Abstract
Two regulatory pathways, bone morphogenetic protein (BMP)/transforming growth factor-β (TGFβ) and the transcription factor RUNX2, are required for bone formation in vivo. Here we show the interdependent requirement of these pathways to induce an osteogenic program. A panel of Runx2 deletion and point mutants was used to examine RUNX2-SMAD protein-protein interaction and the biological consequences on BMP2-induced osteogenic signaling determined in Runx2 null cells. These cells do not respond to BMP2 signal in the absence of Runx2. We established that a triple mutation in the C-terminal domain of RUNX2, HTY (426-428), disrupts the RUNX2-SMAD interaction, is deficient in its ability to integrate the BMP2/TGFβ signal on promoter reporter assays, and is only marginally functional in promoting early stages of osteoblast differentiation. Furthermore, the HTY mutation overlaps the unique nuclear matrix targeting signal of Runx factors and exhibits reduced subnuclear targeting. Thus, formation of a RUNX2-SMAD osteogenic complex and subnuclear targeting are structurally and functionally inseparable. Our results establish the critical residues of RUNX2 for execution and completion of BMP2 signaling for osteoblastogenesis through a mechanism that requires RUNX2-SMAD transcriptional activity.
Skeletal development and bone formation require coordinated activities of multiple signaling pathways that include bone morphogenetic protein 2 (BMP2)5 and transforming growth factor-β (TGFβ). Transduction of these signals results in the activation of target genes that are essential for bone development. Specific receptor-regulated SMADs (R-SMADs) serve as substrates for the BMP and TGFβ/activin/Nodal receptors. SMAD-1, -2, -3, and -5 transduce, whereas Smad-4 serves as a common partner for all R-SMADs to provide the DNA binding property (1, 2). The structural and functional domains of SMAD proteins are well characterized with binding sites for SMAD ubiquitination-related factor (SMURF) ubiquitin ligases, and phosphorylation sites for several classes of protein kinases (3). The MH2 domain mediates interactions with transcriptional activators and repressors for signal transduction; including co-regulators of skeletal development (4-7).
Several studies suggest that the principle activity of BMP and TGFβ SMADs for the control of skeletogenesis is mediated by their interaction with RUNX2 (CBFA1/AML3). This runt-related transcription factor is critical for osteogenic lineage commitment and formation of the skeleton (8-12). Mutations in the human RUNX2 cause cleidocranial dysplasia (13, 14). Targeted disruption of Runx2 in mice results in the maturational arrest of osteoblasts and a complete lack of mineralized bone (15-17). The gene regulatory properties of RUNX factors are mediated not only by DNA binding to cognate elements, but also through the formation of selective co-regulatory protein interactions with co-activator and co-repressor proteins (9, 10). The C terminus of RUNX proteins contains a 31-amino acid nuclear matrix targeting signal (NMTS), an essential well conserved functional domain, required to direct RUNX to distinct nuclear matrix-associated sites within the nucleus that support gene expression (18-20). The tight association of RUNX proteins with the nuclear matrix provides a platform for assembly of multicomponent regulatory complexes that control both activation and repression of genes during cell fate determination and differentiation (9, 21-23). Among the co-regulatory proteins interacting with the RUNX2 C terminus and recruited to RUNX2 subnuclear domains are mediators of developmental signals, which include the TGFβ- and BMP-induced SMADs (13, 24, 25). The biological significance of the RUNX2 C terminus is revealed by a knock-in mutation in which mice lacking the C terminus fail to develop a mineralized skeleton (17), confirming the importance of this region for in vivo osteogenesis.
Particularly relevant to bone formation, early studies showed that BMP2-treated Runx2 null cells could not induce the complete osteoblast differentiation program (15, 26, 27). RUNX2 has been shown to interact with SMAD proteins (13, 28-32) and recruit SMADs to subnuclear sites of active transcription (24). Deletion mutant studies have identified a RUNX2-SMAD interacting domain (SMID) in the C terminus (23) that overlaps the NMTS, but definitive proof that RUNX2-SMAD interaction is essential for osteoblastogenesis has yet to be established. Only by disruption of the RUNX2-SMAD interaction in the context of the entire protein can the hypothesis that RUNX2 is required to mediate the BMP2 osteogenic signal, be tested.
In these studies, by site-directed mutagenesis, we defined the specific RUNX2 amino acids required for physical and functional interaction with either BMP- or TGFβ-responsive SMADs. Three residues (HTY, 426-428) positioned in the carboxyl end of the RUNX2 SMID contribute to the osteogenic activity of RUNX2, formation of the RUNX2-SMAD complex, and integration of the BMP/TGFβ signal. The HTY mutant RUNX2 protein retains DNA binding and transcriptional activity but has impaired subnuclear targeting and no ability to bind SMADs, thus preventing transduction of a BMP2-mediated osteogenic function. These findings of minimal amino acid requirements for both RUNX2-SMAD interactions and the targeting of a RUNX2-SMAD functional complex to subnuclear domains provide compelling evidence for a structural coupling of SMADs with RUNX2 that is essential for execution and completion of BMP2 osteogenic signal.
