Abstract
Synaptic adhesion-like molecules (SALMs) are a newly discovered family of adhesion molecules that play roles in synapse formation and neurite outgrowth. The SALM family is comprised of five homologous molecules that are expressed largely in the central nervous system. SALMs 1–3 contain PDZ-binding domains, whereas SALMs 4 and 5 do not. We are interested in characterizing the interactions of the SALMs both among the individual members and with other binding partners. In the present study, we focused on the interactions formed by the five SALM members in rat brain and heterologous cells. In brain, we found that SALMs 1–3 strongly co-immunoprecipitated with each other, whereas SALMs 4 and 5 did not, suggesting that SALMs 4 and 5 mainly form homomeric complexes. In heterologous cells transfected with SALMs, co-immunoprecipitation studies showed that all five SALMs form heteromeric and homomeric complexes. We also determined if SALMs could form trans-cellular associations between transfected heterologous cells. Both SALMs 4 and 5 formed homophilic, but not heterophilic associations, whereas no trans associations were formed by the other SALMs. The ability of SALM4 to form trans interactions is due to its extracellular N terminus because chimeras of SALM4 N terminus and SALM2 C terminus can form trans interactions, whereas chimeras of SALM2 N terminus and SALM4 C terminus cannot. Co-culture experiments using HeLa cells and rat hippocampal neurons expressing the SALMs showed that SALM4 is recruited to points of contact between the cells. In neurons, these points of contact were seen in both axons and dendrites.
Critical steps in the development of the nervous system, including cell migration, neurite outgrowth, growth cone guidance, and synapse formation, depend on cellular interactions mediated by adhesion molecules (1–4). A number of adhesion molecules that are essential to the proper development of the nervous system have been identified (5–7). The importance of these molecules is illustrated in cases of dysfunctional adhesion molecules that have been associated with specific disorders in humans, including SLITRK1 in Tourette syndrome (8) and neuroligin in autism (9, 10). Whereas our understanding of the role of adhesion molecules in neuronal development is rapidly increasing, little is known about their functions, and it is likely that many remain to be identified.
Synaptic adhesion-like molecules (SALMs)2 are a recently identified class of adhesion molecules that are highly enriched in brain (11–13). All five members of the SALM family contain extracellular leucine-rich repeats (LRR), an immunoglobulin C2-like (IgC2) domain, a fibronectin type III (FNIII) domain, a transmembrane domain, and a cytoplasmic C-terminal tail. SALMs 1–3 contain a C-terminal PDZ-binding domain (PDZ-BD), which associates with the PSD-95 family of proteins (11–13). SALMs are expressed early in the developing nervous system and persist into adulthood. They are present at the synaptic membrane as well as at non-synaptic locations. SALM1 overexpressed in cultured hippocampal neurons 4 days in vitro enhances neurite outgrowth. SALM1 also enhances the surface expression of N-methyl-d-asparatate receptors, and this may involve either a direct SALM/N-methyl-d-asparatate receptor interaction or an indirect interaction through a PDZ protein (11). Overexpression of SALM2 increases the number of excitatory synapses and dendritic spines, whereas knock-down of SALM2 decreases the number of excitatory synapses and spines (12). Thus, SALMs, like other adhesion molecules, have multiple functions in the nervous system.
The function of adhesion molecules depends on their associations with binding partners through their extracellular domains. They can form trans interactions across cellular junctions or cis interactions within the same membrane. These trans interactions can be homophilic as in the case of cadherins (14), heterophilic as in the case of neuroligin/neurexin (15, 16), or mediated by an interaction with a secreted molecule as is the case for Slit/Robo (17). In addition, adhesion molecules can form homomeric cis associations, like the nectins (18), or heteromeric cis associations like axonin-1 with NgCAM (19, 20). In this study, we investigated the associations formed between SALM family members. All five SALMs formed homomeric or heteromeric complexes when co-expressed in heterologous cells. However, only SALMs 4 and 5 formed trans associations.
EXPERIMENTAL PROCEDURES
SALM 1–5 cDNA Constructs—Cloning of the SALM family members was previously described (11). The PCR products were subcloned into the Topo vectors (Invitrogen), and then into the mammalian expression vector pcDNA3.1 (Invitrogen). The SALMs were epitope tagged by inserting a Myc tag (EQKLISEEDL) or hemagglutinin (HA) tag (EYPYDVPDYA) after the predicted signal peptide using the QuikChange site-directed mutagenesis method (Stratagene). The Myc tag was inserted into the sequence of SALM1 before residue 21, the SALM2 Myc tag and HA tag were inserted before residue 33, and SALM4 was Myc-tagged before residue 24. The C-terminal deletion of SALM1 (Myc-SALM1ΔCT) was constructed by inserting a stop codon immediately after the transmembrane domain, resulting in termination at Lys558. The Myc-SALM1Δ4 and Myc-SALM2Δ7 constructs were also generated using QuikChange mutagenesis by inserting a stop codon before the PDZ-binding motif, resulting in a 4-amino acid truncation of SALM1 and 7-amino acid deletion of SALM2. SALM5 pGW1 was a gift from Dr. Eunjoon Kim (KAIST, Korea) (12).
Chimeras of SALM2 and SALM4 were made by creating a BamHI site after the transmembrane domain of SALM2 and SALM4. A single amino acid was changed in the HA-SALM2 (D574S) and the Myc-SALM4 (G566S) constructs for subcloning purposes. The C-terminal region of each construct was then excised and exchanged by utilizing the BamHI and EcoRI sites from the multiple cloning site in the pcDNA3.1 vector.
Anti-SALM Antibody Characterization and Purification—Peptides were synthesized (Princeton Biomolecules, Langhorne, PA) and conjugated to keyhole limpet hemocyanin through a cysteine added to the N terminus of the peptide. Antisera were produced in rabbits (Covance, Denver, PA). Antibodies were purified using antigen affinity columns with peptides coupled to SulfoLink gel (Pierce) following the manufacturer's instructions. The peptide sequence used for generating a SALM1 N-terminal antibody was [NH2]-NSTSRMAPPKSRLS-[COOH] (amino acids (aa) 384–397) and that for the C-terminal antibody was [NH2]-GAGTSSRGHHSDREPL-[COOH] (aa 683–698). SALM2 N-terminal antibody was [NH2]-GSSDIATPGRPGAN-[COOH] (aa 407–420), SALM2 C-terminal antibody was [NH2]-CEETSGEESRAMTGPRR-[COOH] (aa 651–666), SALM3 N-terminal antibody was [NH2]-TSAEGGRPGPSDI-[COOH] (aa 377–389), SALM3 C-terminal antibody was [NH2]-CRGVGGSAERLEESVV-[COOH] (aa 621–636, PDZ binding motif), SALM4 N-terminal antibody was [NH2]-CDPPRDGEPDAGTP-[COOH] (aa 397–410), SALM4 C-terminal antibody was [NH2]-CEPWGPSHEPAGP-[COOH] (aa 614–626), SALM5 N-terminal antibody was [NH2]-NASSSNGDTKMSQDK-[COOH] (aa 406–420), and SALM5 C-terminal antibody was [NH2]-KRKTGTKPSAEPQSE-[COOH] (aa 644–658).
An antibody that recognizes the leucine-rich repeats of the SALMs was made by expressing the SALM2 LRR (residues 38–297) as a glutathione S-transferase fusion protein. The LRR region of SALM2 was subcloned into the BamHI site of pGEX4T-1 (GE Healthcare), which contained an N-terminal glutathione S-transferase tag. The construct was transformed into chemically competent BL21 DE3 star Escherichia coli (Invitrogen) according to the manufacturer's instructions. Protein expression was induced with isopropyl β-d-thiogalactoside, cultures were centrifuged and each isolated cell pellet was resuspended in Tris-buffered saline (TBS) (15 mm Tris-Cl, 150 mm NaCl, pH 7.4), supplemented with protease inhibitors (Complete tablets, Roche Applied Sciences), and lysed with 100 mg/ml lysozyme (Sigma). Then 15 mm dithiothreitol, 10 mm EDTA, and 1.5% Sarkosyl were added. After high-speed centrifugation (125,000 × g for 60 min at 4 °C), the Sarkosyl was neutralized with a final concentration of 2–4% Triton X-100. The protein was purified on a glutathione-Sepharose column (GE Healthcare) and eluted with 15 mm glutathione. Eluate was concentrated using a Centricon filter (Amicon/Millipore, Bedford, MA) and dialyzed against PBS overnight at 4 °C.
