Abstract
There is accumulating evidence that mammalian target of rapamycin (mTOR)-activated pathways play important roles in cell growth and survival of BCR-ABL-transformed cells. We have previously shown that the mTOR/p70 S6 kinase (p70 S6K) pathway is constitutively activated in BCR-ABL transformed cells and that inhibition of BCR-ABL kinase activity by imatinib mesylate abrogates such activation. We now provide evidence for the existence of a novel regulatory mechanism by which BCR-ABL promotes cell proliferation, involving p70 S6K-mediated suppression of expression of programmed cell death 4 (PDCD4), a tumor suppressor protein that acts as an inhibitor of cap-dependent translation by blocking the translation initiation factor eIF4A. Our data also establish that second generation BCR-ABL kinase inhibitors block activation of p70 S6K and downstream engagement of the S6 ribosomal protein in BCR-ABL transformed cells. Moreover, PDCD4 protein expression is up-regulated by inhibition of the BCR-ABL kinase in K562 cells and BaF3/BCR-ABL transfectants, suggesting a mechanism for the generation of the proapoptotic effects of such inhibitors. Knockdown of PDCD4 expression results in reversal of the suppressive effects of nilotinib and imatinib mesylate on leukemic progenitor colony formation, suggesting an important role for this protein in the generation of antileukemic responses. Altogether, our studies identify a novel mechanism by which BCR-ABL may promote leukemic cell growth, involving sequential engagement of the mTOR/p70 S6K pathway and downstream suppression of PDCD4 expression.
Chronic myeloid leukemia (CML)2 is a clonal myeloproliferative disorder, whose hallmark is the presence of the BCR-ABL oncoprotein, which results from the abnormal bcr-abl fusion oncogene (1-3). bcr-abl is created by a reciprocal translocation involving chromosomes 9 and 22, and its abnormal protein product plays a key and essential role in the pathogenesis of CML (1-4). The constitutive tyrosine kinase activity of BCR-ABL mediates phosphorylation of multiple downstream substrates and engagement of various mitogenic pathways that promote cell growth and survival (1-7). The introduction of imatinib mesylate in the management of chronic myelogenous leukemia has had a dramatic impact on the natural history of the disease (reviewed in Refs. 8-11). This agent induces long lasting hematologic and cytogenetic responses in patients with early and late phase CML (8, 12, 13, 14, 16-18) by its ability to block activation the BCR-ABL kinase and generation of mitogenic responses (8-11).
Although the introduction of imatinib mesylate was a major breakthrough in the management of CML, there has been emerging evidence for resistance to its antileukemic properties in vitro and in vivo (19-22). Such resistance involves a variety of cellular mechanisms, including mutations of the bcr-abl gene, amplification of the bcr-abl gene locus, and activation of downstream signaling elements, such as Src-kinase-dependent pathways (19, 23-28). Research efforts aimed to overcome development of imatinib resistance have led to the development of the second generation BCR-ABL tyrosine kinase inhibitors that include nilotinib (AMN-107) and dasatinib, both of which are active against various BCR-ABL kinase mutations associated with refractoriness to imatinib mesylate (29, 31-42). The development and application of such agents in the targeting of BCR-ABL mutations has been a major research breakthrough, although there is recent evidence indicating that resistance to their activities can also develop (43, 44).
We and others have recently shown that mTOR-dependent pathways are constitutively activated in BCR-ABL-transformed cells (45-48). These studies have raised the possibility that regulation of mTOR may be a mechanism by which BCR-ABL promotes cell growth and survival of leukemic cells, but the mTOR effectors mediating such responses remain unknown. In the present study, we demonstrate that BCR-ABL-mediated mTOR activation results in suppression of expression of programmed cell death 4 (PDCD4), a tumor suppressor protein that acts as an inhibitor of cap-dependent translation by blocking the translation initiation factor eIF4A (49-51). In studies using the K562, CML-derived, cell line we demonstrate that inhibition of the BCR-ABL/mTOR/p70 S6K pathway results in enhanced protein expression of PDCD4. Similarly, inhibition of BCR-ABL kinase activity in BaF3 cells stably expressing BCR-ABL or various imatinib-resistant BCR-ABL mutants also results in up-regulation of PDCD4. Importantly, siRNA-mediated knockdown of PDCD4 reverses the suppressive effects of nilotinib (AMN-107) or imatinib mesylate on BCR-ABL-transformed cells, indicating an important role for this protein in the generation of antileukemic responses.
MATERIALS AND METHODS
Cells and Reagents—The CML-derived K562 cell line was grown in RPMI 1640 medium supplemented with 10% fetal bovine serum and antibiotics. Imatinib mesylate (STI571) and nilotinib (AMN-107) were provided by Novartis. Dasatinib was purchased from American Custom Chemicals Corp. (San Diego, CA). Cycloheximide was purchased from Sigma. Actinomycin D was obtained from Calbiochem. Antibodies against the phosphorylated forms of S6 ribosomal protein on Ser-235/236 and Ser-240/244, against the p70 S6 kinase on Thr-389 and against the Cleaved form of poly(ADP-ribose) polymerase Asp-214 were obtained from Cell Signaling Technology (Beverly, MA). An antibody against PDCD4 was obtained from Rock-land, Inc. (Gilbertsville, PA). An antibody against HA was obtained form Covance (Princeton, NJ). The retroviral plasmids MIGR1, MIG P210, and MIG P210 KI were provided from Dr. Rhavi Bhatia (Oregon Health and Sciences University, Portland, OR). The PCDNA3 empty vector, PCDNA3 HA-tagged wild type PDCD4 or HA-tagged PDCD4 (S67A/S71A) were a gift from Dr. Michele Pagano (Department of Pathology, New York University). The various Ba/F3 transfectants with different BCR-ABL mutants that are known to confer resistance to Imatinib mesylate have been described elsewhere (52).
Cell Lysis and Immunoblotting—Cells were treated with nilotinib, imatinib mesylate, or dasatinib for the indicated times and lysed as previously described (53, 54). Immunoblotting using an enhanced chemiluminescence method was performed as previously described (53, 54).
Quantitative RT-PCR (TaqMan)—Cells were treated with the indicated concentrations of nilotinib for the indicated times, and RNA was isolated using the RNeasy kit (Qiagen). 1 μg of total cellular mRNA was reverse transcribed into cDNA using the Omniscript RT kit and oligo(dT) primer (Qiagen). Real time reverse transcriptase PCR for the Pdcd4 gene was carried out by ABI7900 Sequence detection system (Applied Biosystems) using fluorescein amidite-labeled probes and primers. Relative quantitation of mRNA levels was plotted as -fold increase as compared with untreated samples. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used for normalization (55). ΔCt values (target gene Ct minus GAPDH Ct) for each triplicate sample were averaged, and ΔΔCt was calculated as previously described. mRNA amplification was determined by the formula 2-ΔΔCt (55).
siRNA-mediated Knockdown of PDCD4 in Leukemic Progenitors—For the studies to assess the effects of PDC4 knockdown in the induction of nilotinib responses, Ba/F3 cells expressing native BCR-ABL were transfected with mouse pdcd4 siRNA (Qiagen) using TransIT-TKO (Mirus, Madison, WI). The cells were subsequently cultured in a methylcellulose assay system (56, 57) (MethoCult SFBITM3236 (StemCell Technologies), supplemented with 1% fetal bovine serum, and leukemic CFU-blast (CFU-L) colony formation was scored on day 5 of culture. To assess the specificity of the effect of the siRNA-mediated knockdown of PDCD4 in leukemic progenitors, Ba/F3 cells expressing native BCR-ABL were transfected with mouse pdcd4 siRNA (Qiagen) using TransIT-TKO (Mirus) in the presence or absence of PCDNA3 empty vector or PCDNA3 PDCD4-WT using TransIT-LT1 (Mirus). The cells were subsequently cultured in a methylcellulose assay system (56, 57) (MethoCult SFBITM3236), and leukemic CFU-L colony formation was scored on day 5 of culture.
