells express only IFNAR. rhIFNαA stands for recombinant human interferon α A.
Activity of IFNα-2a-EGF Chimeric Activators Depends on EGFR—We next tested whether the IFNα-2a-EGF chimeric proteins could signal through IFNAR in an EGFR-dependent manner by comparing their activity with the activity of the corresponding IFNα-2a (mutant) protein in cells expressing both EGFR and IFNAR. We used two different epithelial cell lines with varying levels of EGFR expression: A431 cells, which show high expression of EGFR (∼2 × 106 EGFR/cell), and HeLa cells, which show much lower expression of EGFR (2 × 104 EGFR/cell) (21). Quantification by fluorescence-activated cell sorter analysis confirmed the expression levels of EGFR and IFNAR (supplemental Table 1). The results indicated that an IFNα-2a-EGF chimeric activator (without a mutation) and IFNα-2a alone induced interferon signaling with about equal efficiency (Fig. 3, A and B, lanes 4–9; see also supplemental Fig. 1) and that the IFNα-2a mutant proteins K133A, R144A, and R149A, which respectively are reduced by 10×, 40×, and 200× for IFNAR binding (Table 1), show correspondingly low levels of IFNα signaling (Fig. 3, A and B, lanes 13–15, 19–22, and 26–29).
The chimeric activators with EGF fused to the mutant IFNα-2a showed higher levels of IFNα signaling than the corresponding IFNα-2a mutants alone, however. For example, in HeLa and A431 cells, pSTAT1 was activated when cells were treated with CA-R144A and CA-R149A (Fig. 3, A and B, lanes 16–18 and 23–25) but not when the cells are treated with the corresponding IFNα-2a mutants, even at higher concentrations (Fig. 3, A and B, lanes 22 and 29). No synergistic effect was seen when cells were treated with a combination of EGF and any of the various IFNα-2a mutants (data not shown).
As a second way to test whether the enhancement of IFNα signaling depends on binding to EGFR, we introduced EGFR into the Daudi cell line, which does not normally express this receptor (supplemental Fig. 2) and compared IFNα signaling in this engineered line with that seen in the parent cell line. The Daudi cell line is derived from a human Burkitt lymphoma cell, and its proliferation and survival are inhibited by IFNα (22). The function of the chimeric activators on Daudi and Daudi-EGFR cells clearly depended on the presence of the receptor for the targeting element (Fig. 3, C and D). The CA-R149A protein stimulated STAT1 phosphorylation only in Daudi-EGFR cells (Fig. 3D, lanes 23–25) and not in the Daudi parental cell line (Fig. 3C, lanes 26–29). The CA-WT and IFNα-2a proteins were active in both cell lines, as expected, whereas the IFNα-2a(R149A) mutant showed only slight activity at high concentrations. Chimeric activators containing the intermediate strength mutations CA-K133A and CA-R144A gave intermediate levels of signaling in Daudi cells, consistent with their predicted properties, and showed quantitative enhancements in signaling relative to IFNα-2a(K133A) and IFNα-2a(R144A) in Daudi-EGFR cells (Fig. 3D, lanes 10–22).
The activities of the chimeric activators differed on the various cell lines (Fig. 3). We hypothesize that these cell type differences were due to differences at the level of receptor expression, although the HeLa, Daudi, and A431 cell lines have different tissue origins and oncogenic mutations so that any comparison of results between these lines can only be suggestive. Daudi cells have 10–20 times more IFNAR molecules at the cell surface than do HeLa and A431 cells (supplemental Table 2), which may explain the greater level of IFNAR activation in Daudi cells, for example, in response to CA-R144A and IFNα-2a(R144A).
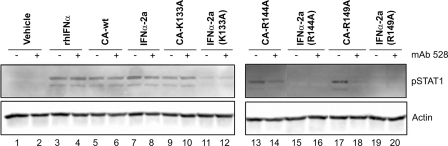
As an additional control, we tested whether signaling by IFNα-2a(mutant)-EGF chimeric activators could be inhibited by an antibody against the receptor for the targeting element (Fig. 4). Pretreatment of HeLa cells for 2 h with the mouse monoclonal anti-EGFR antibody 528 inhibits the activation of the STAT1 signaling pathway by the K144A and R149A mutant chimeric activators (Fig. 4, lanes 13 and 14 and lanes 17 and 18) but not the activation caused by the non-chimeric IFNα-2a and mutants. Neither the wild-type IFNα-2a nor the CA-WT showed inhibition (Fig. 4, lanes 5 and 6 and lanes 9 and 10), as expected. The 528 antibody does not itself activate the EGF or the IFNα signal cascades. Treatment with mAb 528 appeared to have no effect on signaling induced by the CA-K133A protein. Since the IFNα element retains significant binding to the IFNAR, it is possible that this chimera was able to displace the antibody from the EGFR during the 30-min course of the experiment.