Transient Transfection Studies—HEK293 fibroblasts were plated at 20–30% confluence and maintained at 50–90% confluence in Dulbecco's modified Eagle's medium (Invitrogen) containing heat-inactivated fetal bovine serum (Invitrogen), 2 mm l-glutamine, and 1 mm sodium pyruvate. Cells were transiently transfected with expression constructs using a calcium phosphate precipitation protocol (21). To enhance transfection efficiency, an equimolar amount of pAdVantage (Promega, Madison, WI) was present during transfection. Cells were collected 48 h post-transfection and lysed with RIPA buffer (10% glycerol, 1% Nonidet P-40, 0.4% deoxycholate, 0.05% SDS, 150 mm NaCl, 5 mm EGTA, and 5 mm EDTA in 50 mm Tris, pH 7.4, plus protease inhibitors). Lysates were cleared by ultracentrifugation (110,000 × g for 30 min at 4 °C) and 500 μg was used for immunoprecipitations. Protein concentration was quantified using the BCA assay (Pierce).
Immunoprecipitation and Western Blotting—Rat brain tissues were homogenized in 50 mm Tris-HCl, pH 7.5, with protease inhibitors (Complete tablets, Roche). The tissue was stored as homogenate at -80 °C or solubilized with 1% sodium deoxycholate. The solubilized protein was incubated with detergent at 37 °C for 45 min, and then centrifuged to isolate the soluble fraction (110,000 × g for 30 min at 4 °C). For immunoprecipitations, 5 μg of antibody was added to 500 μl of the membrane fraction and incubated with prewashed protein A/G-agarose beads (Pierce) for 4 h at 4 °C. Immunoprecipitations were carried out with SALM antibodies or the corresponding nonspecific control antibodies, such as mouse or rabbit IgG (Jackson ImmunoResearch Laboratories or Zymed Laboratories Inc., respectively). Resin pellets from immunoprecipitations were washed extensively with 500 mm NaCl/TBS, 0.1% Triton X-100/TBS, and TBS, resuspended in 2× SDS loading buffer, heated at 95 °C for 5 min, then resolved by SDS-PAGE on 10 or 4–20% Tris glycine gels (Invitrogen) and transferred to polyvinylidene difluoride membranes. Immunoblotting was performed using peroxidase-coupled secondary antibodies (GE Healthcare) and chemiluminescence (Amersham Biosciences ECL Plus). Antibodies were stripped by incubating the membrane in buffer containing 62.5 mm Tris-Cl, pH 6.7, 2% SDS, and 20 mm dithiothreitol when reprobing was necessary.
Immunocytochemistry—HeLa cells were transiently transfected with expression constructs using calcium phosphate precipitation. For surface staining, HeLa cells were washed with cold PBS (supplemented with 1 mm magnesium and 0.1 mm calcium) (PMC), and incubated on ice with primary antibodies for 1 h. Cells were washed, blocked with 10% normal goat serum and 1% bovine serum albumin in PMC, and incubated with Alexa 568 secondary antibody (Invitrogen) on ice for 30 min. Cells were then fixed with 4% paraformaldehyde at room temperature, washed with PBS, and processed for total staining. For total staining, cells were permeabilized with 0.1% Triton X-100/PBS for 5 min, blocked with 10% normal goat serum in PBS for 1 h, incubated with primary antibody, and stained with Alexa 488 or 568 secondary antibodies (Invitrogen). Total staining was performed at room temperature. Coverslips were mounted onto slides using ProLong Antifade mounting media (Invitrogen). For co-culture experiments in HeLa cells, SALMs were transiently transfected using calcium phosphate precipitation and incubated overnight at 37 °C. Cells were then washed with PBS, trypsinized, and replated with HeLa cells expressing a different family member. Cells were fixed and permeabilized for total staining after 24 h of incubation. For co-culture experiments with hippocampal neurons and HeLa cells, neurons were cultured as previously described (11, 22); co-culture methodologies are reviewed by Biederer and Scheiffele (23). Neurons were transfected with SALM cDNA at 7 days in vitro using calcium phosphate precipitation (Clontech, Palo Alto, CA). Twenty-four hours after transfection, HeLa cells transfected with the same SALM family member were plated on top of the neurons. Cells were incubated overnight at 37 °C and then processed for total staining. Images were acquired using the ×60 or ×40 oil objective of an E-1000 Nikon microscope or ×63 objective of an LSM 510 confocal microscope.
Antibody Assay—HeLa cells that were transfected with Myc-SALM4 were treated for 24 h with anti-LRR, anti-SALM4-CT, or anti-Myc antibodies (9E10, American Type Culture Collection, Manassas, VA). Myc-SALM2-transfected cells were treated with anti-Myc antibodies as a control. After 24 h, the cells were surface stained with anti-Myc antibodies, fixed, and processed for total staining. Antibodies directed toward the C termini of SALM2 and SALM4 (anti-SALM2-CT and anti-SALM4-CT) were used for total staining.
Calcium Dependence Adhesion Assay—To determine the effect of calcium on SALM trans interactions, we followed previously published protocols (24, 25). HeLa cells were transfected with SALM4 using Lipofectamine 2000 (Invitrogen/GIBCO) to minimize the presence of calcium. Cells were either switched to calcium-free Dulbecco's modified Eagle's medium (Invitrogen) supplemented with heat-inactivated fetal bovine serum, 2 mm l-glutamine, and 1 mm sodium pyruvate 24 h after transfection or maintained in normal Dulbecco's modified Eagle's medium (containing heat-inactivated fetal bovine serum, 2 mm l-glutamine, and 1 mm sodium pyruvate). Twenty hours after addition of calcium-free media, cells were processed for total staining (24). Cells that were maintained in normal media were treated with 0.1 mm EGTA for 30 min at 37 °C and processed for total staining (25). All cells were stained with an antibody directed toward the C terminus of SALM4.
Light and Electron Microscopy—Pre-embedding light/electron microscope methods for immunoperoxidase labeling have been described previously (26–28). HeLa cells were fixed and processed for immunoperoxidase labeling using the Vectastain ABC kit and 3,3′-diaminobenzidine peroxidase substrate kit (Vector Laboratories, Burlingame, CA). After the 3,3′-diaminobenzidine reaction, cultures were re-fixed in 2% glutaraldehyde in 0.1 m cacodylate buffer and then in 1% osmium tetroxide in the same buffer, then dehydrated in an alcohol series and embedded in epon. The glass coverslip was dissolved with hydrofluoric acid prior to thin sectioning for electron microscopy. Two separate experiments were conducted. In the first experiment, a series of HeLa cells cultures, transfected with Myc-SALM4, were fixed via three different protocols: 4% paraformaldehyde, 4% paraformaldehyde + 0.1% glutaraldehyde, and 2% glutaraldehyde. All cultures were labeled with the Myc antibody and processed for 3,3′-diaminobenzidine as noted above. Labeled cells and adjacent control cells that lacked labeling were photographed with a light microscope and then cut out of the epon block and examined with electron microscopy. Structures visible with electron microscopy were matched to the corresponding structures seen in the light microscope images. We also ran another experiment in which HeLa cells were plated at three different densities and transfected and processed as above, except that we used the SALM4 C-terminal antibody instead of the Myc antibody. These cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% saponin (26). Control coverslips that lacked the primary antibody showed no labeling. Results of this latter study are not shown, but were similar to those described for the study done with the Myc antibody.
For immunogold labeling, sections of the rat brain were processed as described previously (28, 29). Rats were perfused with 4% paraformaldehyde plus 0.5% glutaraldehyde, then frozen sections were freeze-substituted in a Leica AFS (Vienna, Austria), infiltrated with Lowicryl HM-20 resin, and thin sections were labeled with primary antibody followed by immunogold (Ted Pella, Redding, CA). Sections of hippocampus from P10 and P35 rats were labeled with SALM4 C-terminal antibody at 3.8 μg/ml and 10 nm immunogold. In another study, sections from hippocampus, cerebellum, and olfactory bulb of two P37 rats were labeled with the antibody at 10 μg/ml and 10 nm immunogold. Control sections (hippocampus) lacking the primary antibody showed only rare gold. All figures were processed in Adobe Photoshop with minimal use of levels, brightness, and contrast features, which were employed uniformly over the images. All animal procedures were done in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (National Institutes of Health publication 85–23) under National Institute on Deafness and Other Communication Disorders protocol 1167-07.