PDCD4 Overxpression in Leukemic Progenitors—Ba/F3 cells expressing native BCR-ABL were transiently transfected with PCDNA3 empty vector or HA-tagged PDCD4 (S67A/S71A) using TransIT-LT1 (Mirus). The cells were subsequently cultured in a methylcellulose assay system (56, 57) (MethoCult SFBITM3434 (StemCell Technologies)), and leukemic CFU-L colony formation was scored on day 5 of culture.
RESULTS
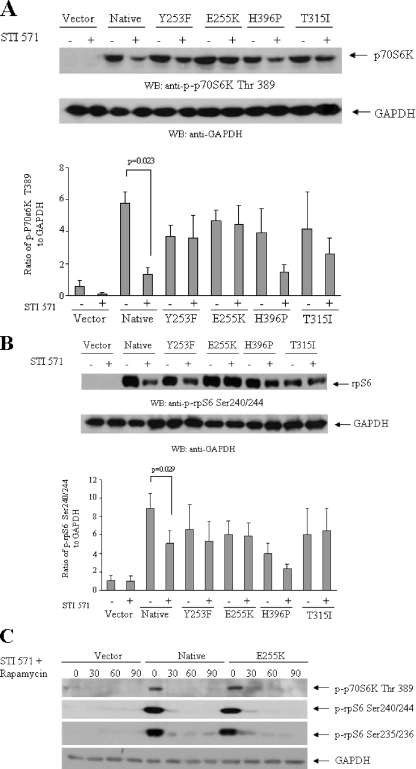
In previous studies, we had demonstrated that mTOR is constitutively activated in BCR-ABL-transformed cells and that imatinib mesylate blocks phosphorylation/activation of mTOR and its downstream effector p70 S6K (47). We sought to determine whether the mTOR pathway is activated in Ba/F3 cells transformed with various BCR-ABL kinase domain mutants, known to exhibit resistance to imatinib mesylate (52). Constitutive phosphorylation/activation of p70 S6K was observed in native BCR-ABL-transformed cells as well as in cells transformed with the ATP binding loop mutants Y253F and E255K or the H296P and T315I mutants (Fig. 1A). Consistent with this, the S6 ribosomal protein was also constitutively phosphorylated in cells transformed with either native BCR-ABL or the various mutants (Fig. 1B). Treatment of Ba/F3 cells expressing native BCR-ABL with imatinib mesylate resulted in a significant decrease in the phosphorylation/activation of both the p70 S6K (paired p = 0.023) (Fig. 1A) and its substrate, S6 ribosomal protein (paired p = 0.029) (Fig. 1B). On the other hand, phosphorylation/activation of p70 S6K and rpS6 was not decreased in cells expressing E255K and T315I mutants, consistent with the well established resistance of these mutants to the effects of imatinib mesylate (52). Some minimal effects in the also resistant (52) mutant H396P and Y253F transfectants were noticeable but were not statistically significant when several experiments were quantitated and analyzed (Fig. 1, A and B). When cells were treated with imatinib mesylate in the presence of rapamycin, there was complete abrogation of phosphorylation of p70 S6K on Thr-389 and the S6 ribosomal protein on Ser-235/236 and Ser-240/244 in both native and E255K BCR-ABL transfectants (Fig. 1C), indicating that rapamycin blocks mTOR activation in cells transformed with imatinib-resistant BCR-ABL kinase mutants.
FIGURE 1.
Effects of imatinib mesylate on the phosphorylation/activation status of elements of the mTOR pathway in Ba/F3 cells expressing BCR-ABL-resistant mutants. A (top), Ba/F3 cells stably transfected with empty vector, wild type (native) BCR-ABL, or the Y253F, E255K, H396P, or T315I mutants were incubated with imatinib mesylate (STI571) (1 μm) for 2 h, as indicated. Total cell lysates were resolved by SDS-PAGE and immunoblotted with antibodies against the phosphorylated form of the p70 S6 kinase on Thr-389 or against GAPDH, as indicated. Bottom, the signals were quantitated by densitometry. Data are expressed ratios of phosphorylated p70 S6 kinase on Thr-389 to GAPDH levels and represent means ± S.E. of three independent experiments. Paired t test analysis for the phosphorylation of p70 S6 kinase on Thr-389 in wild type BCR-ABL-transfected cells treated with imatinib mesylate versus control untreated cells showed a p value of 0.023. Similar analysis for Y253F and H396P transfected cells showed p = 0.475 and p = 0.107, respectively. B (top), Ba/F3 cells stably transfected with empty vector, wild type (native) BCR-ABL, or the Y253F, E255K, H396P, or T315I mutants were incubated with imatinib mesylate (STI571) (1 μm) for 2 h, as indicated. Total cell lysates were resolved by SDS-PAGE and immunoblotted with antibodies against the phosphorylated form of the S6 ribosomal protein on Ser-240/244 or against GAPDH, as indicated. The signals were quantitated by densitometry. Data are expressed ratios of phosphorylated S6 ribosomal protein on Ser-240/244 to GAPDH and represent means ± S.E. of three independent experiments. Paired t test analysis for the phosphorylation of Ser-240/244 in wild type BCR-ABL-transfected cells treated with imatinib mesylate versus control untreated cells showed a p value of 0.029. Similar analysis for Y253F and H396P transfected cells showed p = 0.113 and p = 0.160, respectively. C, Ba/F3 cells stably transfected with empty vector, wild type BCR-ABL, or E255K mutant were incubated with imatinib mesylate (STI571) (1 μm) in the presence or absence of rapamycin (20 nm) for 30, 60, and 90 min, as indicated. Total cell lysates were resolved by SDS-PAGE and immunoblotted with antibodies against the phosphorylated forms of the S6 ribosomal protein on Ser-235/236 or Ser 240/244 or against the phosphorylated form of the p70 S6 kinase on Thr-389 or with an antibody against GAPDH, as indicated.
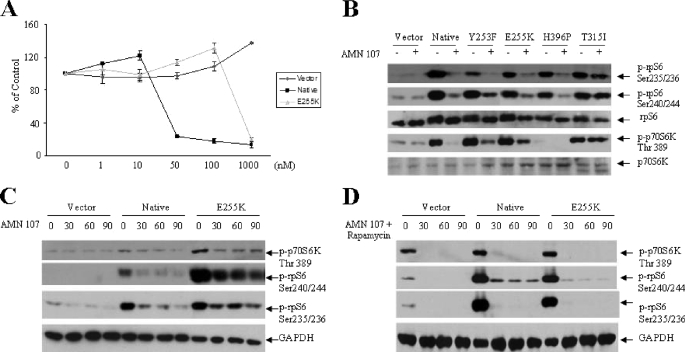
Nilotinib (AMN-107) is a second generation BCR-ABL kinase inhibitor with known activity against imatinib-resistant BCR-ABL mutants in vitro and in vivo (34-40). We examined the effects of this BCR-ABL kinase inhibitor on the activation of components of the mTOR pathway in cells transformed with various BCR-ABL kinase mutants. As expected, treatment of either Ba/F3-BCR-ABL or Ba/F3-BCR-ABL-E255K cells with nilotinib resulted in suppression of cell growth, whereas cells transfected with empty vector were not sensitive to its growth-suppressive effects (Fig. 2A). Nilotinib blocked BCR-ABL-dependent phosphorylation of p70 S6K on Thr-389 and rpS6 phosphorylation on Ser-235/236 and Ser-240/244, in the Y253F, E255K, and H296P mutants (Fig. 2B). On the other hand, nilotinib did not inhibit phosphorylation/activation of p70 S6K in Ba/F3 cells transfected with the T315I mutant (Fig. 2B), consistent with previous work that has established that this mutant is resistant to the effects of nilotinib in vitro and in vivo (30, 42, 58). The suppression of activation of the p70 S6K pathway was fast, occurring within 30 min of treatment of Ba/F3 transfectants (Fig. 2C). When the cells were treated concomitantly with the BCR-ABL inhibitor and the mTOR inhibitor, rapamycin, the suppression of 70 S6K activity and phosphorylation of the S6 ribosomal protein in different sites was almost complete (Fig. 2D). Thus, both imatinib mesylate and nilotinib block phosphorylation/activation of the mTOR/p70 S6K in cells expressing native BCR-ABL, whereas nilotinib also blocks such phosphorylation in cells expressing imatinib-resistant mutants, firmly establishing that the engagement of this signaling cascade is BCR-ABL-dependent.