FIGURE 4.
Neutralization of EGFR. HeLa cells were pretreated for 2 h with 1 μg/ml mAb 528, a mouse monoclonal antibody that prevents EGF from binding to EGFR (even-numbered lanes) followed by treatment for 30 min with vehicle (lanes 1 and 2) or 1.5 nm commercial IFNα A(lanes 3 and 4), IFNα·EGF chimeric activator proteins containing wild-type or mutant IFNα-2a (CA-WT, lanes 5 and 6; CA-K133A, lanes 9 and 10; CA-R144A, lanes 13 and 14; or CA-R149A, lanes 17 and 18), or the corresponding wild-type or mutant IFNα-2a proteins (lanes 7 and 8, 11 and 12, 15 and 16, and 19 and 20). rhIFNα stands for recombinant human interferon α.
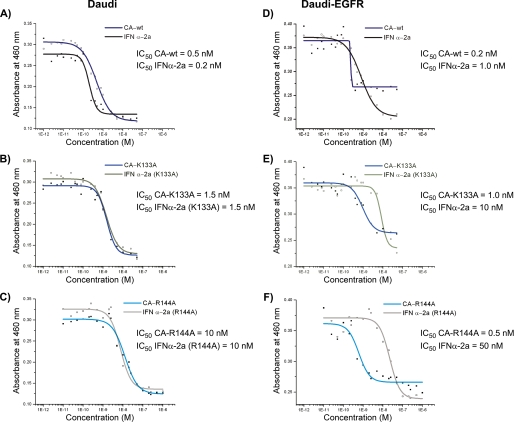
EGFR-dependent Inhibition of Cell Proliferation by Chimeric Activators—The ability of the chimeric proteins to suppress the proliferation of Daudi and Daudi-EGFR cells is shown in Fig. 5. In both cell lines, a dose-dependent growth inhibition was observed. When Daudi cells were treated with the chimeric activators and their IFNα-2a counterparts, essentially no difference in their IC50 values was found (Fig. 5, A–C). Furthermore, the ability to inhibit proliferation diminished according to the ability to bind to IFNAR and activate STAT1.
FIGURE 5.
EGFR-dependent anti-proliferative activity of IFNα-2a-EGF chimeric proteins. Daudi and Daudi-EGFR cells were grown for 60 h in the presence of various concentrations of IFNα-2a-EGF chimeric proteins or the corresponding IFNα-2a proteins purified from Pichia. The relative number of viable cells was determined by the production of formazan, which absorbs at A460, from tetrazolium (see “Experimental Procedures”). Curves were fit to the data using a four-parameter fit (Microcal Origin 5.0).
The chimeric activators, relative to their IFNα counterparts, showed enhanced inhibition of cell proliferation and survival of Daudi-EGFR cells (Fig. 5, E and F). The chimeric activators containing the mutations K133A and R144A were about 10- and 100-fold more potent, respectively, than the corresponding IFNα-2a mutants alone. CA-R149A weakly inhibited proliferation in a pattern that could not be fit to a sigmoid curve (data not shown), and its activity relative to the corresponding IFNα-2a mutant therefore could not be quantified.
The behavior of the Daudi-EGFR cells was somewhat affected by the presence of EGFR, which may contribute prosurvival signals. The IC50 values of wild-type and mutant IFNαs were about 5–6-fold higher in Daudi-EGFR (Fig. 5). The maximal extent of proliferation inhibition was also less than for cells treated with IFNα mutants alone. These results are consistent with previous observations that EGF can antagonize the anti-proliferative and pro-death effects of IFNα (23).
DISCUSSION
In the experiments described here, we addressed whether the cell type specificity of an extracellular signaling protein could be altered by a combination of genetic fusion followed by quantitative modulation. The results have implications for understanding the natural design of multisubunit proteins and ligand-receptor interactions, as well as for the design of artificial proteins targeted to cancer cells and other types of disease-causing cells.
The goal of our particular design was to alter the cell type specificity of IFNα so that it would only activate its receptor on cells bearing EGFR. We attached EGF to IFNα-2a with a flexible linker so that both modules could simultaneously bind to their receptors. We also mutated IFNα-2a so that its ability to bind to its receptor would be significantly reduced, using a set of previously defined mutations that reduce the equilibrium binding of IFNα-2a for its receptor by 10-, 40-, and 200-fold (11). We hypothesized that the resulting chimeric protein would be essentially unable to bind directly to IFNARs but would bind to EGFR with high affinity. By virtue of the high local concentration of the chimera on the cell surface, the IFNα-2a module would then be able to bind to its receptor.