RESULTS
SALM Antibody Characterization—A series of antibodies to the individual SALMs were used to characterize the interactions between SALMs 1–5. To determine antibody specificity, the family members were subcloned into mammalian expression vectors and transfected into HEK293 cells. SALM lysate was examined on an SDS-PAGE gel, and Western blots were probed with anti-SALM1–5 antibodies (supplemental Fig. S1). All of the SALM antibodies used in this study were specific to the individual SALM to which the antibody was directed, except the SALM3 C-terminal PDZ-BD antibody, which recognized both SALM1 and SALM3. The calculated molecular masses of the SALMs range between 65 and 85 kDa. However, the SALMs were detected at higher molecular masses between 82 and 116 kDa on Western blots, suggesting that they undergo extensive post-translational modification. Digestion with N-glycosidase indicated that the SALMs are N-linked glycosylated (supplemental Fig. S1; data not shown).
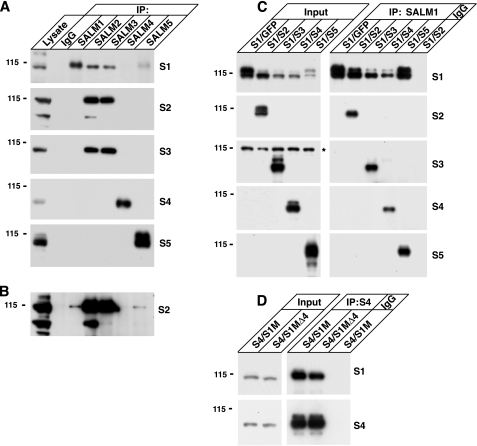
SALMs Form Homomeric and Heteromeric Complexes—To examine SALM complex formation in the brain, we performed immunoprecipitations from rat brain solubilized in 1% deoxycholate (Fig. 1A). SALMs 1–3 formed heteromeric complexes with each other. Whereas SALM2 and SALM3 strongly associated in brain, a longer exposure time of the second panel in Fig. 1A showed that SALM2 also weakly co-immunoprecipitated with SALMs 1 and 5 (Fig. 1B). However, SALMs 4 and 5 mostly did not co-immunoprecipitate with other SALMs, suggesting that they may function mainly as homomeric complexes in the brain. On the other hand, co-immunoprecipitation studies from HEK293 cells transiently transfected with the SALMs showed that all five SALM family members can form heteromers (Fig. 1C). SALMs 2–5 co-immunoprecipitated with SALM1 when SALM2, SALM3, SALM4, or SALM5 were co-transfected with SALM1 in HEK293 cells. We also performed immunoprecipitations from HEK293 lysate expressing other combinations of SALMs, such as SALM5 with SALMs 1–4, with similar results (data not shown). In all combinations, SALM family members co-immunoprecipitated with each other. As a control, we tested if an interaction occurred after cell lysis. The SALMs did not co-immunoprecipitate together if lysate from cells expressing one of the SALMs alone was mixed with lysate expressing another (data not shown).
FIGURE 1.
SALMs form complexes in brain and heterologous cells. A, rat forebrain (P15) was solubilized with 1% deoxycholate. Proteins were immunoprecipitated with anti-SALM 1–5 antibodies or control IgG and probed with anti-SALM antibodies (SALM1–SALM5 = S1–S5). SALMs 1–3 form heteromeric complexes in brain. SALMs 4 and 5 do not strongly co-immunoprecipitate with SALMs 1–3 from rat brain lysate, suggesting that SALMs 4 and 5 may function as homomeric complexes in brain. B, a longer exposure time of the second panel in A indicates that SALM2 also co-immunoprecipitates with SALM1 and SALM5, but not as abundantly as its association with SALM3. C, SALM1 was co-transfected with SALMs 2–5 in HEK293 cells (transfections shown in columns above the panel; input = immunoblots of the lysates). Proteins were solubilized with RIPA buffer and immunoprecipitated with anti-SALM1 antibodies or control rabbit IgG. Blots were then probed with anti-SALM 1–5. SALMs 2–5 co-immunoprecipitate with SALM1, indicating that all the SALMs can form heteromeric complexes when expressed together in HEK293 cells (asterisk indicates a nonspecific band). D, Myc-SALM1 (S1M) or Myc-SALM1Δ4 (S1MΔ4), which we have previously shown is not expressed on the cell surface (11), were co-transfected with SALM4. Both co-immunoprecipitate with SALM4, suggesting that SALM1 can form a complex with SALM4 within intracellular compartments.
We have shown previously that Myc-SALM1Δ4 is not expressed on the surface of heterologous cells (11). Co-expression of Myc-SALM1Δ4 with other SALM family members also does not result in its surface expression (data not shown). Both Myc-SALM1 and Myc-SALM1Δ4 co-immunoprecipitated with SALM4 when co-transfected in HEK293 cells (Fig. 1D), indicating that the SALMs can form heteromeric complexes within intracellular compartments before they reach the cell surface. These results suggest that SALMs on the cell surface are present in cis configurations.
The SALM Family Members Associate through Their Extracellular N Termini—The SALMs also formed homomeric complexes. HA-tagged SALM2 co-immunoprecipitated with Myc-tagged SALM2 when both constructs were co-transfected in HEK293 cells (Fig. 2A). Therefore, the SALMs were able to form homomeric complexes, as well as heteromeric complexes with other SALM family members. To examine whether the SALMs associate through an extracellular interaction, a SALM1 construct with a C-terminal truncation was expressed in HEK293 cells with full-length SALMs 1–4 (Fig. 2B). Deletion of the SALM1 C terminus did not disrupt the ability of full-length SALMs 1–4 to associate with SALM1 (Fig. 2B), demonstrating that SALM1 can form homomeric and heteromeric complexes through its extracellular domain. When SALMs 2–4 were immunoprecipitated, SALM1ΔCT was co-immunoprecipitated (Fig. 2C).
FIGURE 2.
SALM family members associate through their extracellular N termini. A, full-length HA-tagged SALM2 was co-transfected with full-length Myc-SALM2. Proteins were solubilized and immunoprecipitated (IP) with anti-HA or anti-Myc. HA-SALM2 co-immunoprecipitates with Myc-SALM2. B, Myc-SALM1ΔCT (S1MΔCT) was co-transfected with full-length SALMs 1–4. Proteins were solubilized with RIPA buffer. Myc-SALM1ΔCT was immunoprecipitated with anti-Myc antibodies and blots were probed with antibodies to SALMs 1–4. All four SALMs co-immunoprecipitate with Myc-SALM1ΔCT (arrowhead indicates full-length SALM1). C, immunoprecipitation of SALMs 2–4 from the same lysate also shows that Myc-SALM1ΔCT co-immunoprecipitates with the full-length SALMs. These results suggest that the SALMs interact with each other through their extracellular N-terminal domains.
