Abstract
Three methyl-CpG-binding domain (MBD) proteins in Arabidopsis, AtMBD5, AtMBD6, and AtMBD7, are functional in binding methylated CpG dinucleotides in vitro and localize to the highly CpG-methylated chromocenters in vivo. These proteins differ, however, in their subnuclear localization pattern; AtMBD5 and AtMBD6, each containing a single MBD motif, show preference for two perinucleolar chromocenters, whereas AtMBD7, a naturally occurring poly-MBD protein containing three MBD motifs, localizes to all chromocenters. Here we studied the significance of multiple MBD motifs for subnuclear localization and mobility in living cells. We found that the number of MBD motifs determines the subnuclear localization of the MBD protein. Furthermore, live kinetic experiments showed that AtMBD7-green fluorescent protein (GFP) has lower mobility than AtMBD5-GFP and AtMBD6-GFP, which is conferred by cooperative activity of its three MBD motifs. Thus, the number of MBD motifs appears to affect not only binding affinity and mobility within the nucleus, but also the subnuclear localization of the protein. Our results suggest that poly-MBD proteins can directly affect chromatin structure by inducing intra- and inter-chromatin compaction via bridging over multiple methylated CpG sites.
Cytosine methylation is a common epigenetic modification of most eukaryotic nuclear DNA. Intensive studies demonstrated the central role played by cytosine methylation in genome organization, gene expression, and growth and development. The way by which the methyl group is interpreted into a functional state has begun to be unraveled with the isolation and characterization of methylated DNA-binding proteins. This group of proteins includes an evolutionary conserved protein family, initially described in animals, termed methyl-CpG-binding domain (MBD)2 proteins (1). Biochemical and molecular analyses suggest that mammalian MBDs induce the formation of a repressive chromatin structure at methylated CpG (mCpG) sites by the recruitment of various chromatin factors including histone deacetylases, histone methyltransferases, and ATPase-dependent nucleosome remodeling factors (2, 3). Thus far, of the 13 AtMBD proteins found in Arabidopsis, only AtMBD5, AtMBD6, and AtMBD7 were found to bind mCpG sites in vitro and to localize in vivo to the highly methylated chromocenters (ChCs) (reviewed in Refs. 4 and 5). This chromocenter localization is CpG methylation-dependent inasmuch as it was disrupted in the hypomethylated mutants ddm1 and met1 (6). Yet, these three AtMBDs display different localization patterns within Arabidopsis nuclei; AtMBD7 is predominantly localized at all ChCs, whereas AtMBD5 and AtMBD6 show preference for two perinucleolar ChCs adjacent to the ribosomal DNA clusters (6). A major structural feature distinguishing AtMBD7 from AtMBD5 and AtMBD6 is the presence of three MBD motifs within its protein sequence. Notably, naturally occurring poly-MBD proteins appear to be unique to plants as to date no such protein has been found in animals. Here we studied the significance of having multiple MBD motifs for AtMBD7 subnuclear localization and mobility. Using fluorescence recovery after photobleaching (FRAP), we found that most of the AtMBD7 protein is less mobile than AtMBD5 and AtMBD6 and that the number of MBD motifs controls both subnuclear localization and mobility. We also found that AtMBD5 and AtMBD6 can interact with each other. We discuss the possible function of poly-MBD proteins in controlling chromatin structure.