FIGURE 2.
Effects of nilotinib on the phosphorylation/activation status of elements of the mTOR pathway in Ba/F3 cells expressing BCR-ABL-resistant mutants. A, growth-inhibitory effects of nilotinib (AMN107) in Ba/F3 cells stably transfected with empty vector, wild type BCR-ABL, or the E255K mutant. Cells were treated for 7 days with solvent control (Me2SO) or with the indicated concentrations of nilotinib (AMN-107). Results are expressed as percentage of control (Me2SO-treated) cells. B, Ba/F3 cells stably transfected with empty vector, wild type BCR-ABL, or the Y253F, E255K, H396P, or T315I BCR-ABL mutants were incubated with nilotinib (AMN107) (1 μm) for 2 h, as indicated. Total cell lysates were resolved by SDS-PAGE and immunoblotted with antibodies against the phosphorylated forms of the S6 ribosomal protein on Ser-235/236 or Ser-240/244 or against the phosphorylated form of the p70 S6 kinase on Thr-389 or antibodies against the S6 ribosomal protein or the p70 S6K, as indicated. C, Ba/F3 cells stably transfected with empty vector, wild type BCR-ABL, or the E255K BCR-ABL mutant were incubated with nilotinib (AMN107) (1 μm) for 30, 60, and 90 min as indicated. Total cell lysates were resolved by SDS-PAGE and immunoblotted with antibodies against the phosphorylated forms of the S6 ribosomal protein on Ser-235/236 or Ser-240/244 or against the phosphorylated form of the p70 S6 kinase on Thr-389 or against GAPDH, as indicated. D, Ba/F3 cells stably transfected with empty vector, wild type BCR-ABL, or the E255K BCR-ABL mutant were incubated with nilotinib (AMN107) (1 μm), in the presence or absence of rapamycin (20 nm) for 30, 60, and 90 min, as indicated. Total cell lysates were resolved by SDS-PAGE and immunoblotted with antibodies against the phosphorylated forms of the S6 ribosomal protein on Ser-235/236 or Ser-240/244 or against the phosphorylated form of the p70 S6 kinase on Thr-389 or an antibody against GAPDH, as indicated.
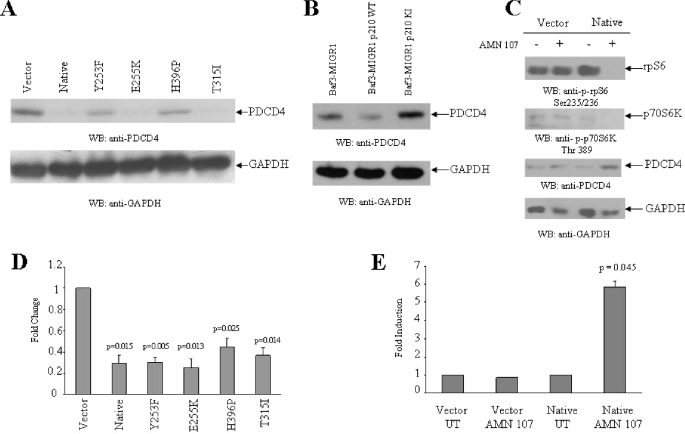
PDCD4 is a tumor suppressor protein that inhibits cap-dependent translation by blocking the activity of eIF4A (49-51), an RNA helicase that is a component of the translation initiation complex (50, 51). Recent studies have shown that PDCD4 is phosphorylated by p70 S6K and that such phosphorylation ultimately results in its degradation by the ubiquitin ligase βTRCP (59). This prompted us to perform studies to determine whether BCR-ABL engagement of the mTOR/p70 S6K pathway results in suppression of PDCD4 expression. In initial studies, the expression of PDCD4 was examined in Ba/F3 cells transfected with either empty vector or wild type BCR-ABL or the various BCR-ABL kinase mutants. As shown in Fig. 3A, PDCD4 protein expression was severely suppressed in all BCR-ABL transfectants as compared with cells transfected with the empty vector alone. Such suppression was more prominent in cells transfected with native BCR-ABL and the E255K or T315I mutants than the Y253F and H396P mutants (Fig. 3A). To examine the requirement for BCR-ABL tyrosine kinase activity in PDCD4 suppression, we used BA/F3 cells stably expressing wild type BCR-ABL or kinase-inactive BCR-ABL. As shown in Fig. 3B, PDCD4 protein expression was clearly defective in cells expressing wild type BCR-ABL as compared with cells transfected with the empty vector or the kinase-inactive BCR-ABL (Fig. 3B). Importantly, when Ba/F3 cells transfected with native BCR-ABL were treated with nilotinib, there was up-regulation of PDCD4 expression, associated with concomitant suppression of phosphorylation of p70 S6K and rpS6 (Fig. 3C). We also noticed that the Pdcd4 mRNA expression was suppressed in the various BCR-ABL transfectants (Fig. 3D), whereas treatment of wild type BCR-ABL-transfected cells with nilotinib resulted in up-regulation of Pdcd4 mRNA expression (Fig. 3E), suggesting a negative regulatory effect of the kinase activity of BCR-ABL on Pdcd4 gene transcription.
FIGURE 3.
Effects of nilotinib on the phosphorylation/activation status of elements of the mTOR pathway and PDCD4 expression in Ba/F3 cells expressing BCR-ABL-resistant mutants.A, cell lysates from Ba/F3 cells stably transfected with empty vector, wild type BCR-ABL, or the Y253F or E255K or H396P or T315I BCR-ABL mutants were resolved by SDS-PAGE and immunoblotted with antibodies against PDCD4 or GAPDH, as indicated. B, cell lysates from Ba/F3 cells stably transfected with MIGR1 empty vector, MIGRI wild type BCR-ABL, or MIG P210 KI mutant were resolved by SDS-PAGE and immunoblotted with antibodies against PDCD4 or GAPDH, as indicated. C, Ba/F3 cells stably transfected with empty vector or wild type BCR-ABL were incubated with nilotinib (AMN107) (1 μm) for 12 h, as indicated. Total cell lysates were resolved by SDS-PAGE and immunoblotted with antibodies against the phosphorylated forms of the S6 ribosomal protein on Ser-235/236, against the phosphorylated form of the p70 S6 kinase on Thr-389, against PDCD4, or against GAPDH, as indicated. D, basal expression of mRNA for the pdcd4 gene was evaluated by quantitative RT-PCR (TaqMan) in Ba/F3 cells stably transfected with empty vector, wild type BCR-ABL, or the Y253F or E255K or H396P or T315I BCR-ABL mutants. GAPDH was used for normalization. Data are expressed as -fold change over Ba/F3 cells stably transfected with empty vector and represent means ± S.E. of three independent experiments. Two-tailed paired t test analysis for the -fold changes of the various transfectants showed the indicated p values. E, Ba/F3 cells stably transfected with empty vector or wild type BCR-ABL were incubated for 12 h at 37 °C in the absence or presence of AMN107 (100 nm). Expression of mRNA for the pdcd4 gene was evaluated by quantitative RT-PCR (TaqMan). GAPDH was used for normalization. Data are expressed as -fold increase over AMN107-untreated samples and represent means ± S.E. of two independent experiments. Paired two-tailed t test analysis showed a two-tailed p = 0.045 for AMN107-treated versus untreated wild type BCR-ABL-expressing cells.