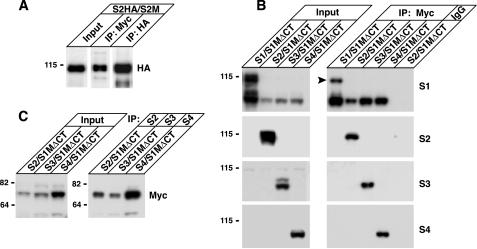
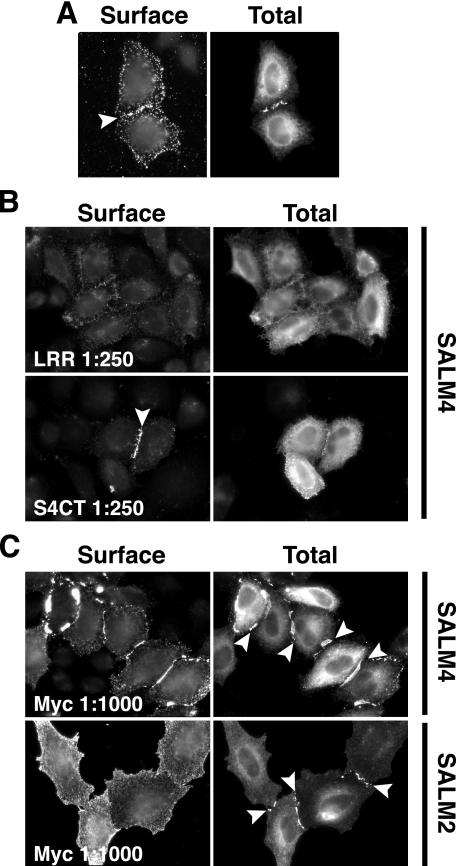
SALMs 4 and 5 Form Homophilic Complexes Across Cell Junctions—We next examined whether the SALMs interacted with each other in trans, across cellular junctions. SALMs were singularly transfected in HeLa cells that were plated on coverslips. The cells were fixed, detergent permeabilized, and stained for SALMs 1–5 to determine whether the SALMs associated between neighboring cells (Fig. 3A). SALMs 1–3 did not appear to form any interactions across cell junctions. Similar to our results, the ectodomain of SALM2 fused to alkaline phosphatase did not bind SALM2 expressed on the surface of heterologous cells (12). However, clusters formed between the cell membranes of adjacent cells expressing SALM4 or SALM5 (Fig. 3A, arrowheads). This clustering was also detected between cells when an N-terminal SALM4 antibody was used to stain for surface expression (Fig. 3B). Immunoperoxidase staining and electron microscopy showed clusters between SALM4-transfected HeLa cells as well (Fig. 3C). A major difference between SALMs 4 and 5 and SALMs 1–3 is the presence of PDZ-BDs on the C termini of SALMs 1–3. To determine whether the absence of a PDZ-BD in SALMs 4 and 5 was responsible for their forming trans-cellular links, we deleted the PDZ-BDs of SALMs 1 and 2. Deletion of the SALM1 or SALM2 PDZ-BDs did not cause the appearance of clusters between cells (Fig. 3D).
FIGURE 3.
SALMs 4 and 5 form homophilic complexes across cell junctions. A, HeLa cells were transfected with SALMs 1–4 individually, then permeabilized and stained. Polyclonal SALM antibodies were used to detect the intracellular expression of the SALMs. Transcellular clusters form between cells containing SALMs 4 or 5 (arrowheads), but not between cells transfected with other SALMs. B, surface staining of HeLa cells containing SALM4 with anti-SALM4-NT shows clustering between cells. C, HeLa cells were transfected with Myc-SALM4 and surface labeled with mouse Myc antibody and immunoperoxidase. i, light microscope view of cluster of transfected HeLa cells and two non-transfected cells (n; bottom). Arrow indicates the intercellular junction examined with electron microscopy at low magnification in ii, and high magnification in iii. Note in iii the places where stained structures traverse the junction between the cells (arrowheads). Fine unstained filaments (some the size of actin; asterisk) are associated with the plasma membranes of the cells. Scale bar is 35 μmin i, 0.9 μm in ii, and 100 nm in iii. D, total staining of cells transfected with Myc-SALM1Δ4 or Myc-SALM2Δ7 alone do not show clustering at cell contacts, suggesting that the SALM4 and SALM5 homophilic complexes are not due to the absence of a PDZ binding motif in the C termini of SALM4 and SALM5 of SALM4. HeLa cells were then transfected with one SALM family member, trypsinized, mixed with cells containing a second SALM, and replated. Color pictures show SALM1 or SALM5 in green and SALMs 2–4 in red. SALMs 1–5 do not form heterophilic interactions between cells (arrow). However, SALM4 and SALM5 form clusters between cells in a homophilic manner (arrowheads and insets).
Because SALMs can form heteromeric as well as homomeric complexes, we determined if SALMs could also form trans-cellular heterophilic associations. We transfected HeLa cells with a single SALM family member and incubated the cells for 24 h. Cells were then trypsinized and replated with cells expressing another SALM family member. Again, cell clustering occurred only between adjacent cells that contained SALM4 or SALM5 (Fig. 3D, arrowheads and insets). No clustering was present between cells containing any of the other SALMs. SALM4 and SALM5 did not form clusters with each other between adjacent cells. Therefore, SALMs 4 and 5 appear to be unique in their ability to form trans homophilic complexes across cell junctions and do not form such interactions with each other.
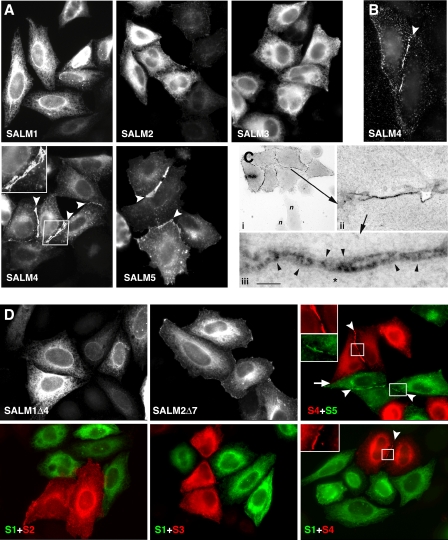
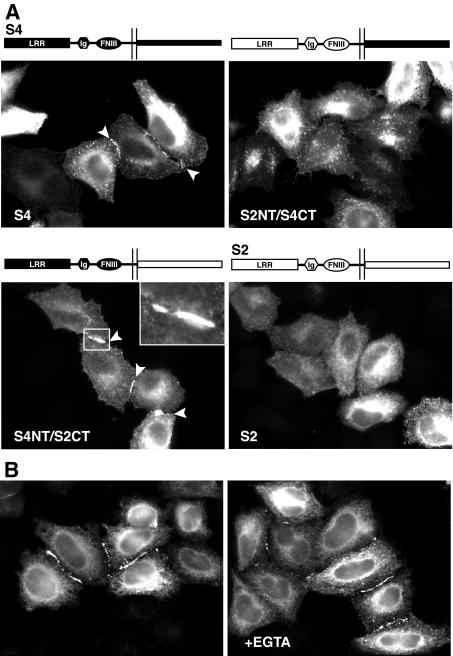
SALM4 Trans-cellular Complex Formation Is Mediated by Its Extracellular Domain—Whereas the homomeric and heteromeric interactions between the SALMs occur through their N-terminal domains, the ability of SALMs 4 and 5 to form trans-cellular complexes could be regulated by either their N- or C-terminal domains. The SALMs are highly homologous in their N termini but very different in their C termini, raising the possibility that either domain is responsible for the trans associations of SALMs 4 and 5. To determine which domains were involved, we constructed chimeras of SALM2, which does not form trans associations, and SALM4, which does form trans associations. By switching the extracellular domains of SALM2 and SALM4, we found that a chimera containing the SALM4 N terminus and the SALM2 C terminus formed clusters, but that a chimera of the SALM2 N terminus and SALM4 C terminus did not (Fig. 4A). These results indicate that the SALM4 N terminus is needed for cluster formation, and that the N termini of the SALMs determine the type of extracellular associations in which they participate. The ability of SALMs 4 and 5 to form trans associations is not dependent on calcium. Both SALMs still formed trans-cellular interactions when HeLa cells expressing SALM4 (Fig. 4B) or SALM5 (data not shown) were incubated in calcium-free media or treated with EGTA. These treatments disrupt calcium-dependent intercellular interactions of presenilin-1 and the cadherins (24, 25).
FIGURE 4.
SALM4 trans-cellular interactions are mediated by its extracellular domain. A, HeLa cells were transiently transfected with SALM2 and SALM4 chimeras tagged with HA or Myc at their N termini. Total staining was detected with antibodies to the C termini of SALM2 and SALM4. Exchanging the extracellular domain of SALM2 with the N terminus of SALM4 induces the clustering of the SALM2 C-terminal region (inset). However, replacing the extracellular domain of SALM4 with the N terminus of SALM2 prevents the clustering seen with SALM4, suggesting that clustering is mediated by the SALM4 extracellular domain. B, HeLa cells transiently transfected with SALM4 were incubated in calcium-free media for 20 h (left panel) or treated with 0.1 mm EGTA for 30 min (right panel). In the absence of calcium, SALM4 expressing cells still form clusters between cells, suggesting that SALM4 trans-cellular interactions are not mediated by a calcium-dependent mechanism.