EXPERIMENTAL PROCEDURES
Cell Cultures—Human cervical carcinoma HeLa cells were cultured and maintained as previously described (23). Runx2 null cells were isolated from calvarial tissue of 17.5-day-old mouse embryos. Cells were maintained in modified Eagle's medium (αMEM) containing 10% fetal bovine serum, and penicillin G (100 units/ml), and streptomycin (100 μg/ml) at 37 °C in a humidified atmosphere of 5% CO2 in air. Cells were stably transfected with expression vector of mouse telomerase using FuGENE 6 regents (Roche Applied Science) and selected against G418 for 2 weeks. Detailed description and morphological characteristic of the established Runx2 null cell lines are reported elsewhere (27).
Plasmids and Adenoviral Constructs—The BMP2- and TGFβ-responsive FLAG-tagged receptor SMAD (SMAD2 and 3) constructs, and the 3×TBRE-Luciferase and 6×Runx-Luciferase plasmids are reported previously (23). Expression construct of hemagglutinin (HA)-tagged RUNX2, deletion Δ391, Y428A, and R398A&Y428A are described earlier (20, 23).
HA-tagged RUNX2 point mutants H426A, T427A, HT426, 427AA, FTY405-407AAA, HTY426-428AAA, and FTYHTY405-407,426-428AAAAAA were generated with a two-step PCR approach. In the first step two independent but overlapping PCR products were generated using wild-type Runx2 cDNA as template. For PCR product 1, the common forward primer contained an ApaI site, 5′-GAACTGGGCCCTTTTTCAGACCCCAG-3′, whereas the mutant specific reverse primers were 5′-GTGGTGGCAGGTACGTGGCGTAGTGAGTG-3′ for H426A, 5′-GTGGTGGCAGGTACGCGTGGTAGTGAGTG-3′ for T427A, 5′-GTGGTGGCAGGTACGCGGCGTAGTGAGTG-3′, for HT426,427AA, 5′-GTGGTGGCAGTGCCGCGGCGTAGTGAGTG-3′ for HTY426-428AAA, and 5′-CTGGCGGGGTTGCTGCAGCGGTGGCTGGG-3′ for FTY405-407AAA. For PCR product 2, the mutant-specific forward primers were 5′-CACTCACTACGCCACGTACCTGCCACCAC-3′ for H426A, 5′-CACTCACTACCACGCGTACCTGCCACCAC-3′ for T427A, 5′-CACTCACTACGCCGCGTACCTGCCACCAC-3′ for HT426,427AA, 5′-CACTCACTACGCCGCGGCACTGCCACCAC-3′ for HTY426-428AAA, and 5′-CCCAGCCACCGCTGCAGCAACCCCGCCAG-3′ for FTY405-407AAA with the common reverse primer containing an XhoI site and stop codon, 5′-TTTCTCGAGTCAATATGGCCG-3′. Both PCR products for each mutant plasmid were purified and combined as template to generate a full-length PCR product containing the respective mutation. For the full-length PCR reactions the forward primer with ApaI site and the reverse primer with XhoI site are the same as above. The final products carrying the respective mutations were sequentially digested with ApaI/XhoI and ligated into the similarly digested HA-RUNX2 vector. A similar two step PCR approach was implied to generate RUNX2 expression plasmid carrying six alanine mutations (FTYHTY405-407,426-428AAAAAA). FTY405-407AAA plasmid was used as template, and the primer pairs are as described above for HTY426-428AAA. The presence of mutated sequences and the in-frame ligation of all the plasmids were confirmed by automated sequencing using an internal primer.
Adenovirus expressing wild-type and C-terminal deletion mutant (Δ391) of RUNX2 are described previously (23). The Y428A and HTY426-428AAA mutant RUNX2 adenoviruses were generated by using the Adenovator system (Qbiogene, Carlsbad, CA). Briefly, HA-tagged mutant Runx2 cDNA were amplified by PCR using a common forward primer, 5′-CTTGGAAGATCTTTACCATGGATCTGTACGACGATGACGATAAG-3′, and a common reverse primer, 5′-CCTGAAAGATCTGCCCTCTAGATCAATATGG-3′, containing an engineered BglII restriction site. PCR products were digested with BglII enzyme and cloned into similarly digested pAdenoVator-CMV-IRES-GFP (Qbiogene). Presence of mutation and the integrity of the reading frame of the positive clone were confirmed by automated DNA sequencing. The expression and transcriptional activity of all pAdeno Vator constructs were examined in HeLa cells. The plasmids were then linearized with PmeI digestion and co-transformed into BJ5183 Escherichia coli cells along with the viral DNA plasmid pAdenoVator ΔE1/ΔE3. Recombinants were selected for kanamycin resistance and screened by restriction enzyme analysis. The recombinant adenoviral constructs were subsequently cleaved with PacI to expose its inverted terminal repeats and transfected into QBI-293A cells to produce viral particles. The virus particles were recovered from cells by three freeze/thaw cycles and further amplified. Fourth amplification containing high titer viral particles was purified by CsCl2 gradient and used for subsequent infections. The following multiplicity of infections (m.o.i.) was used for wild-type and point mutants of Runx2 adenovirus to maintain an equal level of expression and low cytotoxicity in Runx2 null cells (wild type, 50 m.o.i.; HTY, 60 m.o.i.; Y428A, 70 m.o.i.; and Δ391, 37 m.o.i.).