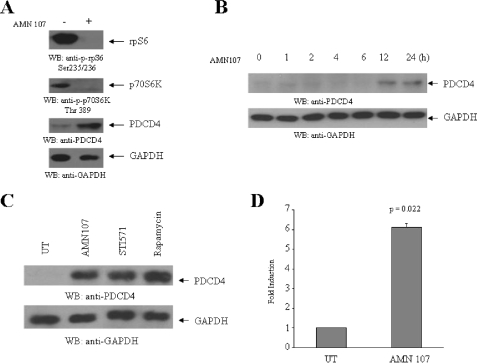
To further determine the potential role of the mTOR/p70 S6K pathway in regulation of PDCD4 protein expression in BCR-ABL-transformed cells, we performed similar studies using K562 cells that express endogenous BCR-ABL. Similarly to what we observed in the Ba/F3 transfectants, nilotinib treatment resulted in strong up-regulation of PDCD4 protein expression, associated with suppressive effects on the phosphorylation/activation of the p70 S6K and rpS6 (Fig. 4A). The up-regulation of PDCD4 protein expression was time-dependent and was clearly seen at 12-24 h of treatment of the cells with the BCR-ABL kinase inhibitor (Fig. 4B). When cells were treated in parallel with nilotinib or imatinib mesylate, we noticed up-regulation of PDCD4 in response to both inhibitors (Fig. 4C), consistent with the previously demonstrated responsiveness of these cells to imatinib mesylate (47). Importantly, treatment of cells with the mTOR inhibitor rapamycin alone also resulted in strong up-regulation of expression of PDCD4 (Fig. 4C), indicating that inhibition of the mTOR pathway is the primary event that determines the up-regulation of protein expression, apparently reflecting its decreased degradation by βTRCP (59). It should also be pointed out that, as in the case of Ba/F3-BCR-ABL transfectants, BCR-ABL inhibition also resulted in enhanced Pdcd4 gene mRNA expression (Fig. 4D).
FIGURE 4.
Up-regulation of PDCD4 protein expression by BCR-ABL kinase inhibitors. A, K562 cells were incubated with nilotinib (AMN107) (100 nm) for 12 h as indicated. Total cell lysates were resolved by SDS-PAGE and immunoblotted with antibodies against the phosphorylated form of the S6 ribosomal protein on Ser-235/236 or the phosphorylated form of the p70 S6 kinase on Thr-389 or PDCD4 or GAPDH, as indicated. B, K562 cells were incubated with nilotinib (AMN107 100 nm) for 1, 2, 4, 6, 12, and 24, h as indicated. Total cell lysates were resolved by SDS-PAGE and immunoblotted with antibodies against PDCD4 or GAPDH, as indicated. C, K562 cells were incubated with nilotinib (100 nm), STI 571 (1 μm), or rapamycin (20 nm) for 12 h as indicated. Total cell lysates were resolved by SDS-PAGE and immunoblotted with antibodies against PDCD4 or the GAPDH, as indicated. D, K562 cells were incubated for 12 h at 37 °C in the absence or presence of AMN107 (100 nm). Expression of mRNA for the pdcd4 gene was evaluated by quantitative RT-PCR (TaqMan). GAPDH was used for normalization. Data are expressed as -fold increase over AMN107-untreated samples and represent means ± S.E. of two experiments. Paired two-tailed t test analysis showed a two-tailed p = 0.022.
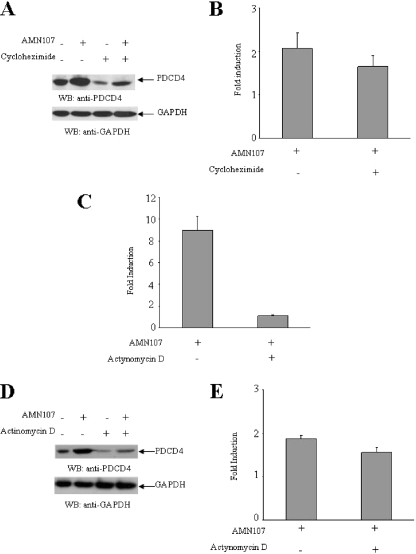
To further determine whether the AMN 107-dependent increase in PDCD4 protein levels reflects negative effects on mTOR/p70 S6K, followed by an increase in PDCD4 protein stability, we examined if AMN 107 increases PDCD4 protein expression under conditions in which protein synthesis is blocked by an inhibitor, such as cycloheximide. As shown in Fig. 5, A and B, although cycloheximide by itself reduced PDCD4 protein levels, there was no significant effect on nilotinib-induced PDCD4 up-regulation. Similar studies were also performed using actinomycin D to inhibit transcription. Treatment of Ba/F3 cells expressing native BCR-ABL with actinomycin D resulted in down-regulation of Pdcd4 mRNA expression (Fig. 5C). However, actinomycin D had only a minimal effect on niolotinib-induced PDCD4 protein expression (Fig. 5, D and E), although there was some decrease at the basal levels of expression. Altogether, these data strongly suggest that the primary mechanism of nilotinib-dependent PDCD4 up-regulation results from mTOR/p70 S6K-inhibition and blocking of protein degradation, whereas induction of PDCD4 transcription appears to constitute a secondary mechanism.
FIGURE 5.
Effect of cycloheximide and actinomycin D on up-regulation of PDCD4 protein expression by AMN107. A, Ba/F3-BCR-ABL cells were incubated with nilotinib (AMN107) (100 nm) in the presence or absence of cycloheximide (20 μm) for 12 h as indicated. Total cell lysates were resolved by SDS-PAGE and immunoblotted with antibodies against PDCD4 or GAPDH, as indicated. B, the signals were quantitated by densitometry, and the ratios of PDCD4 over GAPDH were calculated. Data are expressed as -fold induction for PDCD4 levels normalized to GAPDH, over untreated samples in response to the indicated treatments and represent means ± S.E. of three independent experiments. C, Ba/F3-BCR-ABL cells were incubated with nilotinib (AMN107) (100 nm) in the presence or absence of actinomycin D (2 μg/ml) for 12 h as indicated. Expression of mRNA for the pdcd4 gene was evaluated by quantitative RT-PCR (TaqMan). GAPDH was used for normalization. Data are expressed as -fold increase over AMN107-untreated samples and represent means ± S.E. of two experiments. D, Ba/F3-BCR-ABL cells were incubated with nilotinib (AMN107) (100 nm) in the presence or absence of actinomycin D (2 μg/ml) for 12 h as indicated. Total cell lysates were resolved by SDS-PAGE and immunoblotted with antibodies against PDCD4 or GAPDH, as indicated. E, the signals for the PDCD4 protein were quantitated by densitometry, and the ratios of PDCD4 over GAPDH were calculated. Data are expressed as -fold induction for PDCD4 levels normalized to GAPDH over untreated samples in response to the indicated treatments and represent means ± S.E. of three independent experiments.
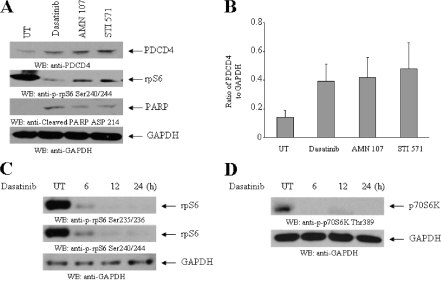
We also performed experiments to determine whether dasatinib, a dual specificity inhibitor that targets both the Abl kinase domain and Src family kinases regulates the mTOR pathway in CML cells and induces PDCD4 up-regulation. Treatment of K562 cells with dasatinib resulted in an increase in the protein levels of PDCD4 that was comparable with what was seen in response to nilotinib or imatinib mesylate (Fig. 6, A and B). Such PDCD4 up-regulation was associated with inhibition of p70 S6K activity/rpS6 phosphorylation and generation of a proapoptotic state (Fig. 6, A-D). Thus, all different BCR-ABL kinase inhibitors target the p70 S6 kinase pathway and induce PDCD4 expression, suggesting a key role for this pathway in the generation of antileukemic responses in CML.
FIGURE 6.