To further show that the extracellular region was involved in the formation of SALM4 trans-cellular complexes, we made an antibody to the LRR region of the SALMs and compared treatment of transfected SALM4 cells with this antibody to treatment with a C-terminal antibody. If the N terminus is involved in the trans-cellular associations of SALMs 4 and 5, such an antibody may disrupt the interaction. The anti-LRR antibody interacted with the extracellular domains of all SALMs expressed in heterologous cells (data not shown) and showed enriched surface staining at regions of SALM4 clustering between cells (Fig. 5A, arrowhead). HeLa cells transfected with Myc-SALM4 were treated for 24 h with anti-LRR or anti-SALM4-CT antiserum, added directly to the culture medium. Cells were surface stained with a Myc antibody and total staining was detected with anti-SALM4-CT antibodies (Fig. 5B). The anti-LRR antibody inhibited trans associations between SALM4 expressing cells. In contrast, antiserum to the intracellular, C-terminal region of SALM4 had no effect on the trans-cellular associations of SALM4. On the other hand, treatment of cells with anti-Myc antibodies enhanced Myc-SALM4 clustering between cells (Fig. 5C). A similar treatment of cells expressing Myc-SALM2 induced clustering of SALM2. Thus, Myc-tagged SALM2 can be induced to form trans-cellular associations with the application of anti-Myc antibodies.
FIGURE 5.
Inhibition of SALM4 clustering by an antibody to LRR. A, Myc-SALM4 was transfected into HeLa cells, and surface expression was detected using an antibody directed against the SALM leucine-rich repeats (anti-LRR). Intracellular SALM4 was detected with anti-Myc (see Total). Surface staining indicates that the LRR antibody recognizes SALM4 intercellular clusters (arrowhead). B, HeLa cells transiently transfected with Myc-SALM4 were treated for 24 h with antisera directed against the SALM LRR or SALM4 C terminus (anti-S4CT), which served as a control. SALM4 surface staining was detected by anti-Myc, and intracellular SALM4 was detected by anti-S4CT antibodies. SALM4 cluster formation is largely inhibited by anti-LRR (1:250). However, SALM4 surface clustering is not disrupted by application of anti-S4CT (1:250). C, treatment of Myc-SALM4-transfected HeLa cells with anti-Myc antibodies (1:1000) for 24 h enhances SALM4 clustering (arrowheads). Anti-Myc antibodies also induce clustering of the SALM2 C terminus (arrowheads, bottom panel) in HeLa cells transfected with full-length Myc-SALM2 (detected with anti-S2CT).
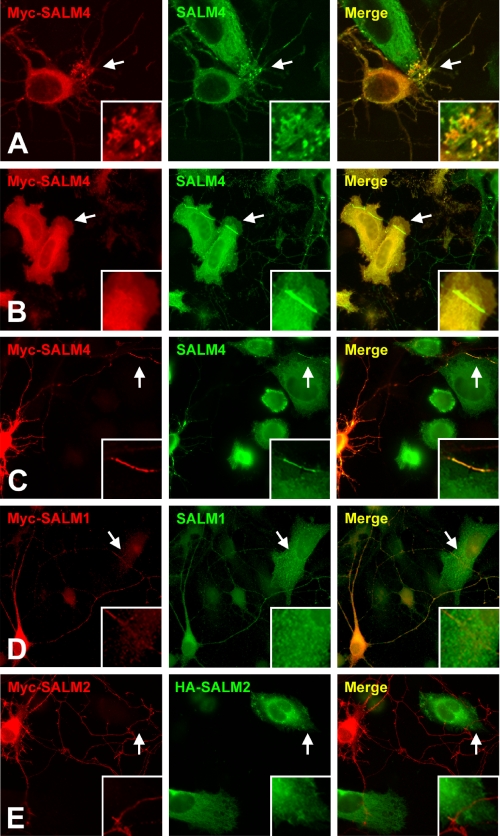
SALM4 Trans-cellular Clustering in Neurons—Our results in heterologous cells suggest that SALMs 4 and 5 may form homophilic trans-cellular associations in neurons. To address this, HeLa cells and hippocampal neurons were transfected with SALM4 and co-cultured. Enrichment of SALM4 immunoreactivity in neurons was clearly present at points of contact between transfected HeLa cells and neuronal processes (Fig. 6, A–C). Based on morphological characteristics, these processes appear to include both dendrites and axons (30). The accumulation of staining appeared to occur when SALM4-transfected neuronal processes (Fig. 6, A–C) associated with SALM4-transfected HeLa cells. Neurons and HeLa cells were also transfected with SALM1 or SALM2 and co-cultured. No indication of an accumulation of either SALM1 or SALM2 was seen at points of contact between neurons and HeLa cells (Fig. 6, D and E), consistent with our finding that these SALMs did not form trans-cellular associations in heterologous cells. We did not see an accumulation of SALM4 immunoreactivity at points of contact between transfected neurons and untransfected HeLa cells or between untransfected neurons and transfected HeLa cells (data not shown). Whereas SALM4 is present in untransfected hippocampal neurons, the level may be insufficient to detect accumulations at points of contact with heterologous cells.
FIGURE 6.
Recruitment of SALM4 in SALM-transfected heterologous cells and hippocampal neurons. A–E, hippocampal neurons (7 days in vitro) were transfected with SALMs 1–4 using calcium phosphate precipitation. Twenty-four hours after transfection, HeLa cells transfected with the same SALM were plated on the neurons and allowed to incubate overnight. Cells were then fixed and permeabilized for total staining. SALM4 expressing cells were transfected with Myc-tagged or untagged SALM4 and stained with anti-Myc (red) or anti-S4CT (green). A, near the soma of neurons, SALM4-transfected in HeLa cells induces an accumulation of labeling in putative dendrites of Myc-SALM4-transfected neurons. B, along putative axons, contact between neurons and HeLa cells containing SALM4 induces enrichment of SALM4 at the point of contact between SALM4-transfected neurons and Myc-SALM4-transfected HeLa cells (arrows). C, a similar enrichment occurs between the point of contact between Myc-SALM4-transfected neurons and SALM4-transfected HeLa cells (arrows). D–E, clustering is not present in cells transfected with SALM1 (D) or SALM2 (E).
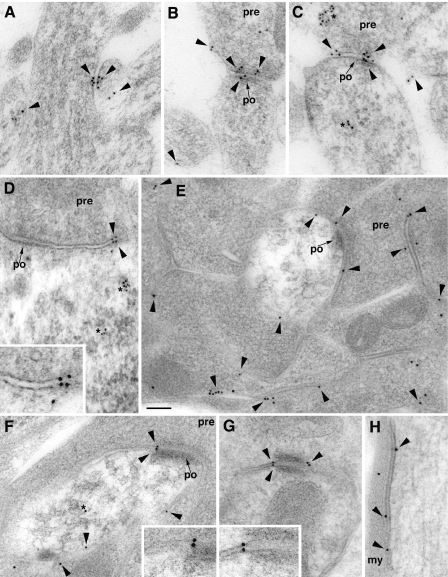
Ultrastructure of Native SALM4 in Rat Brain—In immunogold-labeled sections from the hippocampus of P10 rats, and hippocampus, cerebellum, and olfactory bulb of young adult rats, SALM4 was localized largely to the cell membrane of neuronal processes (Fig. 7). At synapses, these included both the synaptic and extrasynaptic membranes on both presynaptic and postsynaptic sides, as well as the cell membrane of other structures such as neuron somas (Fig. 7D) and myelinated axons (Fig. 7H). In many cases, plasma membrane-associated gold could be found on both sides of adjacent membrane appositions (Fig. 7, A–D, F, and G). Although gold was also commonly seen in vesicular and tubulovesicular structures within the cytoplasm, a high prevalence of surface labeling was evident from qualitative analysis; for example, in Fig. 7E, about 80% of the gold in the micrograph labeled cell membranes between cell processes (within 20 nm, considering the theoretical spatial resolution of the method). These results suggest that SALM4 occurs widely in and near cell junctions as well as at non-specialized contacts formed between many neuronal processes.
FIGURE 7.