Transient Transfection and Luciferase Assays—All transient transfections were performed using SuperFect transfection reagent (Qiagen Inc). For promoter reporter assays, HeLa cells plated in 6-well dishes were transfected at 40% confluency with varying concentrations (250-500 ng) of Runx2 expression vectors, 0.5 μg of receptor SMADs, 500 ng of the multimerized Runx (6xRunx) or 3xTBRE promoter fused with luciferase gene, and 100 ng of Renilla luciferase reporter construct. Briefly, DNA-lipid complexes were formed in serum-free Dulbecco's modified Eagle's medium by mixing the indicated DNA amount with 7 μl of SuperFect reagent and incubating for 15 min at room temperature. The complexes were diluted in 1 ml of complete Dulbecco's modified Eagle's medium and overlaid on pre-washed cells. After 3 h, DNA-lipid complexes were removed, washed once with 1× phosphate-buffered saline, fed with fresh medium, and cultured for an additional 18 h in the presence or absence of 100 ng/ml BMP2. For TGFβ-responsive Smads, treatment with TGFβ (5 ng/ml) was carried out 6 h prior to harvesting. Cells were lysed in reporter lysis buffer, and luciferase activity was determined using a Dual Luciferase Reporter Assay kit (Promega, Madison, WI). Results were obtained from at least three independent experiments with triplicate samples.
Adenoviral Infections—Runx2 null cells were plated at a density of 0.3 × 106 cells per well of a 6-well plate. Cultures at 70% confluence were infected with the Runx2 (WT, Y428A, HTY, and Δ391) or β-galactosidase expressing control adenovirus in serum-free αMEM. After 90 min, cells were washed in αMEM without serum followed by addition of complete αMEM. Cells were cultured in the presence or absence of BMP2 (100 ng/ml) for an additional 12 days. Two days post infection cells were fed with osteogenic media (αMEM containing β-glycerophosphate and ascorbic acid). Media were changed every other day, and cells were subsequently harvested at indicated days for isolation of RNA or fixed in 2% formaldehyde in cacodylic buffer for ALP histology.
RNA Isolation and Quantitative PCR Analysis—RNA was isolated from control and adenovirus-reconstituted Runx2 null cells at different days of culture in the presence and absence of BMP2. Cells were briefly washed with phosphate-buffered saline and lysed with TRIzol reagent (Invitrogen) to isolate total RNA according to the manufacturer's protocol. RNA was treated with DNase I to remove any DNA contamination and purified using a DNA-free RNA column kit (Zymo Research, Orange, CA). SuperScript first strand synthesis kit (Invitrogen) was used to reverse transcribed 1 μg of RNA. cDNA was then subjected to real-time PCR to measure relative transcript levels using SYBR Green Master Mix (Bio-Rad) and gene-specific primers. Specificity of primers was initially verified by dissociation/melting curve for the amplicons. Transcript levels were normalized to glyceraldehyde-3-phosphate dehydrogenase levels in respective samples. The primers used for amplification were reported previously (27).
ALP Histology—Cells infected with indicated adenoviruses and cultured in the presence or absence of BMP2 for 4, 8, and 12 days were washed twice in 0.1 m cacodylic buffer and subsequently fixed in 2% formaldehyde in cacodylic buffer for 10 min at room temperature. After washing with 0.1 m cacodylic buffer, the following reagents all purchased from Sigma-Aldrich were added for the histological stain: Napthol AS-Mx phosphate disodium salt (25 mg), N,N-dimethyl formamide (1.4 ml), 0.2 m Tris maleate buffer (25 ml), and Fast red salt (50 mg). The total volume was made up to 50 ml by the addition of distilled water, and the solution was filter-sterilized and immediately added to the wells. The plates were incubated at 37 °C for 30 min or until color development was observed. The wells were then rinsed with distilled water, and stained cells were visualized using an inverted microscope.