EXPERIMENTAL PROCEDURES
Construction of AtMBD-GFP/mRFP Plasmids—Fusion of AtMBD5, AtMBD6, and AtMBD7 to GFP was described previously (6). To generate AtMBD5 and AtMBD7 derivatives fused to GFP, we amplified the various fragments by PCR using the following primers: for AtMBD5-(1-92), MBD5-S (5′-TGATATCAGATCTATGTCGAACGGCACGGATCAG-3′) and MBD5(1-92aa)-AS (5′-TCTCCCCGGGTTCCGCTGTCTTGACGGAC-3′); for AtMBD7-(1-231), MBD7-S (5′-GAGAAGATCTAGAATGCAGACGAGATCCTCTTCCTCTC-3′) and MBD7(231aa)-AS (5′-TCTCCCCGGGGTCAAGTGTAATGTTCCCG-3′); for AtMBD7-(103-231), MBD7(103aa)-S (5′-GAGGGATCCATGGATCACTGCGGAGTTGAATATG-3′) and MBD7(231aa)-AS; for AtMBD7-(173-231), MBD7(173aa)-S (5′-GAGAAGATCTATGCGCCGTGGCCATTCCAAA-3′) and MBD7(231aa)-AS; and for AtMBD7-(103-175), MBD7(103aa)-S (5′-GAGGGATCCATGGATCACTGCGGAGTTGAATATG-3′) and MBD7(175aa)-AS (5′-TCTCCCCGGGGCCACGGCGATTCTGTAAAAC-3′). The various AtMBD PCR products were digested either with BamHI and SmaI or with BglII and SmaI and ligated into BglII-SmaI sites of pUC19-35S-GFP (6). AtMBD5 × 2-GFP was constructed by PCR of AtMBD5-GFP using p35S-MBD5-GFP plasmid as a template and primers MBD5-S and GFP-AS (5′-TGCCTGCAGTCAGGTCGACTTGTATAGTTC-3′). The AtMBD5-GFP PCR product was digested with EcoRV and PstI and ligated into SmaI and PstI sites of p35S-MBD5. AtMBD5-mRFP was constructed by PCR of mRFP1 using pRSETB-mRFP1 plasmid (23) as a template and primers mRFP-S (5′-GAGCCCGGGGCCTCCTCCGAGGACGTCATC-3′) and mRFP-AS (5′-TCTCATGCATTTAGGCGCCGGTGGAGTGGCGG-3′). The mRFP1 PCR product was digested with SmaI and NsiI and ligated into SmaI and PstI sites of p35S-MBD5. The integrity of each construct was verified by sequencing. These constructs were used for protoplast transformation experiments.
Protoplast Transformation—Arabidopsis thaliana (“Columbia” ecotype) plants were grown under short-day conditions at 20 °C. At 4-6 weeks after germination, rosette leaves were collected for the isolation and transformation of protoplasts essentially as described (7). The GFP signal was detected 24 h after transfection using a laser confocal microscope (Olympus IX70, Fluoview FV500, Hamburg, Germany). Images for GFP and RFP were obtained using excitation wavelengths of 488 and 543 nm and collection through 505-525- and 610-700-nm filters, respectively.
Fluorescence Recovery after Photobleaching—FRAP experiments were carried out using the laser confocal microscope. GFP fluorescence images were obtained using an excitation wavelength of 488 nm, and signals were collected through 505-525-nm filters. In the FRAP experiments, images of 196 × 96 pixels were collected at maximum speed of ∼180 ms using 1% power laser for 2 s before the bleach and up to 10 s afterward. The bleach pulse was produced by 100% power laser for 0.1 s in a rectangular area (∼1 μm2) of the nucleus. The relative fluorescence intensity was normalized to the nonbleached signal after subtraction of the background signal. Values are averages of at least five cells from three independent experiments. Fluorescence recovery plots were drawn using Excel software (Microsoft). Statistical significance was determined using Student's t test.
Construction of HisAtMBD6 Plasmid, Nuclear Extraction, and Glutathione S-Transferase (GST) Pulldown Assays—His-AtMBD6 was constructed by PCR using pGEX-MBD6 (8) as a template and primers MBD6-S (5′-GAGGGATCCTCAGATTCTGTGGCCGGCG-3′) and MBD6-AS (5′-CTCCCCGGGTCAAGCCGACACTTTACTA-3′). The PCR product was digested with BamHI and SmaI and ligated into the same sites of pQE30 (Qiagen). Expression in Escherichia coli bacteria (BL21) and purification using nickel-agarose columns of HisAt-MBD6 were performed according to the manufacturer's protocol (Qiagen).