Ultrastructure of native SALM4 interactions in the brain using 10-nm immunogold labeling. Labeling for SALM4 is found on the cell membrane of processes in the brain (arrowheads). A–C, P10 CA1 stratum radiatum of the hippocampus. Note labeling at contacts between a dendrite and two unidentified small processes in A and between postsynaptic spines and presynaptic terminals (pre) of immature synapses in B and C. Labeling of internal, vesicular, and tubular vesicular structures (asterisks) is also evident. D–H, labeling in the adult brain. In the olfactory bulb, labeling in D is shown between a mitral cell body and the extrasynaptic membrane (also see inset) of an inhibitory synaptic terminal (identified by the pleomorphic vesicles and symmetric densities of the synapse) and in E and F on extrasynaptic membranes either between two olfactory nerve terminals or between an olfactory nerve terminal and the postsynaptic dendrite in the center of the micrographs. Whereas all plasma membrane-associated gold in E is extrasynaptic, F also shows an example of labeling between the membranes within the edge of a synapse (also see inset). Examples of cell membrane labeling from the granular layer of the cerebellar cortex include labeling on both sides of an attachment plaque between two granule cell dendrites (G; also see inset) and on the cell membrane of a large myelinated axon (H). Small arrows indicate the postsynaptic membrane (po) of synapses; my, myelin sheath. Scale bar is 100 nm (50 nm for insets).
DISCUSSION
SALMs 1–5 comprise a family of adhesion molecules whose expression is limited largely to the central nervous system. All SALMs are present early in development and they have been linked to neurite outgrowth and synapse formation (11–13). These properties suggest that the SALMs play critical roles in the formation and maintenance of the central nervous system. To determine more closely how the individual SALMs may function, we investigated whether SALMs can form heteromeric and homomeric complexes and whether they interact in trans configurations. Our results show that all five SALMs can form heteromeric complexes when co-expressed in heterologous cells, but, in brain, heteromeric complexes appear to be largely limited to SALMs 1–3. This difference in SALM association between brain and overexpressing cells may be due to a difference in expression of the SALMs in brain cell types or location, resulting in a lack of contact, or a difference of SALM localization within the same cell population, such as synaptic versus extrasynaptic populations or different adhesion sites. Interestingly, only two members of the SALM family form trans associations; in heterologous cells, SALMs 4 and 5 form homophilic trans associations, whereas such associations are not seen with SALMs 1–3. These results suggest that SALMs can be divided into two groups, those that form heteromeric complexes in brain but do not form trans associations (SALMs 1–3) and those that mainly do not form heteromeric complexes in brain but do form trans associations (SALMs 4 and 5). These findings suggest different functional roles and mechanisms for these two populations.
The SALMs are members of a larger family of adhesion molecules that are characterized by extracellular LRRs, and either Ig or FNIII domains or both. These include, for example, AMIGO/Alivin (31, 32), LINGO (33, 34), NGL (35), FLRT (36), NLRR (37–39), and PAL (40). Structurally, NLRR and PAL, a retinal adhesion molecule, are most similar to the SALMs in that they contain both Ig and FNIII domains in addition to the LRRs. With the exception of NGL, these molecules do not contain PDZ-BDs. All of these proteins are reported to play a role in neurite outgrowth (41). Similar to the proteins in this large family of molecules are the Slits and Robos, which contain LRR domains or both FNIII and Ig repeats in their ectodomains, respectively. Slits are large secreted proteins involved in axonal guidance, which requires their LRR region for repellent signaling (17, 42–45). Interestingly, the Slits and Robos have been shown to interact through the LRR region of the Slits and the Ig domains of Robo (44–47). The Slits have been shown to dimerize through their LRR domain (46), whereas Robo family members interact homophilically, as well as heterophilically (47, 48).
Our results show that SALMs form homomeric or heteromeric complexes in brain and heterologous cells, although SALMs 4 and 5 may largely form homomeric complexes in brain. This suggests that the first step in the formation of functional adhesion complexes is the formation of dimers, or possibly higher order multimers. For SALMs 4 and 5, these complexes would then form trans complexes with SALMs at apposing plasma membranes. Whereas defining the precise nature of the complexes formed requires further investigation, dimers are likely formed through extracellular domains because SALMs can assemble into heteromeric complexes in the absence of their C termini. SALM4 also interacts with SALM1Δ4, which does not reach the cell surface, indicating that it is possible for SALMs to interact with each other in an intracellular compartment before they reach the plasma membrane. We presume that the complexes formed in intracellular compartments are precursors to SALM complexes on the cell surface and that cis interactions between the SALMs are present at the cell surface.
We find that SALMs 4 and 5 form homophilic trans associations when expressed in HeLa cells. By expressing epitope-tagged SALM4 in neurons and HeLa cells in co-cultures, we show that SALM4 is concentrated at points of contact between axons or dendrites and HeLa cells. This suggests that SALM4 (and 5) may form homophilic interactions between neurons. Our EM immunocytochemical data support this by showing immunogold labeling on both axonal and dendritic membranes, in some cases with clear apposition of gold particles. The selectivity for homophilic trans interactions of SALMs 4 and 5 is not unprecedented. Classical cadherins exhibit a much higher affinity for trans homophilic interactions than for trans heterophilic interactions (14, 49). The specificity of these interactions is also dependent largely on the extracellular domains of cadherins (49).
Whereas SALM4 is associated with pre- and postsynaptic membranes, our immunocytochemical results show that SALM4 does not appear to be enriched at the synapse. The lack of enrichment of SALM 4 at the synapse may be due to the absence of a PDZ-BD. SynCAM, an Ig domain adhesion molecule that forms homophilic and heterophilic interactions between pre- and postsynaptic membranes, recruits synaptic proteins in co-cultures (50, 51). Like SynCAM, SALMs 1–3 contain intracellular PDZ-BDs that could play a role in their recruitment and retention at the synapse. Members of the PSD-95 family of membrane-associated guanylate kinases, and particularly PSD-95 itself, are highly enriched at the postsynaptic density and are thought to play a role in clustering proteins with PDZ-BDs at the synapse. However, whereas SALMs 1–3 are present at the PSD, their distributions are more general than that of PSD-95 (11),3 indicating a wider distribution in neurons and functions at both synaptic and non-synaptic locations. Whereas the differential synaptic localization of the SALMs could indicate different roles at the synapse, all five SALMs are present at growth cones and enhance neurite outgrowth when overexpressed.4
The SALMs are highly homologous in their extracellular domains but have few similarities in their intracellular domains. By constructing chimeras of SALMs 2 and 4, we show that the ability of SALM4 to form trans associations is due to its extracellular domain. Our data using an antibody to the LRR domain support the idea that the LRR of SALMs is responsible for the trans associations; however, it is possible that the Ig and FNIII domains of SALMs are also involved in the trans associations and/or that this antibody disrupts the extracellular interactions of other adhesion molecules. We find that the trans interaction formed by SALMs 4 and 5 is not dependent on calcium. Calcium dependence is variable among neuronal adhesion molecules. The LRR domain of FLRT has been reported to be involved in homophilic cell sorting in a calcium-dependent manner (52). Presenilin-1, cadherin, neurexin, and neuroligin are also involved in calcium-dependent trans-cellular interactions (24, 25, 53). However, similar to SynCAM (50), calcium does not appear to regulate SALM4 or SALM5 trans associations.
Our results also do not suggest a direct role for the intracellular domain in the formation of trans associations. Although not directly involved in trans interactions, intracellular domains of adhesion molecules have been shown to influence extracellular interactions, as seen with integrins (54, 55). The role of the intracellular domains of the SALMs remains to be determined, but it is likely that they will be involved in functions related to intracellular signaling and forming associations with other proteins.
The enhanced clustering of SALM4 in response to application of the Myc antibody is most likely due to the bivalent interactions of a monoclonal antibody linking together SALM4 epitope-tagged extracellular domains. Alternatively, the antibody association may mimic ligand binding, causing a conformational change of the extracellular domain. A change in conformation might explain why SALM2 does not form trans-cellular interactions under normal conditions, but treatment of epitope-tagged SALM2 transfected cells with a Myc antibody induces clustering of the SALM2 C terminus at cell junctions. A similar change in ectodomain conformation has been suggested for the Nogo receptor, a LRR containing receptor involved in the inhibition of neurite outgrowth (56, 57). The crystal structure of the Nogo receptor ectodomain (56, 58), as well as binding assays (57), suggest that the LRR can form multimers. In addition, the Nogo receptor LRR domain has been implicated in trans interactions. Similar to our data indicating that an antibody to the SALM LRR domain inhibits SALM4 trans-cellular associations, a monoclonal antibody to the third LRR of Nogo receptor reverses the inhibition of neurite outgrowth by myelin and blocks binding of the Nogo receptor ligands Nogo, myelin-associated glycoprotein, and oligodendrocyte-myelin glycoprotein (59).