Several lines of evidence indicated that the resulting chimerized, mutated proteins induced IFNα signaling in an EGFR-dependent manner. First, a comparison of IFNα-2a(mutant)-EGF chimeras with their unchimerized IFNα-2a(mutant) counterparts in HeLa and A431 cells, which express both EGFR and IFNAR, showed that the chimeric proteins were more potent in inducing STAT1 phosphorylation, which results specifically from the activation of IFNAR (Fig. 3). As expected, the differential effect was particularly pronounced for chimeric activators carrying mutations that significantly reduce IFNα binding to its receptor. In contrast, IFNα-2a-EGF (CA-WT) was essentially indistinguishable from wild-type IFNα-2a; this result is expected because the binding of IFNα to its receptor is quantitatively similar to that for EGF. Second, the activities of the chimerized and unchimerized proteins were compared on Daudi cells and Daudi cells engineered to express EGFR. These results indicated that the enhanced stimulation of STAT1 phosphorylation by the EGF chimeras depended on the presence of EGFR on the cell surface. The improved selectivity of the mutant chimeric activators seen with HeLa and A431 cells was reproduced in Daudi-EGFR cells but not in parental Daudi cells. Third, the stimulation of STAT1 phosphorylation could be inhibited by an anti-EGFR antibody.
The chimeric activator proteins described here induced a biological response in an EGFR-dependent manner. The proliferation and survival of Daudi cells is inhibited by IFNα-2a. We found that Daudi cells expressing EGFR were more sensitive than parental Daudi cells to IFNα-2a(mutant)-EGF chimeric activators by up to more than an order of magnitude (Fig. 5). In a therapeutic context, the Daudi-EGFR cells may be considered to represent target cells, the parental Daudi cells may represent non-target cells where receptor activation results in side effects, and the differential effect corresponds to the therapeutic index of a protein drug. Thus, by reducing the binding of a given activator and attaching it to a targeting element, we can improve the therapeutic index by more than an order of magnitude.
A natural extension of our approach would be to use a tumor-specific antibody as a targeting element. The molecular designs reported here depended in part on structural models of both the IFNα·IFNAR2 and the EGF·EGFR complexes. Use of antibody V regions in chimeric activators will become more feasible as more structures of antibody-receptor complexes are solved (24, 25).
In summary, we have constructed artificial signaling molecules based on quantitative principles that may reproduce the design of natural systems. We envision that further analysis of these principles may allow the improvement of protein therapeutics and a deeper understanding of the forces that shape natural biological system design.
Supplementary Material
Acknowledgments
We thank Dr Rebecca Ward and Jessica Hurt for careful proofreading and helpful comments.
This work was supported by in part by grants from National Institutes of Health and funds from Harvard University (to P. A. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains two supplemental figures and a supplemental table.
Footnotes
The abbreviations used are: EGF, epidermal growth factor; EGFR, EGF receptor; CA, chimeric activator; WT, wild type; IFNα-2a, interferon-α-2a; IFNAR, IFNα receptor; Mut+, methanol utilization wild-type; MutS, methanol utilization slow; STAT-1, signal transducer and activator 1; mAb, monoclonal antibody.