To isolate Arabidopsis (Columbia) nuclei, 1 g of leaves (4-week-old plants) was ground in 10 ml of nuclear isolation buffer (10 mm Tris-HCl, pH 8.0, 1.14 m sucrose, 5 mm MgCl2, 2 mm dithiothreitol). After 30 min of shaking, the mixture was filtered using a 50-μm nylon filter and centrifuged for 10 min at 500 × g. The pellet was washed once with nuclear isolation buffer, and a small fraction was taken for 4′,6-diamidino-2-phenylindole staining to test nucleus concentration and quality. For nuclear extraction, isolated nuclei were resuspended in 0.4 ml of NETN buffer (100 mm NaCl, 1 mm EDTA, 20 mm Tris, pH 8, and 0.5% Nonidet P-40) supplemented with protease inhibitor mixture (Sigma) followed by sonication three times for 20 s at 50 watts (XL-2000, Misonix). After centrifugation at 11,000 × g for 5 min, the supernatant was tested for protein concentration by the Bradford reagent. All steps (except 4′,6-diamidino-2-phenylindole staining and Bradford assay) were done at 4 °C.
Purified HisAtMBD6 (5 μg) or nuclear extract (100 μg) derived from transgenic Arabidopsis plants expressing AtMBD5-GFP was subjected to GST pulldown assay (9) using GST alone or the indicated GST-AtMBDs described previously (6, 8). Precipitated proteins were resolved by SDS-PAGE and immunoblotted using either anti-His6 monoclonal antibody (Covance) or anti-GFP (B-2) monoclonal antibody (Santa Cruz Biotechnology).
RESULTS AND DISCUSSION
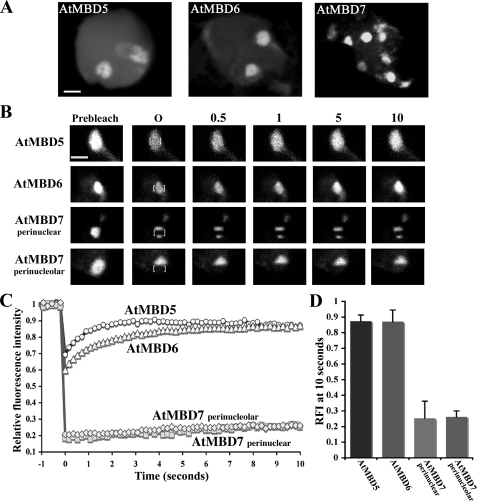
In Arabidopsis, only AtMBD5, AtMBD6, and AtMBD7 were shown to bind mCpG sites in vitro and to localize in vivo to ChCs in a CpG methylation-dependent manner (4, 5). Yet AtMBD7 is localized predominantly to all ChCs, whereas AtMBD5 and AtMBD6 show preference for two perinucleolar ChCs adjacent to the rRNA gene clusters (Ref. 6; see also Fig. 1A). Because AtMBD7 is unique in having three MBD motifs, we tested the significance of this feature for localization and binding affinity to its chromosomal sites. Recently, it has been shown that a recombinant poly-MBD protein made of the MBD motif of the mammalian Mbd1 has higher binding affinity to mCpG sites than the monomeric Mbd1 protein (10). This finding suggested that the naturally occurring poly-MBD protein AtMBD7 might display higher binding affinity for methylated sites than the monomeric AtMBD5 and AtMBD6 proteins. This proposition was tested by measuring the mobility of AtMBD proteins fused to GFP using FRAP analysis following transient expression in Arabidopsis cells (Fig. 1). Accordingly, the GFP signal was bleached by high power laser light in a rectangular area within individual chromocenters, after which fluorescence recovery of the bleached area was measured by sequential imaging scans at intervals of 0.18 s (Fig. 1, B and C). We compared the mobility of AtMBD7 at two distinct compartments, namely perinucleolar and perinuclear ChCs. FRAP analyses showed that the recovery rate of AtMBD7 is less than those of AtMBD5 and AtMBD6 (Fig. 1C). Whereas AtMBD5 and AtMBD6 were highly mobile at perinucleolar ChCs, reaching their maximal relative fluorescence intensity (RFI) of 0.9 within 5 s, AtMBD7 displayed significantly lower mobility at perinucleolar ChCs, reaching an RFI of 0.25 within 10 s (Fig. 1, C and D). The mobility of AtMBD7 at perinucleolar ChCs was indistinguishable from that of AtMBD7 at perinuclear ChCs (Fig. 1, C and D). These results confirm that AtMBD7 binds more tightly to ChCs than either AtMBD5 or AtMBD6 and that the low mobility of AtMBD7 is not a characteristic of the type of ChC but rather of the AtMBD7 protein itself. Hence, besides differences in their subnuclear localization patterns, functional AtMBDs differ in their subnuclear mobility as well. Furthermore, similar to many eukaryotic nuclear proteins, AtMBD5 and AtMBD6 are highly mobile proteins with a residence time in the range of seconds (11, 12), whereas AtMBD7 is less mobile with a residence time of minutes, similar to that displayed by the linker histone H1 (13, 14). A common feature of low mobility nuclear proteins is their interaction with chromatin via multiple binding domains (15). For example, histone H1 uses its highly basic N- and C-terminal domains together with the central globular domain to bind chromatin (16, 17).