The cis associations formed by the SALMs may also influence their ability to form trans complexes. For example, the L1 chick homolog NgCAM forms homophilic trans interactions across the extracellular space through its N-terminal Ig domains 1–4, but also forms a heteromeric cis complex with axonin-1 through its Ig domains 2–4 and the third FNIII domain (60). This interaction prevents the formation of trans axonin-1/axonin-1 associations (60, 61), and enhances neurite fasciculation (60, 62). Another example of cis interactions regulating trans associations includes the neurexins, which were recently reported to be postsynaptic, in addition to being presynaptic (63). When postsynaptic neurexins form cis complexes with postsynaptic neuroligin-1, the interaction inhibits the ability of neuroligin-1 to interact with presynaptic neurexins (63). Therefore, SALM4 and SALM5 trans-cellular interactions may also be influenced and regulated by their cis interactions with other SALM family members, and vice versa, similar to other cell adhesion molecules.
Our results showing differences between the two groups of SALM proteins, SALMs 1–3 and SALMs 4 and 5, raise additional questions of the binding partners of SALMs 1–3. Several possibilities exist. SALMs 1–3 may form trans complexes, but under conditions not examined. Alternatively, SALMs 1–3 may interact with soluble molecules or their N termini may be cleaved and the soluble cleavage products may themselves serve as signaling molecules, as seen with the Slits. The N-terminal, LRR-containing fraction of Slit can induce the branching of DRG axons, after Slit is cleaved; however, this effect is antagonized by the presence of uncleaved Slit (42, 64). Furthermore, there are multiple adhesion molecules with LRR, Ig, and FNIII domains that could interact with SALMs through one or more of these domains. Finally, we cannot rule out the possibility that SALMs 4 and 5, in addition to forming trans complexes, also interact with other molecules or are themselves cleaved, releasing diffusible products that may interact with other adhesion molecules. Therefore, the two main functions of the SALMs, neurite outgrowth and synapse formation, may involve several different interacting proteins besides the SALM associations discussed in this paper.
Supplementary Material
Acknowledgments
We thank members of the Laboratory of Neurochemistry for helpful comments on the manuscript.
This work was supported by the National Institutes of Health NIDCD Intramural Research Program. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and methods.
Footnotes
The abbreviations used are: SALM, synaptic adhesion-like molecule; LRR, leucine-rich repeat; FN, fibronectin; PDZ-BD, PDZ-binding domain; HA, hemagglutinin; aa, amino acid(s); TBS, Tris-buffered saline; PBS, phosphate-buffered saline.
R. S. Petralia, G. K. Seabold, Y.-X. Wang, and R. J. Wenthold, unpublished data.
P. Y. Wang, G. K. Seabold, and R. J. Wenthold, unpublished data.
References
- 1.Benson, D. L., Colman, D. R., and Huntley, G. W. (2001) Nat. Rev. Neurosci. 2 899-909 [DOI] [PubMed] [Google Scholar]
- 2.Yamagata, M., Sanes, J. R., and Weiner, J. A. (2003) Curr. Opin. Cell Biol. 15 621-632 [DOI] [PubMed] [Google Scholar]
- 3.Craig, A. M., Graf, E. R., and Linhoff, M. W. (2006) Trends Neurosci. 29 8-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerrow, K., and El-Husseini, A. (2006) Front. Biosci. 11 2400-2419 [DOI] [PubMed] [Google Scholar]
- 5.Dalva, M. B., McClelland, A. C., and Kayser, M. S. (2007) Nat. Rev. Neurosci. 8 206-220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maness, P. F., and Schachner, M. (2007) Nat. Neurosci. 10 19-26 [DOI] [PubMed] [Google Scholar]
- 7.Washbourne, P., Dityatev, A., Scheiffele, P., Biederer, T., Weiner, J. A., Christopherson, K. S., and El-Husseini, A. (2004) J. Neurosci. 24 9244-9249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abelson, J. F., Kwan, K. Y., O'Roak, B. J., Baek, D. Y., Stillman, A. A., Morgan, T. M., Mathews, C. A., Pauls, D. L., Rasin, M. R., Gunel, M., Davis, N. R., Ercan-Sencicek, A. G., Guez, D. H., Spertus, J. A., Leckman, J. F., Dure, L. S., IV, Kurlan, R., Singer, H. S., Gilbert, D. L., Farhi, A., Louvi, A., Lifton, R. P., Sestan, N., and State, M. W. (2005) Science 310 317-320 [DOI] [PubMed] [Google Scholar]
- 9.Philibert, R. A., Winfield, S. L., Sandhu, H. K., Martin, B. M., and Ginns, E. I. (2000) Gene (Amst.) 246 303-310 [DOI] [PubMed] [Google Scholar]
- 10.Chih, B., Afridi, S. K., Clark, L., and Scheiffele, P. (2004) Hum. Mol. Genet. 13 1471-1477 [DOI] [PubMed] [Google Scholar]
- 11.Wang, C. Y., Chang, K., Petralia, R. S., Wang, Y. X., Seabold, G. K., and Wenthold, R. J. (2006) J. Neurosci. 26 2174-2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ko, J., Kim, S., Chung, H. S., Kim, K., Han, K., Kim, H., Jun, H., Kaang, B. K., and Kim, E. (2006) Neuron 50 233-245 [DOI] [PubMed] [Google Scholar]
- 13.Morimura, N., Inoue, T., Katayama, K., and Aruga, J. (2006) Gene (Amst.) 380 72-83 [DOI] [PubMed] [Google Scholar]
- 14.Tepass, U., Truong, K., Godt, D., Ikura, M., and Peifer, M. (2000) Nat. Rev. Mol. Cell Biol. 1 91-100 [DOI] [PubMed] [Google Scholar]
- 15.Ichtchenko, K., Hata, Y., Nguyen, T., Ullrich, B., Missler, M., Moomaw, C., and Sudhof, T. C. (1995) Cell 81 435-443 [DOI] [PubMed] [Google Scholar]
- 16.Scheiffele, P., Fan, J., Choih, J., Fetter, R., and Serafini, T. (2000) Cell 101 657-669 [DOI] [PubMed] [Google Scholar]
- 17.Brose, K., and Tessier-Lavigne, M. (2000) Curr. Opin. Neurobiol. 10 95-102 [DOI] [PubMed] [Google Scholar]
- 18.Takai, Y., and Nakanishi, H. (2003) J. Cell Sci. 116 17-27 [DOI] [PubMed] [Google Scholar]
- 19.Stoeckli, E. T., Ziegler, U., Bleiker, A. J., Groscurth, P., and Sonderegger, P. (1996) Dev. Biol. 177 15-29 [DOI] [PubMed] [Google Scholar]
- 20.Buchstaller, A., Kunz, S., Berger, P., Kunz, B., Ziegler, U., Rader, C., and Sonderegger, P. (1996) J. Cell Biol. 135 1593-1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kingston, R. E. (1997) Current Protocols in Molecular Biology, John Wiley & Sons, Inc., New York
- 22.Sans, N., Prybylowski, K., Petralia, R. S., Chang, K., Wang, Y. X., Racca, C., Vicini, S., and Wenthold, R. J. (2003) Nat. Cell. Biol. 5 520-530 [DOI] [PubMed] [Google Scholar]
- 23.Biederer, T., and Scheiffele, P. (2007) Nat. Protoc. 2 670-676 [DOI] [PubMed] [Google Scholar]
- 24.Georgakopoulos, A., Marambaud, P., Efthimiopoulos, S., Shioi, J., Cui, W., Li, H. C., Schutte, M., Gordon, R., Holstein, G. R., Martinelli, G., Mehta, P., Friedrich, V. L., Jr., and Robakis, N. K. (1999) Mol. Cell 4 893-902 [DOI] [PubMed] [Google Scholar]