Effects of dasatinib on PDCD4 expression in BCR-ABL-expressing cells. A, K562 cells were incubated with nilotinib (AMN107) (100 nm), STI 571 (1 μm), or dasatinib (10 nm) for 12 h, as indicated. Total cell lysates were resolved by SDS-PAGE and immunoblotted with antibodies against PDCD4 or Ser-240/244 rpS6 or a cleaved form of poly(ADP-ribose) polymerase (Asp-214) or GAPDH, as indicated. B, the signals for PDCD4 were quantitated by densitometry. Data are expressed ratios of PDCD4 protein to GAPDH levels and represent means ± S.E. of two independent experiments. C, K562 cells were incubated with dasatinib (10 nm) for 6, 12, and 24 h as indicated. Total cell lysates were resolved by SDS-PAGE and immunoblotted with antibodies against the phosphorylated forms of the S6 ribosomal protein on Ser-235/236 or Ser-240/244 or GAPDH, as indicated. D, K562 cells were incubated with dasatinib (10 nm) for 6, 12, and 24 h, as indicated. Total cell lysates were resolved by SDS-PAGE and immunoblotted with antibodies against the phosphorylated form of the p70 S6 kinase on Thr-389 or GAPDH, as indicated.
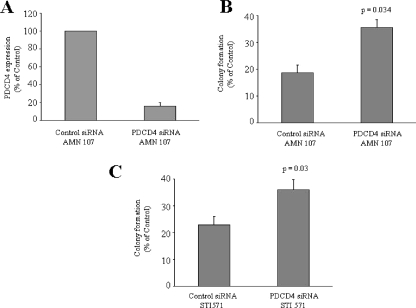
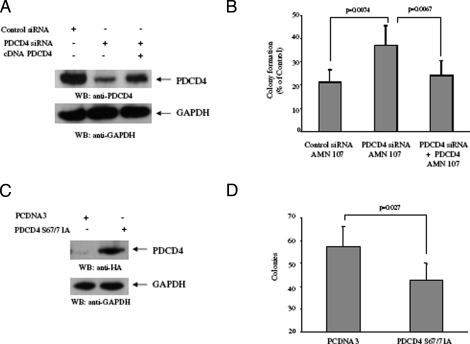
To directly assess the functional relevance of PDCD4 up-regulation in the generation of the antileukemic effects of BCR-ABL kinase inhibitors, we determined whether siRNA-mediated knockdown of the protein reverses the suppressive effects of nilotinib on leukemic progenitor colony formation. BaF3/BCR-ABL cells were transfected with siRNAs specific for PDCD4, and leukemic CFU-L colony formation was assessed. As expected, the PDCD4 siRNA blocked nilotinib-dependent Pdcd4 mRNA expression (Fig. 7A). Interestingly, the ability of nilotinib to suppress leukemic progenitor colony formation was partially reversed by PDCD4 knockdown (Fig. 7B), and such effects were statistically significant (two-tailed p = 0.034, n = 5). Similarly, siRNA-mediated knockdown of PDCD4 also resulted in partial reversal of the effects of imatinib mesylate (Fig. 7C), demonstrating that PDCD4 plays a key role in the generation of the antileukemic effects of BCR-ABL kinase inhibitors. To further establish the specificity of the effects of siRNA-mediated knockdown of the protein on leukemic progenitor colony formation, BaF3/BCR-ABL cells were transfected with mouse siRNAs specific for PDCD4 in presence or absence of the human PCDNA3-PDCD4-wild type (resistant to the mouse siRNA knockdown; Fig. 8A), and leukemic CFU-L colony formation was assessed. As expected, the ability of nilotinib to suppress leukemic progenitor colony formation was partially reversed by endogenous PDCD4 knockdown (Fig. 8B). However, the effects of nilotinib on leukemic progenitor colony formation were restored to its maximum in the presence of HA-tagged wild type human PDCD4 (Fig. 8B).
FIGURE 7.
Effects of PDCD4 knockdown on nilotinib-mediated suppression of leukemic progenitor (CFU-L). A, Ba/F3-BCR-ABL cells were transfected with control siRNA or PDCD4 siRNA (targeting mouse PDCD4) and were incubated for 12 h, in the presence of nilotinib. Expression of mRNA for the pdcd4 gene was evaluated by quantitative RT-PCR (TaqMan), using Gapdh for normalization. Data are expressed as percent control siRNA-transfected samples and represent means ± S.E. of two independent experiments. B, Ba/F3-BCR-ABL cells were transfected with control siRNA or PDCD4 siRNA and subsequently incubated in methylcellulose, in the presence or absence of nilotinib (100 nm), and leukemic CFU-L colony formation was assessed. Data are expressed as percentage of control colony formation of untreated samples for each condition and represent means ± S.E. of five independent experiments. Paired t test analysis comparing the effects of nilotinib in the presence of PDCD4 siRNA versus control siRNA showed a two-tailed p = 0.034. C, Ba/F3-BCR-ABL cells were transfected with control siRNA or PDCD4 siRNA and subsequently incubated in methylcellulose, in the presence or absence of imatinib mesylate (STI571) (1 μm), and leukemic CFU-L colony formation was assessed. Data are expressed as percentage of control colony formation of untreated samples for each condition and represent means ± S.E. of five independent experiments. Paired t test analysis comparing the effects of imatinib mesylate in the presence of PDCD4 siRNA versus control siRNA showed a two-tailed p = 0.03.
FIGURE 8.
Reversal of nilotinib-mediated suppression of leukemic progenitor (CFU-L) growth by PDCD4 knockdown and negative regulatory effects of PDCD4 on BCR-ABL-expressing leukemic progenitor growth. A, Ba/F3-BCR-ABL cells were transfected with control siRNA or mouse PDCD4 siRNA and were or were not concomitantly transfected with a construct for human PDCD4 cDNA, as indicated. After nilotinib treatment, total cell lysates were resolved by SDS-PAGE and immunoblotted with antibodies against PDCD4 or GAPDH, as indicated. B, Ba/F3-BCR-ABL cells were transfected with control siRNA or PDCD4 siRNA (targeting mouse PDCD4) and were or were not concomitantly transfected with a construct for human PDCD4 cDNA, as indicated. The cells were subsequently plated in methylcellulose, in the presence or absence of nilotinib (100 nm), and leukemic CFU-L colony formation was assessed. Data are expressed as percentage of control colony formation of untreated samples for each condition and represent means ± S.E. of five independent experiments. Paired t test analysis comparing the effects of nilotinib in the presence of PDCD4 siRNA versus control siRNA showed a p value of 0.0074 and a p value of 0.0067 for the effects of nilotinib in the presence of PDCD4 siRNA and human PDCD4 cDNA versus PDCD4 siRNA alone. C, Ba/F3-BCR-ABL cells were transfected with PCDNA3 vector control or PDCD4 S67A/S71A mutant. Total cell lysates were resolved by SDS-PAGE and immunoblotted with antibodies against HA tag and GAPDH as indicated. D, Ba/F3-BCR-ABL cells were transfected with control empty vector (pcDNA3) or a pcDNA3-PDCD4-S67A/S71A construct. The cells were subsequently plated in methylcellulose, and leukemic CFU-L colony formation was assessed. Data are expressed as colony numbers for each condition and represent means ± S.E. of five independent experiments. Paired t test analysis comparing the effects of PDCD4 overexpression versus control empty vector showed a p value of 0.027.
In other studies, we determined the effects of ectopic PDCD4 overxpression (Fig. 8C) on leukemic CFU-L colony formation. BaF3/BCR-ABL cells were transfected with HA-tagged degradation-resistant (59) PDCD4 (S67A/S71A) cDNA or control empty vector, and leukemic CFU-L colony formation was assessed. Overexpression of PDCD4 S67A/S71A mutant induced partial, but consistent and statistically significant (p value = 0.027, n = 5), suppression of growth of BCR-ABL-expressing leukemic progenitors (Fig. 8D), consistent with inhibitory effects of the protein on BCR-ABL mediated transformation and/or induction of mitogenic responses.