Immunoprecipitation and Western Blots—HeLa cells were plated at a density of 0.7 × 106 cells per 100-mm plates. Cultures at 60% confluence were transfected with 5 μg each of RUNX2 (WT or mutant protein) and SMAD1 or -3 expression plasmids using SuperFect reagent. BMP2 (100 ng/ml) treatments were carried out 3 h post-transfection, whereas those for TGFβ (5 ng/ml) were carried out 6 h before harvesting. Cell were harvested 20-24 h following transfection in phosphate-buffered saline containing 25 μm proteasome inhibitor MG132 (Sigma-Aldrich) and Complete protease inhibitor mixture (Roche Applied Science). Cells were lysed by sonication at 10% for 10 s, and this process was repeated 5 times. Lysates clear of cellular debris were collected by centrifugation at 3000 rpm for 4 min, and 10% of the total lysate was saved as input. Remaining lysates were combined with 15 μg of species matched normal IgG, rabbit polyclonal Runx2 (M70, Santa Cruz Biotechnology) or mouse monoclonal FLAG (Sigma-Aldrich) antibody, and incubated on a rotating wheel for 1 h at 4 °C. Immunoprecipitates were collected with Protein A/G-agarose beads (Santa Cruz Biotechnology) and washed four times with phosphate-buffered saline containing 25 μm MG132 and protease inhibitor mixture. Total cell lysates or immunoprecipitates were fractionated by electrophoresis on a 12% SDS-polyacrylamide gel, transferred onto Immobilon P membranes (Millipore). Western blots were probed with monoclonal antibodies against Runx2 and FLAG and visualized with appropriate horseradish peroxidase-conjugated secondary antibody and chemiluminescence.
Isolation of Nuclear Extracts and Electrophoretic Mobility Shift Assay—HeLa cells plated in 100-mm culture dishes were transfected with 5 μg of wild-type and mutant Runx2 expression plasmid and pcDNA3.1 as empty vector control. Cells were harvested 30 h later for isolation of nuclear extracts essentially as described previously (22). Concentrations of nuclear protein were determined by Bradford assay, and vials were flash frozen and stored at -75 °C till required. Oligonucleotides representing a consensus Runx binding site was used for electrophoretic mobility shift assay and are reported earlier (22). Preparation and radioactive labeling of the oligonucleotide probe is essentially as described previously (22). Nuclear extracts (10 μg) from wild-type and mutated RUNX2 were incubated with labeled probe for 20 min at 22 °C and loaded onto a 4% nondenaturing polyacrylamide gel. Dried gels were exposed to film for autoradiography.
In Situ Immunofluorescence—HeLa cells were plated at a density of 8 × 104 cells/well on gelatin-coated coverslips in 6-well culture dishes. Cells at 40% confluency were transfected with 1 μg each of Runx2 (WT or mutant), FLAG-tagged BMP2-responsive Smad1 and cultured for 18 h. BMP2 (100 ng/ml) was added 3 h post-transfection. Cells were processed for in situ immunofluorescence as whole cell preparations as described previously (19). RUNX2 was detected by a rabbit polyclonal antibody (Santa Cruz Biotechnology) and SMAD with a mouse monoclonal antibody against FLAG epitope (Sigma-Aldrich) at a dilution of 1:400 each. Secondary antibodies used were anti-rabbit Alexa 488 and anti-mouse Alexa 568 (Molecular Probes) at a dilution of 1:800. Images were captured using a Zeis Axioplan microscope interfaced with a charge-coupled device camera and analyzed with MetaMorph software (Universal Imaging Inc).
For analysis of in situ co-localization, confocal microscopy was performed. A Leica TCS-SP inverted confocal microscope equipped with argon-helium and neon lasers with specific excitation lines was used (Bannockburn, IL). Ten randomly selected cells (five each from two independent cover slips) with equal expression levels of both proteins were used. Cell images with 0.25-μm section in respective wavelength were captured using similar exposure time. Two-dimensional progression was obtained, and numbers of Runx2 and SMAD foci per nucleus were counted using Leica Lite 2.0 and MetaMorph Imaging Software. The percent overlap was counted from total foci expressing WT and HTY mutant proteins. Average numbers of foci pooled from ten independent cells that exhibited colocalization are presented.
RESULTS
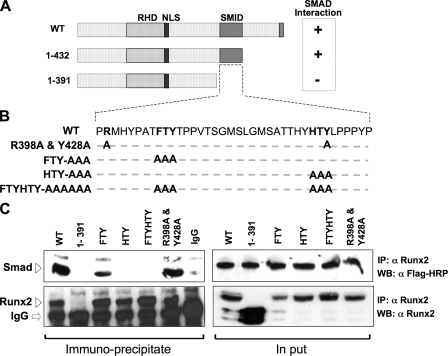
Specific Residues within the SMID/NMTS Domain Define RUNX2-SMAD Interactions—Studies were initiated to test the hypothesis that a critical role of RUNX2 in inducing bone formation and in regulating the progression of osteoblast differentiation depends on its interaction with SMAD proteins in response to BMP2 and TGFβ signaling. We previously documented by C-terminal deletion studies the presence of a SMID in RUNX2, which overlaps the well characterized nuclear matrix targeting signal (NMTS) as illustrated in Fig. 1A. We first mutated the FTY and HTY residues that form the tips of loops 1 and 2 in the NMTS/SMID, respectively. These residues are conserved among other family members and species (18) (Fig. 1B). For reference points, the N- and C-terminal amino acids of the RUNX2 SMID (Arg-398 and Tyr-428) were also mutated. Co-immunoprecipitation studies showed a decreased interaction of SMAD1 with the triple mutations of FTY residues in loop 1, while the HTY mutation in loop 2 resulted in complete loss of interaction. The combined mutation of both sequences (FTY-HTY) also results in complete loss of the RUNX2-SMAD interaction, similar to the Δ391-mutant RUNX2, which is shown as negative control (Fig. 1C). In contrast, the paired mutation (R398A and Y428A) had no effect on SMAD interaction. Expression levels of SMAD1 and mutant RUNX2 proteins are shown in input samples (Fig. 1C). RUNX2 interaction also occurs with the TGFβ-responsive FLAG-SMAD3 and reciprocal co-immunoprecipitation studies using FLAG antibody (data not shown). Thus, our data show the HTY motif in loop 2 of the NMTS (18) is required for formation of a RUNX2-SMAD complex.