- 25.Hirano, S., Nose, A., Hatta, K., Kawakami, A., and Takeichi, M. (1987) J. Cell Biol. 105 2501-2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eshhar, N., Petralia, R. S., Winters, C. A., Niedzielski, A. S., and Wenthold, R. J. (1993) Neuroscience 57 943-964 [DOI] [PubMed] [Google Scholar]
- 27.Petralia, R. S., Wang, Y. X., Mayat, E., and Wenthold, R. J. (1997) J. Comp. Neurol. 385 456-476 [DOI] [PubMed] [Google Scholar]
- 28.Petralia, R. S., and Wenthold, R. J. (1999) Methods Mol. Biol. 128 73-92 [DOI] [PubMed] [Google Scholar]
- 29.Petralia, R. S., Sans, N., Wang, Y. X., and Wenthold, R. J. (2005) Mol. Cell. Neurosci. 29 436-452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaech, S., and Banker, G. (2006) Nat. Protoc. 1 2406-2415 [DOI] [PubMed] [Google Scholar]
- 31.Kuja-Panula, J., Kiiltomaki, M., Yamashiro, T., Rouhiainen, A., and Rauvala, H. (2003) J. Cell Biol. 160 963-973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ono, T., Sekino-Suzuki, N., Kikkawa, Y., Yonekawa, H., and Kawashima, S. (2003) J. Neurosci. 23 5887-5896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carim-Todd, L., Escarceller, M., Estivill, X., and Sumoy, L. (2003) Eur. J. Neurosci. 18 3167-3182 [DOI] [PubMed] [Google Scholar]
- 34.Mi, S., Lee, X., Shao, Z., Thill, G., Ji, B., Relton, J., Levesque, M., Allaire, N., Perrin, S., Sands, B., Crowell, T., Cate, R. L., McCoy, J. M., and Pepinsky, R. B. (2004) Nat. Neurosci. 7 221-228 [DOI] [PubMed] [Google Scholar]
- 35.Lin, J. C., Ho, W. H., Gurney, A., and Rosenthal, A. (2003) Nat. Neurosci. 6 1270-1276 [DOI] [PubMed] [Google Scholar]
- 36.Lacy, S. E., Bonnemann, C. G., Buzney, E. A., and Kunkel, L. M. (1999) Genomics. 62 417-426 [DOI] [PubMed] [Google Scholar]
- 37.Taguchi, A., Wanaka, A., Mori, T., Matsumoto, K., Imai, Y., Tagaki, T., and Tohyama, M. (1996) Brain Res. Mol. Brain Res. 35 31-40 [DOI] [PubMed] [Google Scholar]
- 38.Haines, B. P., Gupta, R., Jones, C. M., Summerbell, D., and Rigby, P. W. (2005) Dev. Biol. 281 145-159 [DOI] [PubMed] [Google Scholar]
- 39.Bando, T., Sekine, K., Kobayashi, S., Watabe, A. M., Rump, A., Tanaka, M., Suda, Y., Kato, S., Morikawa, Y., Manabe, T., and Miyajima, A. (2005) Mol. Cell. Biol. 25 4166-4175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gomi, F., Imaizumi, K., Yoneda, T., Taniguchi, M., Mori, Y., Miyoshi, K., Hitomi, J., Fujikado, T., Tano, Y., and Tohyama, M. (2000) J. Neurosci. 20 3206-3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen, Y., Aulia, S., Li, L., and Tang, B. L. (2006) Brain Res. Rev. 51 265-274 [DOI] [PubMed] [Google Scholar]
- 42.Brose, K., Bland, K. S., Wang, K. H., Arnott, D., Henzel, W., Goodman, C. S., Tessier-Lavigne, M., and Kidd, T. (1999) Cell 96 795-806 [DOI] [PubMed] [Google Scholar]
- 43.Li, H. S., Chen, J. H., Wu, W., Fagaly, T., Zhou, L., Yuan, W., Dupuis, S., Jiang, Z. H., Nash, W., Gick, C., Ornitz, D. M., Wu, J. Y., and Rao, Y. (1999) Cell 96 807-818 [DOI] [PubMed] [Google Scholar]
- 44.Battye, R., Stevens, A., Perry, R. L., and Jacobs, J. R. (2001) J. Neurosci. 21 4290-4298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen, J. H., Wen, L., Dupuis, S., Wu, J. Y., and Rao, Y. (2001) J. Neurosci. 21 1548-1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Howitt, J. A., Clout, N. J., and Hohenester, E. (2004) EMBO J. 23 4406-4412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu, Z., Patel, K., Schmidt, H., Andrews, W., Pini, A., and Sundaresan, V. (2004) Mol. Cell. Neurosci. 26 232-240 [DOI] [PubMed] [Google Scholar]
- 48.Hivert, B., Liu, Z., Chuang, C. Y., Doherty, P., and Sundaresan, V. (2002) Mol. Cell. Neurosci. 21 534-545 [DOI] [PubMed] [Google Scholar]
- 49.Shan, W. S., Tanaka, H., Phillips, G. R., Arndt, K., Yoshida, M., Colman, D. R., and Shapiro, L. (2000) J. Cell Biol. 148 579-590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Biederer, T., Sara, Y., Mozhayeva, M., Atasoy, D., Liu, X., Kavalali, E. T., and Sudhof, T. C. (2002) Science 297 1525-1531 [DOI] [PubMed] [Google Scholar]
- 51.Fogel, A. I., Akins, M. R., Krupp, A. J., Stagi, M., Stein, V., and Biederer, T. (2007) J. Neurosci. 27 12516-12530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karaulanov, E. E., Bottcher, R. T., and Niehrs, C. (2006) EMBO Rep. 7 283-290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dean, C., Scholl, F. G., Choih, J., DeMaria, S., Berger, J., Isacoff, E., and Scheiffele, P. (2003) Nat. Neurosci. 6 708-716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O'Toole, T. E., Mandelman, D., Forsyth, J., Shattil, S. J., Plow, E. F., and Ginsberg, M. H. (1991) Science 254 845-847 [DOI] [PubMed] [Google Scholar]
- 55.Ginsberg, M. H., Partridge, A., and Shattil, S. J. (2005) Curr. Opin. Cell Biol. 17 509-516 [DOI] [PubMed] [Google Scholar]
- 56.Barton, W. A., Liu, B. P., Tzvetkova, D., Jeffrey, P. D., Fournier, A. E., Sah, D., Cate, R., Strittmatter, S. M., and Nikolov, D. B. (2003) EMBO J. 22 3291-3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fournier, A. E., Gould, G. C., Liu, B. P., and Strittmatter, S. M. (2002) J. Neurosci. 22 8876-8883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He, X. L., Bazan, J. F., McDermott, G., Park, J. B., Wang, K., Tessier-Lavigne, M., He, Z., and Garcia, K. C. (2003) Neuron 38 177-185 [DOI] [PubMed] [Google Scholar]
- 59.Li, W., Walus, L., Rabacchi, S. A., Jirik, A., Chang, E., Schauer, J., Zheng, B. H., Benedetti, N. J., Liu, B. P., Choi, E., Worley, D., Silvian, L., Mo, W., Mullen, C., Yang, W., Strittmatter, S. M., Sah, D. W., Pepinsky, B., and Lee, D. H. (2004) J. Biol. Chem. 279 43780-43788 [DOI] [PubMed] [Google Scholar]
- 60.Kunz, S., Spirig, M., Ginsburg, C., Buchstaller, A., Berger, P., Lanz, R., Rader, C., Vogt, L., Kunz, B., and Sonderegger, P. (1998) J. Cell Biol. 143 1673-1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sonderegger, P., Kunz, S., Rader, C., Buchstaller, A., Berger, P., Vogt, L., Kozlov, S. V., Ziegler, U., Kunz, B., Fitzli, D., and Stoeckli, E. T. (1998) Prog. Brain Res. 117 93-104 [DOI] [PubMed] [Google Scholar]
- 62.Stoeckli, E. T., and Landmesser, L. T. (1995) Neuron 14 1165-1179 [DOI] [PubMed] [Google Scholar]
- 63.Taniguchi, H., Gollan, L., Scholl, F. G., Mahadomrongkul, V., Dobler, E., Limthong, N., Peck, M., Aoki, C., and Scheiffele, P. (2007) J. Neurosci. 27 2815-2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nguyen Ba-Charvet, K. T., Brose, K., Ma, L., Wang, K. H., Marillat, V., Sotelo, C., Tessier-Lavigne, M., and Chedotal, A. (2001) J. Neurosci. 21 4281-4289 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.