DISCUSSION
The mTOR signaling cascade plays a critical and essential role in the initiation of mRNA translation and mediates signals important for growth and survival of malignant cells (60-66). It is well established by extensive work over the years that two major pathways are engaged downstream of mTOR to control mRNA translation. One cascade involves activation of p70 S6K, which regulates phosphorylation of the S6 ribosomal protein and the initiation factor 4B (60-66). The other mTOR-regulated cascade involves hierarchical phosphorylation of the translational repressor 4E-BP1 on Thr-37/46, Thr-70, and Ser-65, events that ultimately lead to its deactivation and dissociation from the eukaryotic initiation factor 4E, to allow cap-dependent translation (51-57, 67). Activation of mTOR and its effectors is induced by various cytokines, hormones, and receptor tyrosine kinases, and such activation is important for the generation of mitogenic and cell proliferative responses (60-66). Recently, this pathway has been shown to be constitutively activated in BCR-ABL-transformed cells (45-48) and to be inhibited by imatinib mesylate (45-48). Importantly, the mTOR inhibitor rapamycin was shown to enhance the suppressive effects of imatinib mesylate on leukemic progenitors from patients with CML in vitro (47, 48), raising the possibility that combined use of imatinib mesylate with mTOR inhibitors may be more effective in inhibiting the in vivo growth of primary leukemic progenitors than imatinib mesylate alone (47, 48). Despite these observations, there has been no direct evidence for a functional role of this pathway in promoting BCR-ABL-dependent leukemic cell growth and antiapoptotic effects, whereas the precise downstream effector mechanisms for the generation of such effects remain unknown.
In the present study, we provide evidence that second generation BCR-ABL kinase inhibitors, such as nilotinib and dasatinib, target and block engagement of the mTOR pathway in BCR-ABL-expressing cells. Such effects of BCR-ABL kinase inhibitors are also observed in cells transformed by imatinib-resistant BCR-ABL kinase mutants, definitively establishing that the mTOR/p70 S6K pathway plays a critical role in promoting the leukemogenic effects of BCR-ABL. Importantly, we identify a novel target of the BCR-ABL-activated mTOR/p70 S6K pathway as the proapoptotic protein PDCD4. Our data establish that expression of PDCD4 is strongly up-regulated by inhibition of BCR-ABL kinase activity or by mTOR inhibition by rapamycin and correlates with suppression of p70 S6K. In addition, we provide some evidence that the transcriptional activation of this protein is suppressed by BCR-ABL in both K562 cells and BaF3/BCR-ABL cells, although this appears to constitute a secondary/complementary mechanism by which BCR-ABL suppresses PDCD4 expression. The primary mechanism appears to be directly related to p70 S6K-mediated regulation of the protein and its subsequent ubiquitination (59), since beyond the BCR-ABL kinase inhibitors, PDCD4 expression in BCR-ABL-transformed cells by rapamycin alone is inducible.
To directly examine the functional role of PDCD4 in the generation of the antileukemic effects of nilotinib or imatinib mesylate, we used siRNA targeting to knock down its expression in BCR-ABL-transformed cells. Using such an approach, we were able to demonstrate that PDCD4 knockdown reverses the antileukemic effects of nilotinib or dasatinib on leukemic BCR-ABL progenitors, suggesting a critical role of this protein in the induction of antileukemic responses. Thus, our data support a model in which expression of PDCD4 is suppressed by BCR-ABL-mediated activation of mTOR, whereas small molecule inhibitors targeting BCR-ABL generate their antileukemic effects, in part, via restoration of PDCD4 expression.
PDCD4 inhibits cap-dependent translation by negatively regulating the translation initiation factor eIF4A (49-51), an RNA helicase that is a component of the translation initiation complex (50, 51). Several recent studies have established that PDCD4 blocks both the helicase activity of eIF4A and its incorporation into the eIF4F complex (50, 51, 68). Recently, the crystal structure of the C-terminal MA3 (cMA3) domain of Pdcd4 was resolved, and the structural basis for the inhibition of translation was defined (68). Such studies have demonstrated that the cMA3 domain competes with eIF4Gc for binding to eIF4A, resulting in inhibition of translation initiation (68). It should be noted that PDCD4 is expressed in normal tissues, whereas there is evidence that its expression is suppressed in various tumors (69-74), including lung cancer (71), breast cancer (72), hepatocellular carcinoma (73), and gliomas (74). Such suppressed expression may have important consequences in tumorigenesis and neoplastic transformation, since there is accumulating evidence that PDCD4 promotes apoptosis and cell differentiation of neoplastic cells (72-75), whereas it also blocks AP-1 activation, malignant transformation, and neoplastic cell invasion in different systems (15, 76-78).
Our studies provide the first direct evidence linking suppression of PDCD4 to BCR-ABL leukemogenesis. They also directly link restoration of PDCD4 expression and inhibition of leukemic progenitor cell growth by various BCR-ABL kinase inhibitors to suppression of the mTOR/p70 S6K pathway. The fact that PDCD4 expression in BCR-ABL-expressing cells is restored by either BCR-ABL targeting or mTOR inhibition is consistent with recent work that demonstrated that p70 S6K-mediated phosphorylation of PDCD4 results in βTRCP-mediated degradation of PDCD4 (59) and suggests that a similar mechanism exists in BCR-ABL-transformed cells. Importantly, our finding that PDCD4 expression plays a key role in the generation of the antileukemic effects of BCR-ABL kinase inhibitors has important translational-therapeutic implications for the treatment of CML. The introduction of imatinib mesylate and other BCR-ABL kinase inhibitors in the treatment of CML was a major breakthrough that transformed the field and changed the natural history of the disease. Nevertheless, development of leukemic cell resistance remains a problem, and novel approaches to overcome such resistance are necessary. Our findings identify PDCD4 as a potential target for the development of novel therapeutic approaches for CML and, possibly, other leukemias. Since the function of PDCD4 appears to be critical for the generation of antileukemic responses, efforts to develop agents that inhibit the selective targeting and ultimate degradation of PDCD4 by the mTOR/p70 S6K pathway are warranted and could result in important alternative/complementary approaches for the treatment of BCR-ABL-expressing leukemias.
This work was supported by National Institutes of Health Grants CA77816, CA100579, CA121192, and CA94079 and a merit review grant from the Department of Veterans Affairs. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: CML, chronic myeloid leukemia; mTOR, mammalian target of rapamycin; RT, reverse transcription; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HA, hemagglutinin; S6K, S6 kinase; siRNA, small interfering RNA.