W. S. Pear, M. L. Scott, and G. P. Nolan, personal communication.
References
- 1.Lorberboum-Galski, H., FitzGerald, D., Chaudhary, V., Adhya, S., and Pastan, I. (1998) Proc. Natl. Acad. Sci. U. S. A. 85 1922-1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegall, C. B., Xu, Y-H., Chaudhary, V. K., Adhya, S., Fitzgerald, D., and Pastan, I. (1989) FASEB J. 3 2647-2652 [DOI] [PubMed] [Google Scholar]
- 3.Heimbrook, D. C., Stirdivant, S. M., Ahern, J. D., Balishin, N. L., Edwards, G. M., Defeo-Jones, D., FitzGerald, D. J., Pastan, I, and Oliff. A. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 4697-4701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pai, L. H., Gallo, M. G., FitzGerald, D. J., and Pastan, I. (1991) Cancer Res. 51 2808-2812 [PubMed] [Google Scholar]
- 5.Liu, T. F., Tatter, S. B., Willingham, M. C., Yang, M., Hu, J. J., and Frankel, A. E. (2003) Mol. Cancer Ther. 2 783-787 [PubMed] [Google Scholar]
- 6.Kreitman, R. J., and Pastan, I. (1999) Handbook of Experimental Pharmacology: Novel Therapeutics from Modern Biotechnology, pp. 89-110, Springer-Verlag, Berlin, NY
- 7.Todhunter, D. A., Hall, W. A., Rustamzadeh, E., Shu, Y., Doumbia, S. O., and Vallera, D. A. (2004) Protein Eng. Des. Sel. 17 157-164 [DOI] [PubMed] [Google Scholar]
- 8.Adam, G., and Delbrück, M. (1968) in Structural Chemistry and Molecular Biology (Rich, A, and Davidson, N, eds) pp. 198-215, W. H. Freeman and Co., San Francisco
- 9.Reginato, M. J., Mills, K. R., Paulus, J. K., Lynch, D. K., Sgroi, D. C., Debnath, J., Muthuswamy, S. K., and Brugge, J. S. (2003) Nat. Cell Biol. 5 733-740 [DOI] [PubMed] [Google Scholar]
- 10.French, A. R., Tadaki, D. K., Niyogi, S. K., and Lauffenburger, D. A. (1995) J. Biol. Chem. 270 4334-4340 [DOI] [PubMed] [Google Scholar]
- 11.Piehler, J., Roisman, L. C., and Schreiber, G. (2000) J. Biol. Chem. 275 40425-40433 [DOI] [PubMed] [Google Scholar]
- 12.Ogiso, H., Ishitani, R., Nureki, O., Fukai, S., Yamanaka, M., Kim, J. H., Saito, K., Sakamoto, A., Inoue, M., Shirouzu, M., and Yokoyama, S. (2002) Cell 110 775-787 [DOI] [PubMed] [Google Scholar]
- 13.Garrett, T. P., McKern, N. M., Lou, M., Elleman, T. C., Adams, T. E., Lovrecz, G. O., Zhu, H. J., Walker, F., Frenkel, M. J., Hoyne, P. A., Jorissen, R. N., Nice, E. C., Burgess, A. W., and Ward, C. W. (2002) Cell 110 763-773 [DOI] [PubMed] [Google Scholar]
- 14.Roisman, L. C., Piehler, J., Trosset, J-Y., Scheraga, H. A., and Schreiber, G. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 13231-13236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quadt-Akabayov, S. R., Chill, J. H., Levy, R., Kessler, N., and Anglister, J. (2006) Protein Sci. 15 2656-2668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piehler, J., and Schreiber, G. (1999) J. Mol. Biol. 289 57-67 [DOI] [PubMed] [Google Scholar]
- 17.Platanias, L. C. (2005) Nat. Rev. Immunol. 5 375-386 [DOI] [PubMed] [Google Scholar]
- 18.Huston, J. S., Tai, M. S., McCartney, J., Keck, P., and Oppermann, H. (1993) Cell Biophys. 22 189-224 [DOI] [PubMed] [Google Scholar]
- 19.Clare, J. J., Romanos, M. A., Rayment, F. B., Rowedder, J. E., Smith, M. A., Payne, M. M., Sreekrishna, K., and Henwood, C. A. (1991) Gene (Amst.) 105 205-212 [DOI] [PubMed] [Google Scholar]
- 20.Garcia, J. N., Aguiar, J. A., Gill, M., Alvarez, A., Morales, J., Ferrero, J., Gonzalez, B., Padron, G., and Menendez, A. (1995) Biotecnol. Apl. 12 152-155 [Google Scholar]
- 21.Masui, H., Kawamoto, T., Sato, J. D., Wolf, B., Sato, G., and Mendelsohn, J. (1984) Cancer Res. 44 1002-1007 [PubMed] [Google Scholar]
- 22.Evinger, M., and Pestka, S. (1981) Methods Enzymol. 79 362-368 [DOI] [PubMed] [Google Scholar]
- 23.Caraglia, M., Abbruzzese, A., Leardi, A., Pepe, S., Budillon, A., Baldassare, G., Selleri, C., Lorenzo, S. D., Fabbrocini, A., Giuberti, G., Vitale, G., Lupoli, G., Bianco, A. R., and Tagliaferri, P. (1999) Cell Death Differ. 6 773-780 [DOI] [PubMed] [Google Scholar]
- 24.Franklin, M. C., Carey, K. D., Vajdos, F. F., Leahy, D. J., de Vos, A. M., and Sliwkowski, M. X. (1994) Cancer Cell 5 317-328 [DOI] [PubMed] [Google Scholar]
- 25.Cho, H.-S., Mason, K., Ramyar, K. X., Stanley, A. M., Gabelli, S. B., Denney, Jr., D. W., and Leahy, D. J. (2003) Nature 421 756-760 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.