FIGURE 1.
Three residues in the SMID of RUNX2 define physical association with receptor SMADs. A, schematic illustration of the wild-type Runx2 protein with key regulatory domains indicated (RHD, DNA binding runt homology domain; NLS, nuclear localization signal; SMID, Smad interacting domain). SMID was established by deletion mutagenesis and co-immunoprecipitation approach. B, sequence of the 41 amino acids that constitute SMID. Based on SMID, crystal structure residues in two predicted fingers were mutated to alanine. Relative positions of mutated amino acids are indicated in bold. C, HeLa cells were transiently co-transfected with 5 μg of FLAG-tagged receptor Smad1 and HA-tagged wild-type or mutant Runx2 expression plasmid. Cells were treated with 100 ng/ml of recombinant BMP2 3 h post-transfection and harvested 24 h later. Immunoprecipitations were performed on sonicated cell lysates using RUNX2 antibody or species-matched control IgG. Immunoprecipitates and 10% of input samples were resolved on SDS-PAGE, and Western blots were probed with either RUNX2 antibody or FLAG antibody to detect SMAD proteins. Specific bands corresponding to RUNX2 and SMAD 1 are indicated by arrowheads.
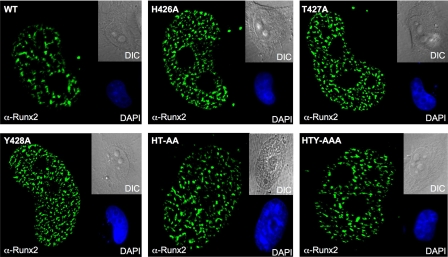
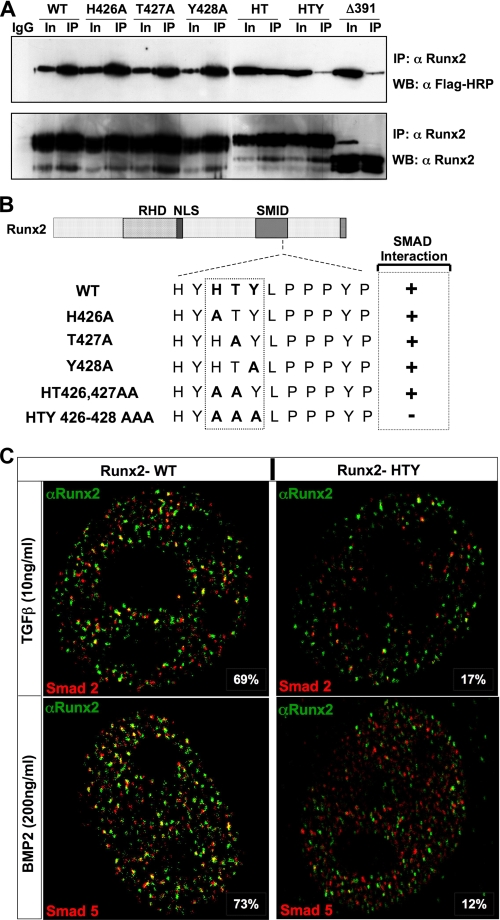
To further define the critical amino acids within the RUNX2 HTY motif for SMAD interaction, we generated a panel of additional RUNX2 mutant proteins. All three residues of the HTY motif were mutated individually and in pairs. We first confirmed the expression and in situ organization of the mutant RUNX2 proteins (Fig. 2). All mutant proteins showed comparable expression and a very punctate nuclear distribution pattern similar to the wild-type RUNX2 protein. Thus, substitution of each HTY residue with alanine does not alter the expression, nuclear import or cellular distribution of the RUNX2 protein. Co-immunoprecipitation studies revealed that physical association with SMAD1 protein is preserved when single or pairs of residues are mutated (Fig. 3). Loss of the RUNX2-SMAD interaction is observed only when all three amino-acids (HTY) are mutated together, equivalent to deletion of the entire C terminus domain (Δ391) of RUNX2 (Fig. 3, A and B). The input sample demonstrates comparable expression levels of all mutant proteins.
FIGURE 2.