FIGURE 1.
AtMBD7 subnuclear mobility is extremely low. A, transient expression showing different patterns of subnuclear localization of AtMBD-GFP fusion proteins in Arabidopsis nuclei. AtMBD7 localized at all ChCs, whereas AtMBD5 and AtMBD6 localized mainly at two perinucleolar ChCs. Scale bar = 2 μm. B, FRAP analyses of AtMBD-GFP proteins. A rectangular area within the ChCs (marked by brackets) was photobleached, after which sequential images were taken at the indicated times (seconds) showing the fluorescence recovery of the examined AtMBD-GFP proteins. Scale bar = 1 μm. C, average fluorescence recovery kinetics of five independent experiments of each of the GFP fusion proteins. Note the significantly lower recovery rate of AtMBD7 compared with those of AtMBD5 and AtMBD6. D, RFI of the indicated proteins after a 10-s recovery.
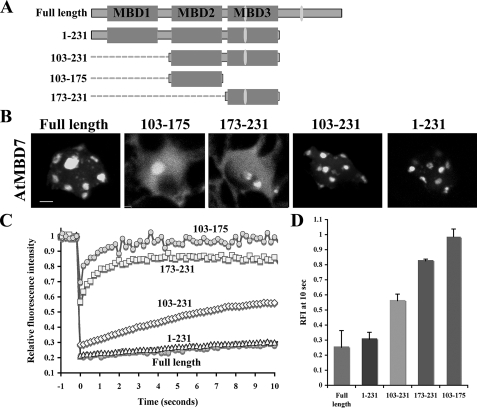
To examine further the significance of the three MBD motifs of AtMBD7 for the protein subnuclear localization and mobility, we generated truncated forms of AtMBD7 containing one (AtMBD7-(173-231)), two (AtMBD7-(103-231)), or three (AtMBD7-(1-231)) MBD motifs (Fig. 2A). Results showed that one functional MBD motif of AtMBD7 (AtMBD7-(173-231)) displayed subnuclear localization similar to that of AtMBD5 and AtMBD6 proteins; it was localized to two perinucleolar ChCs (Fig. 2B). By contrast, truncated AtMBD7 having two (103-231) or three MBD motifs (1-231) showed a subnuclear localization pattern similar to that of the full-length AtMBD7 protein. These results suggest that one MBD motif is sufficient for localization to perinucleolar ChCs and that two or three MBD motifs direct the protein to all ChCs (Fig. 2B). Thus, it appears that the number of functional MBD motifs determines the protein localization within the nucleus. Next, we carried out photobleaching experiments to test the binding capability of each one of the AtMBD7 MBD motifs. FRAP analyses of truncated AtMBD7 proteins showed inverse correlation between the number of MBD motifs and mobility rates, i.e. the highest mobility was found for a protein containing one MBD motif, intermediate mobility for two, and very low mobility for a three MBD motif-containing protein (0.82, 0.56, and 0.3 RFI at 10 s, respectively) (Fig. 2, C and D). Because the low mobility truncated AtMBD7 proteins (1-231 and 103-231) share the internal MBD motif (MBD2 motif) (see Fig. 2A) the possibility existed that their high binding affinity was conferred by this motif rather than by the number of MBD motifs. To test this possibility, we generated AtMBD7-(103-173) containing only the internal MBD2 motif (Fig. 2A) and tested its localization and mobility within the nucleus. Results showed (Fig. 2, B and C) that the internal motif fused to GFP was localized only at perinucleolar ChC and displayed high mobility similarly to other single MBD motif-containing proteins. Together, these results indicate that the number of MBD motifs in a protein determines both localization and mobility within the nucleus. The results also highlight the significance of each of the three MBD motifs and their additive contribution for increasing the binding affinity of AtMBD7 for methylated CpG-containing chromosomal regions.