References
- 1.Groffen, J., Stephenson, J. R., Heisterkamp, N., de Klein, A., Bartram, C. R., and Grosveld, G. (1984) Cell 36 93-99 [DOI] [PubMed] [Google Scholar]
- 2.Heisterkamp, N., Stam, K., Groffen, J., de Klein, A., and Grosveld, G. (1985) Nature 315 758-761 [DOI] [PubMed] [Google Scholar]
- 3.Ben-Neriah, Y., Daley, G. Q., Mess-Mason, A.-M., Witte, O. N., and Baltimore, D. (1986) Science 233 212-214 [DOI] [PubMed] [Google Scholar]
- 4.Daley, G. Q., Van Etten, R. A., and Baltimore, D. (1990) Science 247 824-830 [DOI] [PubMed] [Google Scholar]
- 5.Lugo, T. G., Pendergast, A. M., Muller, A. J., and Witte, O. N. (1990) Science 247 1079-1082 [DOI] [PubMed] [Google Scholar]
- 6.Kelliher, M. A., Mc Laughlin, J., Witte, O. N., and Rosenberg, A. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 6649-6653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verma, A., and Platanias, L. C. (2002) Leuk. Lymphoma 43 703-709 [DOI] [PubMed] [Google Scholar]
- 8.Mauro, M. J., O'Dwyer, M., Heinrich, M. C., and Druker, B. J. (2002) J. Clin. Oncol. 20 325-334 [DOI] [PubMed] [Google Scholar]
- 9.Deininger, M., Buchdunger, E., and Druker, B. J. (2005) Blood 105 2640-2653 [DOI] [PubMed] [Google Scholar]
- 10.Lydon, N. B., and Druker, B. J. (2004) Leuk. Res. 28 S29-38 [DOI] [PubMed] [Google Scholar]
- 11.Druker, B. J. (2004) Adv. Cancer Res. 91 1-30 [DOI] [PubMed] [Google Scholar]
- 12.Druker, B. J., Talpaz, M., Resta, D. J., Peng, B., Buchdunger, E., Ford, J. M., Lydon, N. B., Kantarjian, H., Capdeville, R., Ohno-Jones, S., and Sawyers, C. L. (2001) N. Engl. J. Med. 344 1031-1037 [DOI] [PubMed] [Google Scholar]
- 13.Druker, B. J., Sawyers, C. L., Kantarjian, H., Resta, D. J., Reese, S. F., Ford, J. M., Capdeville, R., and Talpaz, M. (2001) N. Engl. J. Med. 344 1038-1042 [DOI] [PubMed] [Google Scholar]
- 14.O'Brien, S. G., Guilhot, F., Larson, R. A., Gathmann, I., Baccarani, M., Cervantes, F., Cornelissen, J. J., Fischer, T., Hochhaus, A., Hughes, T., Lechner, K., Nielsen, J. L., Rousselot, P., Reiffers, J., Saglio, G., Shepherd, J., Simonsson, B., Gratwohl, A., Goldman, J. M., Kantarjian, H., Taylor, K., Verhoef, G., Bolton, A. E., Capdeville, R., and Druker, B. J. (2003) N. Engl. J. Med. 348 994-1004 [DOI] [PubMed] [Google Scholar]
- 15.Hilliard, A., Hilliard, B., Zheng, S. J., Sun, H., Miwa, T., Song, W., Göke, R., and Chen, Y. H. (2006) J. Immunol. 177 8095-8102 [DOI] [PubMed] [Google Scholar]
- 16.Kurzrock, R., Kantarjian, H. M., Druker, B. J., and Talpaz, M. (2003) Ann. Int. Med. 138 819-830 [DOI] [PubMed] [Google Scholar]
- 17.Druker, B. J. (2003) Semin. Hematol. 40 50-58 [DOI] [PubMed] [Google Scholar]
- 18.Deininger, M. W. N., and Druker, B. J. (2003) Pharmacol. Rev. 55 401-423 [DOI] [PubMed] [Google Scholar]
- 19.Kantarjian, H. M., Talpaz, M., Giles, F., O'Brien, S., and Cortes, J. (2006) Ann. Intern. Med. 145 913-923 [DOI] [PubMed] [Google Scholar]
- 20.Melo, J. V., and Chuah, C. (2007) Cancer Lett. 249 121-132 [DOI] [PubMed] [Google Scholar]
- 21.Shah, N. P., and Sawyers, C. L. (2003) Oncogene 20 7389-7395 [DOI] [PubMed] [Google Scholar]
- 22.Burgess, M. R., and Sawyers, C. L. (2006) Sci. World J. 6 918-930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donato, N. J., Wu, J. Y., Stapley, J., Gallick, G., Lin, H., Arlinghaus, R., and Talpaz, M. (2003) Blood 101 690-698 [DOI] [PubMed] [Google Scholar]
- 24.Dai, Y., Rahmani, M., Corey, S. J., Dent, P., and Grant, S. A. (2004) J. Biol. Chem. 279 34227-34239 [DOI] [PubMed] [Google Scholar]
- 25.Hu, Y., Liu, Y., Pelletier, S., Buchdunger, E., Warmuth, M., Fabbro, D., Hallek, M., Van Etten, R. A., and Li, S. (2004) Nat. Genet. 36 453-461 [DOI] [PubMed] [Google Scholar]
- 26.Lionberger, J. M., Wilson, M. B., and Smithgall, T. E. (2000) J. Biol. Chem. 275 18581-18585 [DOI] [PubMed] [Google Scholar]
- 27.Danhauser-Riedl, S., Warmuth, M., Druker, B. J., Emmerich, B., and Hallek, M. (1996) Cancer Res. 56 3589-3596 [PubMed] [Google Scholar]
- 28.Nimmanapali, R., and Bhalla, K. (2002) Curr. Opin. Oncol. 14 616-620 [DOI] [PubMed] [Google Scholar]
- 29.O'Hare, T., Walters, D. K., Stoffregen, E. P., Jia, T., Manley, P. W., Mestan, J., Cowan-Jacob, S. W., Lee, F. Y., Heinrich, M. C., Deininger, M. W., and Druker, B. J. (2005) Cancer Res. 65 4500-4505 [DOI] [PubMed] [Google Scholar]
- 30.Weisberg, E., Manley, P. W., Breitenstein, W., Bruggen, J., Cowan-Jacob, S. W., Ray, A., Huntly, B., Fabbro, D., Fendrich, G., Hall-Meyers, E., Kung, A. L., Mestan, J., Daley, G. Q., Callahan, L., Catley, L., Cavazza, C., Azam, M., Neuberg, D., Wright, R. D., Gilliland, D. G., and Griffin, J. D. (2005) Cancer Cell 7 129-141 [DOI] [PubMed] [Google Scholar]
- 31.Golemovic, M., Verstovsek, S., Giles, F., Cortes, J., Manshouri, T., Manley, P. W., Mestan, J., Dugan, M., Alland, L., Griffin, J. D., Arlinghaus, R. B., Sun, T., Kantarjian, H., and Beran, M. (2005) (2005) Clin. Cancer Res. 1 4941-4947 [DOI] [PubMed] [Google Scholar]
- 32.Weisberg, E., Manley, P., Mestan, J., Cowan-Jacob, S., Ray, A., and Griffin, J. D. (2006) Br. J. Cancer 94 1765-1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weisberg, E., Catley, L., Wright, R. D., Moreno, D., Banerji, L., Ray, A., Manley, P. W., Mestan, J., Fabbro, D., Jiang, J., Hall-Meyers, E., Callahan, L., Dellagatta, J. L., Kung, A. L., and Griffin, J. D. (2007) Blood 109 2112-2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O' Hare, T., Walters, D. K., Deininger, M. W. N., and Druker, B. J. (2005) Cancer Cell 7 117-119 [DOI] [PubMed] [Google Scholar]
- 35.Tauchi, T., and Ohyashiki, K. (2006) Int. J. Hematol. 83 294-300 [DOI] [PubMed] [Google Scholar]
- 36.Kantarjian, H., Giles, F., Wunderle, L., Bhalla, K., O'Brien, S., Wassmann, B., Tanaka, C., Manley, P., Rae, P., Mietlowski, W., Bochinski, K., Hochhaus, A., Griffin, J. D., Hoelzer, D., Albitar, M., Dugan, M., Cortes, J., Alland, L., and Ottmann, O. G. (2006) N. Engl. J. Med. 354 2542-2551 [DOI] [PubMed] [Google Scholar]
- 37.Shah, N. P., Tran, C., Lee, F. Y., Chen, P., Norris, D., and Sawyers, C. L. (2004) Science 305 399-401 [DOI] [PubMed] [Google Scholar]
- 38.Guilhot, F., Apperley, J., Kim, D. W., Bullorsky, E. O., Baccarani, M., Roboz, G. J., Amadori, S., de Souza, C. A., Lipton, J. H., Hochhaus, A., Heim, D., Larson, R. A., Branford, S., Muller, M. C., Agarwal, P., Gollerkeri, A., and Talpaz, M. (2007) Blood 109 4143-4150 [DOI] [PubMed] [Google Scholar]