Expression and normal cellular distribution are preserved in HTY domain mutant RUNX2 protein. HeLa cells were seeded on gelatin-coated glass coverslips and transfected with 1 μg of HA-tagged wild-type or mutant Runx2 expression plasmid. Cells were processed for in situ immunofluorescence. Whole cells were fixed in 4% formaldehyde, permeabilized, and stained with a polyclonal RUNX2 antibody followed by anti-rabbit Alexa488 secondary antibody. A similar pattern of punctate nuclear signal is observed for all mutant RUNX2 proteins. Nuclei are revealed by 4′,6-diamidino-2-phenylindole staining (DAPI). Differential interference contrast images represent bright field microscopy of respective cells. All magnifications are 63×.
FIGURE 3.
Histidine, tyrosine, and threonine are the minimal contact points required for formation of the SMAD-Runx2 complex. A, HeLa cells were transiently co-transfected with 5 μg each of receptor Smad 1 and either wild-type or indicated Runx2 mutant plasmids using SuperFect transfection reagent. Cells were cultured in the presence of 100 ng/ml of BMP2 and harvested 24 h later. Immunoprecipitations were performed with polyclonal RUNX2 antibody as described under “Experimental Procedures.” HTY and Δ391 RUNX2 protein failed to co-immunoprecipitate SMAD1. B, summary of RUNX2-SMAD complex formation is indicated schematically. Loss of association is observed only for HTY mutant RUNX2 protein. C, HeLa cells were co-transfected with 1 μg of Smad2, Smad5, and wild-type or HTY mutant Runx2 expression plasmids. Cells were cultured in the presence of either 10 ng/ml TGFβ or 100 ng/ml BMP2 and processed 21 h later for in situ immunofluorescence. Ten cells from two independent coverslips with equal expression levels of both proteins were quantified for co-localizations of RUNX2 and SMAD foci by confocal microscopy, and a representative image is shown. The numbers represent the degree of co-localization observed for wild-type (69 and 73%) and HTY mutant (17 and 12%) proteins, respectively.
We further examined the consequences of HTY mutation on in situ organization of RUNX2-SMAD complexes by confocal microscopy. Like wild type, HTY mutant protein is trafficked into the nucleus by the intact NLS but is severely compromised in its ability to co-localize with Smad protein (Fig. 3C). In an intact cell, a high degree of co-localization is observed between wild-type RUNX2 protein and TGFβ-activated SMAD2 (69%) and BMP2-responsive SMAD5 (73%) foci. In contrast HTY mutant protein exhibits a dramatically decreased co-localization of only 17 and 12% with SMAD2 and SMAD5, respectively. Thus HTY mutation disrupts both biochemical and in situ association with receptor SMADs. Taken together these findings establish the HTY motif as a minimal contact point that is required for formation of the RUNX2-SMAD regulatory complex in response to BMP/TGFβ signaling.
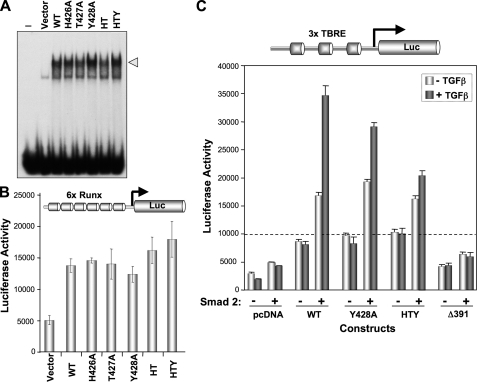
Mutations in SMID Do Not Alter RUNX2-mediated Transcription, but Integration of the BMP/TGFβ Signal on Target Gene Promoters Is Impaired—To understand the functional consequences of disruption of the RUNX2-SMAD interaction, we compared the ability of various SMID mutants to transcriptionally activate RUNX2 target gene promoters. Initially DNA binding properties of each of the mutations of the HTY motif were assessed with electrophoretic mobility shift assay assays (Fig. 4A). Nuclear extracts prepared from transiently transfected HeLa cells with the indicated Runx2 expression vector were incubated with an oligonucleotide containing consensus Runx sequences. Because HeLa cells lack endogenous RUNX2 protein, RUNX2 DNA binding of WT and mutant proteins could be compared. All mutant RUNX2 proteins retain DNA binding activities (Fig. 4A). Similar results were also noted with in vitro transcribed and translated wild-type and SMID mutant RUNX2 proteins (data not shown). Consistent with their similar DNA binding activities, no significant difference in transcriptional activation of a multimerized (6X-Runx-luciferase) promoter reporter was observed (Fig. 4B). WT and all mutant RUNX2 proteins showed enhanced promoter activity (3- to 3.5-fold) when compared with empty vector control. However, when the RUNX2-SMAD transcriptional activation function was examined using a promoter that contains DNA binding sites for both Runx and Smad (3X-TBRE-Luc), differential responses to TGFβ in the presence of SMAD2 were observed (Fig. 4C). When compared with WT, the HTY mutant RUNX2 showed 50-70% reduced TGFβ responsiveness, whereas a complete loss was seen for Δ391 RUNX2 proteins. Impaired TGFβ responsiveness of HTY and Δ391 proteins is consistent with loss of RUNX2-SMAD physical association. Taken together these data demonstrate that mutation of the HTY motif in RUNX2 does not change DNA binding or transcriptional activation but selectively blocks integration of the BMP/TGFβ regulatory signal for target gene expression.