FIGURE 2.
All three MBD motifs contribute to AtMBD7 subnuclear localization and mobility. A, schematic representation of AtMBD7 and its truncated forms fused to GFP. Gray boxes represent MBD motifs; closed ovals represent the predicted nuclear localization signals. B, subnuclear localization of transiently expressed AtMBD7 and its truncated constructs in Arabidopsis protoplasts. Note that the single truncated MBD proteins (173-231 and 103-175) accumulate only at perinucleolar ChCs, whereas the double (103-231) and triple (1-231) truncated MBD proteins are localized at all ChCs, resembling the full-length protein. Scale bar = 2 μm. C, FRAP experiments. The graph indicates the average fluorescence recovery kinetics of three independent experiments of each of the indicated GFP fusion proteins. D, RFI of the indicated proteins after a 10-s recovery.
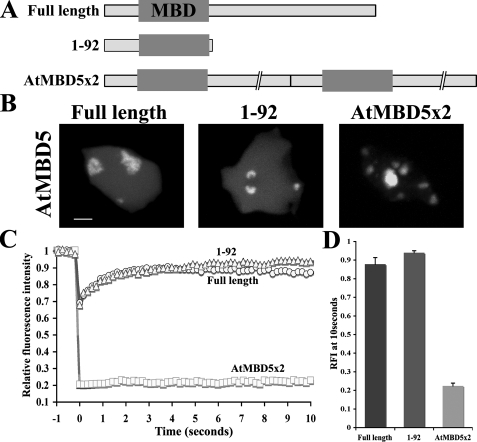
Although the level of cytosine methylation is quite similar in all 10 Arabidopsis ChCs (18, 19), AtMBD5 and AtMBD6 accumulate mostly at only two ChCs adjacent to the nucleolus. This suggests that besides the mCpG-binding activity of the MBD motif, additional factors participate in recruiting AtMBD proteins to mCpG chromosomal sites. As shown here, the chromatin-binding affinity of a single MBD motif is relatively weak. Consequently, it could be influenced more easily and eventually be directed to restricted mCpG chromosomal regions. In contrast, the high binding stringency of AtMBD7 to mCpG sites is less affected by other factors and therefore allows the protein to bind mCpG sites at all ChCs. To corroborate the importance of the number of MBD motifs for chromatin-binding affinity and protein localization, we fused two AtMBD5 (AtMBD5 × 2) in tandem to GFP to generate a single GFP fusion protein containing two MBD motifs (Fig. 3A). The AtMBD5 × 2 subnuclear distribution was extended to all ChCs, similar to that displayed by AtMBD7 (Fig. 3B). In addition, this fusion protein had less mobility (0.22 RFI at 10 s) than wild-type AtMBD5 (Fig. 3, C and D). These results demonstrate that the number of MBD motifs indeed plays an important role in determining localization as well as intranuclear mobility of MBD proteins.
FIGURE 3.