- 39.Hochhaus, A., Kantarjian, H. M., Baccarani, M., Lipton, J. H., Apperley, J. F., Druker, B. J., Facon, T., Goldberg, S. L., Cervantes, F., Niederwieser, D., Silver, R. T., Stone, R. M., Hughes, T. P., Muller, M. C., Ezzeddine, R., Countouriotis, A. M., and Shah, N. P. (2007) Blood 109 2303-2309 [DOI] [PubMed] [Google Scholar]
- 40.Kantarjian, H., Jabbour, E., Grimley, J., and Kirkpatrick, P. (2006) Nat. Rev. Drug Discov. 5 717-718 [DOI] [PubMed] [Google Scholar]
- 41.Talpaz, M., Shah, N. P., Kantarjian, H., Donato, N., Nicoll, J., Paquette, R., Cortes, J., O'Brien, S., Nicaise, C., Bleickardt, E., Blackwood-Chirchir, M. A., Iyer, V., Chen, T. T., Huang, F., Decillis, A. P., and Sawyers, C. L. (2006) N. Engl. J. Med. 354 2531-2541 [DOI] [PubMed] [Google Scholar]
- 42.Druker, B. J. (2006) N. Engl. J. Med. 354 2594-2596 [DOI] [PubMed] [Google Scholar]
- 43.Ray, A., Cowan-Jacob, S. W., Manley, P. W., Mestan, J., and Griffin, J. D. (2007) Blood 109 5011-5015 [DOI] [PubMed] [Google Scholar]
- 44.von Bubnoff, N., Manley, P. W., Mestan, J., Sanger, J., Peschel, C., and Duyster, J. (2006) Blood 108 1328-1333 [DOI] [PubMed] [Google Scholar]
- 45.Ly, C., Arechiga, A. F., Melo, J. V., Walsh, C. M., and Ong, S. T. (2003) Cancer Res. 63 5716-5722 [PubMed] [Google Scholar]
- 46.Mohi, M. G., Boulton, C., Gu, T. L., Sternberg, D. W., Neuberg, D., Griffin, J. D., Gilliland, D. G., and Neel, B. G. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 3130-3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parmar, S., Smith, J., Sassano, A., Uddin, S., Katsoulidis, E., Majchrzak, B., Kambhampati, S., Eklund, E. A., Tallman, M. S., Fish, E. N., and Platanias, L. C. (2005) Blood 106 2436-2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prabhu, S., Saadat, D., Zhang, M., Halbur, L., Fruehauf, J. P., and Ong, S. T. (2007) Oncogene 26 1188-1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cmarik, J. L., Min, H., Hegamyer, G., Zhan, S., Kulesz-Martin, M., Yoshinaga, H., Matsuhashi, S., and Colburn, N. H. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 14037-14042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang, H. S., Knies, J. L., Stark, C., and Colburn, N. (2003) Oncogene 22 3712-3720 [DOI] [PubMed] [Google Scholar]
- 51.Yang, H. S., Jansen, A. P., Komar, A. A., Zheng, X. M., Merrick, W. C., Costes, S., Lockett, S. J., Sonenberg, N., and Colburn, N. H. (2003) Mol. Cell. Biol. 23 26-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Griswold, I. J., MacPartlin, M., Bumm, T., Goss, V. L., O'Hare, T., Lee, K. A., Corbin, A. S., Stoffregen, E. P., Smith, C., Johnson, K., Moseson, E. M., Wood, L. J., Polakiewicz, R. D., Druker, B. J., and Deininger, M. W. (2006) Mol. Cell. Biol. 26 6082-6093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ahmad, S., Alsayed, Y. M., Druker, B. J., and Platanias, L. C. (1997) J. Biol. Chem. 272 29991-29994 [DOI] [PubMed] [Google Scholar]
- 54.Alsayed, Y., Uddin, S., Mahmud, N., Lekmine, F., Kalvakolanu, D. V., Minucci, S., Bokoch, G., and Platanias, L. C. (2001) J. Biol. Chem. 276 4012-4019 [DOI] [PubMed] [Google Scholar]
- 55.Livak, K. J., and Schmittgen, T. D. (2001) Methods 25 402-408 [DOI] [PubMed] [Google Scholar]
- 56.Katsoulidis, E., Li, Y., Yoon, P., Sassano, A., Balasubramanian, L., Parmar, S., Varga, J., Tallman, M. S., Verma, A., and Platanias, L. C. (2005) Cancer Res. 5 9029-9037 [DOI] [PubMed] [Google Scholar]
- 57.Kannan-Thulasiraman, P., Katsoulidis, E., Tallman, M. S., Arthur, J. S., and Platanias, L. C. (2006) J. Biol. Chem. 281 22446-22452 [DOI] [PubMed] [Google Scholar]
- 58.Doggrell, S. A. (2005) Expert Opin. Investig. Drugs 14 1063-1066 [DOI] [PubMed] [Google Scholar]
- 59.Dorrello, N. V., Peschiaroli, A., Guardavaccaro, D., Colburn, N. H., Sherman, N. E., and Pagano, M. (2006) Science 314 467-471 [DOI] [PubMed] [Google Scholar]
- 60.Raught, B., Gingras, A. C., and Sonenberg, N. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 7037-7044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fingar, D. C., and Blenis, J. (2004) Oncogene 23 3151-3171 [DOI] [PubMed] [Google Scholar]
- 62.Jaccinto, E., and Hall, M. N. (2003) Nat. Rev. Mol. Cell Biol. 4 117-126 [DOI] [PubMed] [Google Scholar]
- 63.Hay, N. (2005) Cancer Cell. 8 179-183 [DOI] [PubMed] [Google Scholar]
- 64.Platanias, L. C. (2005) Nat. Rev. Immunol. 5 375-386 [DOI] [PubMed] [Google Scholar]
- 65.Hay, N., and Sonenberg, N. (2004) Genes Dev. 18 1926-1945 [DOI] [PubMed] [Google Scholar]
- 66.Bhaskar, P. T., and Hay, N. (2007) Dev. Cell. 12 487-502 [DOI] [PubMed] [Google Scholar]
- 67.Gingras, A. C., Raught, B., Gygi, S. P., Niedzwiecka, A., Miron, M., Burley, S. K., Polzkiewicz, R. D., Wyslouch-Cieszynska, A., Aebersold, R., and Sonenberg, N. (2001) Genes Dev. 15 2852-2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.LaRonde-LeBlanc, N., Santhanam, A. N., Baker, A. R., Wlodawer, A., and Colburn, N. H. (2007) Mol. Cell. Biol. 27 147-156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Goke, R., Gregel, C., Goke, A., Arnold, R., Schmidt, H., and Lankat-Buttgereit, B. (2004) Ann. N. Y. Acad. Sci. 1014 220-221 [DOI] [PubMed] [Google Scholar]
- 70.Lankat-Buttgereit, B., and Goke, R. (2003) Biol. Cell 95 515-519 [DOI] [PubMed] [Google Scholar]
- 71.Chen, Y., Knösel, T., Kristiansen, G., Pietas, A., Garber, M. E., Matsuhashi, S., Ozaki, I., and Petersen, I. (2003) J. Pathol. 200 640-646 [DOI] [PubMed] [Google Scholar]
- 72.Afonja, O., Juste, D., Das, S., Matsuhashi, S., and Samuels, H. H. (2004) Oncogene 23 8135-8145 [DOI] [PubMed] [Google Scholar]
- 73.Zhang, H., Ozaki, I., Mizuta, T., Hamajima, H., Yasutake, T., Eguchi, Y., Ideguchi, H., Yamamoto, K., and Matsuhashi, S. (2006) Oncogene 25 6101-6112 [DOI] [PubMed] [Google Scholar]
- 74.Gao, F., Zhang, P., Zhou, C., Li, J., Wang, Q., Zhu, F., Ma, C., Sun, W., and Zhang, L. (2007) Oncol. Rep. 17 123-128 [PubMed] [Google Scholar]
- 75.Ozpolat, B., Akar, U., Steiner, M., Zorrilla-Calancha, I., Tirado-Gomez, M., Colburn, N., Danilenko, M., Kornblau, S., and Berestein, G. L. (2007) Mol. Cancer Res. 5 95-108 [DOI] [PubMed] [Google Scholar]
- 76.Jansen, A. P., Camalier, C. E., and Colburn, N. H. (2005) Cancer Res. 65 6034-6041 [DOI] [PubMed] [Google Scholar]
- 77.Palamarchuk, A., Efanov, A., Maximov, V., Aqeilan, R. I., Croce, C. M., and Pekarsky, Y. (2005) Cancer Res. 65 11282-11286 [DOI] [PubMed] [Google Scholar]
- 78.Yang, H. S., Matthews, C. P., Clair, T., Wang, Q., Baker, A. R., Li, C. C., Tan, T. H., and Colburn, N. H. (2006) Mol. Cell. Biol. 26 1297-1306 [DOI] [PMC free article] [PubMed] [Google Scholar]