FIGURE 4.
Mutant RUNX2 proteins exhibit normal DNA binding and functional activation but not the integration of BMP/TGFβ signal. A, nuclear extracts (10 μg) prepared from transfected HeLa cells with indicated Runx2 plasmids were used for electrophoretic mobility shift assay. Comparable DNA binding activities indicated by the arrowhead are noted for all RUNX2 proteins. B, HeLa cells cultured in 6-well dishes were co-transfected with 0.4μg of pcDNA3.1 empty vector, wild-type, and mutant Runx2 plasmid and 1 μg of 6X-Runx-Luciferase reporter plasmids. Cells were harvested 24 h post-transfection, and luciferase activities were determined. Pooled data from two independent experiments (n = 12) are shown. C, cells were co-transfected with 0.5 μg of Smad2, 0.5 μg of indicated Runx2, 0.5 μg of 3X-TBRE-Luciferase reporter plasmids, and 100 ng of Renilla luciferase construct as an internal control. Each TGFβ response element in TBRE-luciferase reporter also contains an overlapping Runx binding element. Cells were treated with 5 ng of TGFβ for 6 h prior to harvest for luciferase assays. TGFβ stimulated enhanced (2- to 2.5-fold) promoter activation is lost by HTY and Δ391 RUNX2 mutant proteins.
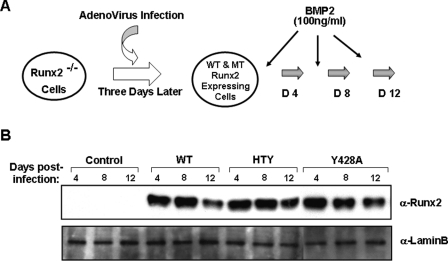
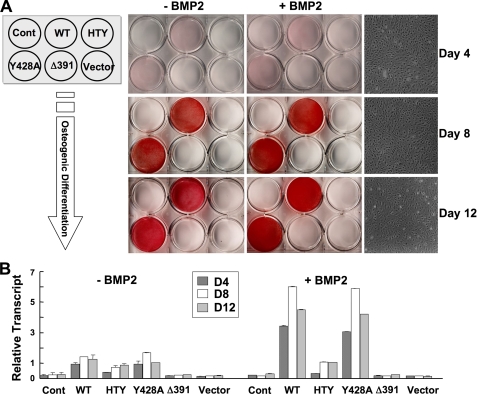
Integrity of the RUNX2-SMAD Molecular Complex Supports BMP2-induced Commitment to the Osteoblast Phenotype and Osteoblast Differentiation—Induction and progression of osteogenesis from mesenchymal stem cells are linked with bone morphogenetic protein signals. Therefore, interaction of BMP responsive SMADs with RUNX2 may represent a key component of the BMP2 signal transduction pathway. To test the hypothesis that osteoblast differentiation is critically dependent on the RUNX2-SMAD molecular complex, we designed a biological assay using a previously characterized immortalized Runx2 null cell line (15, 27). These cells were infected with adenovirus expressing either wild-type or mutant RUNX2 proteins and cultured for 12 days in the presence or absence of BMP2 as shown in Fig. 5. Comparable levels of both wild-type and mutant RUNX2 proteins are expressed for an extended time sufficient to allow cellular differentiation, then slightly decrease by 2 weeks (Fig. 5B). Expression of the bone phenotypic early marker ALP during 12 days of culture in osteogenic media was determined by mRNA levels and cytochemical staining (Fig. 6). Runx2 null cells are incompetent to differentiate in response to BMP2 alone or when infected with control (empty vector) virus, as indicated by the lack of ALP activity in control cells. However, reintroduction of either wild-type RUNX2 or Y428A RUNX2 mutant protein (previously shown to reduce RUNX2 subnuclear targeting (24)) by adenovirus infection restored the osteoblast phenotype with nearly equivalent activity (Fig. 6B). A progressive increase in ALP activity was observed during 12 days, which was further enhanced from 3- to 6-fold upon BMP2 stimulation (Fig. 6A). In sharp contrast, cells reconstituted with either HTY or Δ391 RUNX2 mutant were devoid of ALP enzyme activity in the absence or presence of BMP2 at any time point. These findings are not related to modifications in cell density nor to the loss of protein expression, as comparable levels were seen over 12 days (Fig. 5B, and data not shown). We do observe a slight increase in ALP mRNA levels (2-fold) with HTY RUNX2; however, treatment with BMP2 did not change these levels. The negative controls (EV and RUNX2 Δ391) had undetectable ALP mRNA levels.
FIGURE 5.