Fusion of two AtMBD5 proteins synergistically increases the binding affinity to ChCs. A, schematic representation of AtMBD5 and its truncated/fusion derivatives fused to GFP. Gray boxes represent MBD motifs. B, subnuclear localization of the indicated proteins in Arabidopsis protoplasts. Note the accumulation of AtMBD5 × 2 at all ChCs. Scale bar = 2 μm. C, FRAP analysis of three independent experiments using the indicated GFP fusion proteins. Note the significant reduction in the recovery rate of AtMBD5 × 2 compared with the single AtMBD5 protein. D, RFI of the indicated proteins after a 10-s recovery.
The high mobility displayed by many chromatin-associated proteins was suggested to provide a mechanism for generating plasticity in genome expression (11). Based on these findings, we suggest that the high dynamics of nuclear proteins could have another role in specifying their subnuclear localization. This is especially relevant for protein family members that contain a common chromatin-binding domain but are destined for different chromosomal targets.
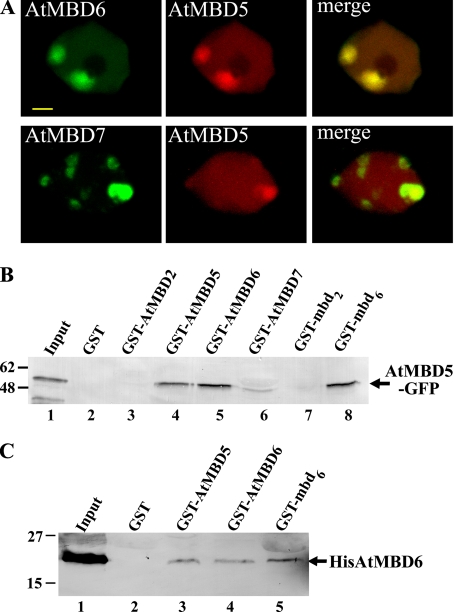
Although the mobility of AtMBD5 and AtMBD6 is high (Fig. 1), neither of them displayed full fluorescence recovery, suggesting that a small portion (∼10%) of these proteins is relatively immobile. We have shown previously that GST-AtMBD5 precipitated AtMBD6-GFP from nuclear extracts derived from transgenic plants expressing AtMBD6-GFP (6), suggesting that both proteins share the same multiprotein complex and bind to one another. The second possibility predicts the formation of a heteroduplex protein complex having two MBD motifs, which could lead to increased chromatin-binding affinity and therefore to reduction in subnuclear mobility. To address a possible physical interaction between AtMBD5 and AtMBD6 proteins, we transiently coexpressed AtMBD5-mRFP in Arabidopsis cells with either AtMBD6-GFP or AtMBD7. Fig. 4A shows that AtMBD5 perfectly colocalizes with AtMBD6 but only partially with AtMBD7. Next, we examined the ability of AtMBD5 and AtMBD6 to precipitate AtMBD5-GFP from nuclear extracts prepared from transgenic Arabidopsis expressing AtMBD5-GFP using GST pulldown assays. Precipitated proteins were resolved by SDS-PAGE and immunoblotted using anti-GFP antibody. Fig. 4B shows that among the GST-AtMBD proteins tested, GST-AtMBD5 (lane 4) and GST-AtMBD6 (lane 5) precipitated AtMBD5-GFP; slight interaction was also evident with the GST-AtMBD7 protein but not with the GST-AtMBD2 protein. Furthermore, the MBD motif of AtMBD6 (GST-mbd6, lane 8), but not of AtMBD2 (GST-mbd2, lane 7), was sufficient to pull down AtMBD5-GFP from nuclear extracts. To learn whether this interaction is direct or mediated by additional nuclear proteins, we performed pulldown experiments using His-tagged AtMBD6 recombinant protein. Purified HisAt-MBD6 was incubated with GST-AtMBDs and precipitated with glutathione-Sepharose beads. Western blot experiments using anti-His6 antibody verified physical interaction of HisAtMBD6 and GST-AtMBD5, GST-AtMBD6, and GST-mbd6 (Fig. 4C). Altogether, these results suggest that AtMBD5 and AtMBD6 can form both homo- and heterodimers through their MBD motifs, although the involvement of additional protein domains in this dimerization cannot be excluded. Therefore, AtMBD5 and AtMBD6 might reside in Arabidopsis nuclei either as monomers with low chromatin-binding affinity and high mobility or as homo- or heterodimers with higher binding affinity and lower mobility. This dimerization may explain the extended localization of a small fraction of AtMBD6 and AtMBD5 from perinucleolar ChCs to additional ChCs at the periphery of the nucleus (6).
FIGURE 4.
Colocalization and interaction between AtMBD5 and AtMBD6. A, colocalization of AtMBD5-mRFP with AtMBD6-GFP or AtMBD7-GFP in Arabidopsis protoplasts. B, GST-AtMBD pulldown assays of AtMBD5-GFP. GST alone and the indicated GST fusion proteins were mixed with nuclear extract derived from transgenic Arabidopsis expressing AtMBD5-GFP. Precipitated proteins were resolved by SDS-PAGE and immunoblotted using anti-GFP antibody to detect AtMBD5-GFP. GST-mbd2 and GST-mbd6 represent GST fusion proteins containing only the MBD motif of AtMBD2 and AtMBD6, respectively. Input lane indicates 12.5% of the input proteins. C, GST-AtMBD pulldown assays of HisAtMBD6. GST alone and the indicated GST fusion proteins were mixed with purified HisAtMBD6 recombinant protein. Precipitated proteins were resolved by SDS-PAGE and immunoblotted using anti-His antibody to detect HisAtMBD6. Input lane indicates 50% of the input HisAtMBD6 protein.
Notably, AtMBD7 represents a unique type of MBD protein not found in animals having more than one MBD motif. The plant chromatin data base (www.chromdb.org) lists such proteins in poplar, maize, and rice. Interestingly, monocots lack homologues to functional AtMBD proteins (20), which raises doubts as to the relevance of monocot MBD proteins in interpreting the cytosine methylation signal. On the other hand, monocots possess multiple poly-MBD proteins, e.g. the rice genome encodes for five putative poly-MBD proteins. It is thus possible that monocots may have evolved a specific requirement for poly-MBD proteins to interpret the CpG methylation signal into a functional state.
The occurrence of a poly-MBD protein such as AtMBD7 predicts that one molecule can potentially bind multiple mCpG sites (4). Accordingly, we propose that one molecule of AtMBD7 or a multidimer composed of AtMBD5 and/or AtMBD6 proteins can bridge over several mCpG sites, bringing DNA strands close to each other and thus enhancing chromatin compaction and gene silencing. This proposition is consistent with a model for nucleosome array compaction by chromatin architectural proteins depicted in a recent review (15). It appears that chromatin architectural proteins involved in chromatin compaction possess several chromatin-binding domains that simultaneously bind two or more chromatin-binding sites (15, 16) or are capable of oligomerizing and cooperatively binding chromatin. Likewise, the effect of the single MBD motif-containing MeCP2, the founder member of the MBD protein family, on chromatin compaction appears to be mediated by multiple chromatin interaction regions in addition to the MBD motif (21, 22). In conclusion, the three MBD motifs in AtMBD7 were found to increase the protein binding affinity for chromatin and to determine its subnuclear localization and could potentially induce chromatin compaction of methylated CpG chromosomal regions.
Acknowledgments
We thank Yigal Avivi for critical reading and editing of the manuscript, and Vladimir Kiss for confocal assistance. The pRSETB-mRFP1 plasmid was kindly provided by Prof. Roger Y. Tsien, University of California San Diego, La Jolla, CA.
This work was supported in part by the Israel Science Foundation and by the Jewish Colonization Association. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: MBD, methyl-CpG-binding domain; AtMBD, A. thaliana MBD; ChCs, chromocenters; FRAP, fluorescence recovery after photobleaching; GFP, green fluorescent protein; mRFP, monomeric red fluorescent protein; mCpG, methylated CpG; RFI, relative fluorescence intensity; GST, glutathione S-transferase.
References
- 1.Hendrich, B., and Bird, A. (1998) Mol. Cell. Biol. 18 6538-6547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klose, R. J., and Bird, A. P. (2006) Trends Biochem. Sci. 31 89-97 [DOI] [PubMed] [Google Scholar]
- 3.Fatemi, M., and Wade, P. A. (2006) J. Cell Sci. 119 3033-3037 [DOI] [PubMed] [Google Scholar]
- 4.Zemach, A., and Grafi, G. (2007) Trends Plant Sci. 12 80-85 [DOI] [PubMed] [Google Scholar]
- 5.Grafi, G., Zemach, A., and Pitto, L. (2007) Biochim. Biophys. Acta 1769 287-294 [DOI] [PubMed] [Google Scholar]
- 6.Zemach, A., Li, Y., Wayburn, B., Ben-Meir, H., Kiss, V., Avivi, Y., Kalchenko, V., Jacobsen, S. E., and Grafi, G. (2005) Plant Cell 17 1549-1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoo, S.-D., Cho, Y.-H., and Sheen, J. (2007) Nat. Protocols 2 1565-1572 [DOI] [PubMed] [Google Scholar]
- 8.Zemach, A., and Grafi, G. (2003) Plant J. 34 565-572 [DOI] [PubMed] [Google Scholar]
- 9.Grafi, G., Burnett, R. J., Helentjaris, T., Larkins, B. A., DeCaprio, J. A., Sellers, W. R., and Kaelin, W. G. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 8962-8967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jorgensen, H. F., Adie, K., Chaubert, P., and Bird, A. P. (2006) Nucleic Acids Res. 34 e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phair, R. D., Scaffidi, P., Elbi, C., Vecerová, J., Dey, A., Ozato, K., Brown, D. T., Hager, G., Bustin, M., and Misteli, T. (2004) Mol. Cell. Biol. 24 6393-6402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zemach, A., Li, Y., Ben-Meir, H., Oliva, M., Mosquna, A., Avivi, Y., Ohad, N., and Grafi, G. (2006) Plant Cell 18 133-145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Misteli, T., Gunjan, A., Hock, R., Bustin, M., and Brown, D. T. (2000) Nature 408 877-882 [DOI] [PubMed] [Google Scholar]
- 14.Lever, M. A., Th'ng, J. P., Sun, X., and Hendzel, M. J. (2000) Nature 408 873-877 [DOI] [PubMed] [Google Scholar]
- 15.McBryant, S. J., Adams, V. H., and Hansen, J. C. (2006) Chromosome Res. 14 39-51 [DOI] [PubMed] [Google Scholar]
- 16.Roque, A., Orrego, M., Ponte, I., and Suau, P. (2004) Nucleic Acids Res. 32 6111-6119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Catez, F., Ueda, T., and Bustin, M. (2006) Nat. Struct. Mol. Biol. 13 305-310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zilberman, D., Gehring, M., Tran, R. K., Ballinger, T., and Henikoff, S. (2007) Nat. Genet. 39 61-69 [DOI] [PubMed] [Google Scholar]
- 19.Zhang, X., Yazaki, J., Sundaresan, A., Cokus, S., Chan, S. W., Chen, H., Henderson, I. R., Shinn, P., Pellegrini, M., Jacobsen, S. E., and Ecker, J. R. (2006) Cell 126 1189-1201 [DOI] [PubMed] [Google Scholar]
- 20.Springler, N. M., and Kaeppler, S. M. (2005) Plant Physiol. 138 92-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Georgel, P. T., Horowitz-Scherer, R. A., Adkins, N., Woodcock, C. L., Wade, P. A., and Hansen, J. C. (2003) J. Biol. Chem. 278 32181-32188 [DOI] [PubMed] [Google Scholar]
- 22.Nikitina, T., Shi, X., Ghosh, R. P., Horowitz-Scherer, R. A., Hansen, J. C., and Woodcock, C. L. (2007) Mol. Cell. Biol. 27 864-877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campbell, R. E., Tour, O., Palmer, A. E., Steinbach, P. A., Baird, G. S., Zacharias, D. A., and Tsien, R. Y. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 7877-7882 [DOI] [PMC free article] [PubMed] [Google Scholar]