Expression of wild-type and mutant RUNX2 proteins in reconstituted null cells is sustained throughout the osteoblast differentiation time course. A, schematic illustration of the reconstitution biological assay. Three days after plating, Runx2 null cells were infected with adenovirus expressing wild-type or mutant RUNX2 protein for 4 h. Cells were then cultured for additional 12 days in osteo-inductive mixture and in the presence and absence of 100 ng/ml BMP2. B, Runx2 null cells were infected with wild-type (50 m.o.i.), Y428A (70 m.o.i.), HTY (60 m.o.i.), and Δ391 (37 m.o.i.) and cultured for 12 days in 6-well dishes. Cells at indicated days were directly lysed in plates as described under “Experimental Procedures.” For comparison of RUNX2 expression levels, equal amounts of cell lysates were resolved on 10% SDS-PAGE for Western blot analysis. Blots were probed with monoclonal RUNX2 antibody, stripped, reprobed with LaminB antigen, and used as loading control.
FIGURE 6.
Loss of osteogenic differentiation and BMP signal integration by SMID mutant RUNX2. A, commitment of reconstituted Runx2 null cells to osteoblast lineages was tested for 12-day time period. Runx2 null cells cultured in 6-well dishes were infected with pre-determined m.o.i. of Runx2 (wild-type, Y428A, HTY, and Δ391) and control adenovirus. Schematic illustration of a 6-well culture dish on the left indicates different adenovirus and their respective wells. Cells with indicated treatments and days were fixed, and immunocytochemistry for early marker alkaline phosphatase was carried out as described under “Experimental Procedures.” Images of scanned ALP-stained plates are presented. ALP activity and BMP2 enhanced signal were seen only in WT- and Y428A-expressing cells. B, cells infected with indicated adenoviruses were cultured in the presence and absence of BMP2 (100 ng/ml) in osteogenic media and harvested at the indicated days for total RNA isolation. Relative expression levels of the ALP early marker gene for osteoblasts were monitored by real-time reverse transcription-PCR analysis.
These data suggest that, although HTY mutant RUNX2 protein exhibits normal DNA binding and transcriptional activation profiles in promoter-reporter assays, loss of SMAD interaction considerably decreased its capacity to induce osteogenic differentiation, as well as resulted in failure of integration of the BMP2 osteogenic signal in a biological assay.
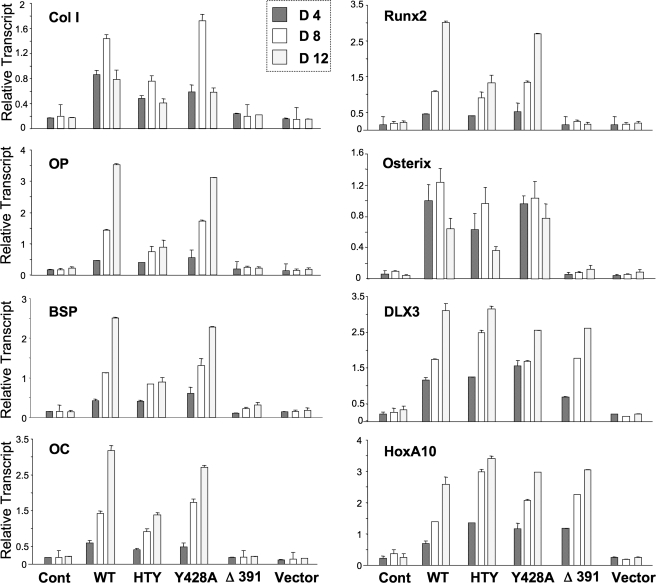
The RUNX2-SMAD Complex Is Required for the Progression of Osteoblast Differentiation—To determine the function of the HTY mutant RUNX2 protein with respect to late stages of osteoblast differentiation, the osteogenic properties of the reconstituted Runx2 null cells were examined by monitoring expression of mid and late osteoblast marker genes (Fig. 7). Histone H4 mRNA expression, which is coupled to DNA synthesis showed a similar profile in all treatment groups with cessation of cell growth observed after day 8 of differentiation (data not shown). Wild-type and the Tyr-428 mutant RUNX2 protein showed a temporal activation of phenotypic markers (collagen type I, osteopontin, bone sialoprotein, Runx2, Osterix, and osteocalcin) to the same extent. In contrast, this expression profile for RUNX2 target genes is reduced to 50% with the HTY mutant, similar to early stage marker ALP. The Δ391 RUNX2 mutant, which lacks the SMID and C terminus, completely failed to activate any RUNX2 target genes that reflect differentiation. Thus Runx2 null cells can only fully progress through stages of osteoblast maturation if reconstituted with RUNX2 protein that retains a RUNX2-SMAD interaction. However, we do find that the homeodomain proteins DLX3 and HOXA10, recently characterized as proteins that contribute to activation of bone-related genes for promoting osteoblast differentiation, independent of RUNX2 (33, 34), are expressed nearly equivalently by wild-type and all mutant RUNX2 proteins. These results indicate their expression during osteoblast differentiation is not dependent on RUNX2 activities (Fig. 7).
FIGURE 7.
Disruption of RUNX2-SMAD interaction results in altered expression of osteoblast markers genes. The osteogenic capacity of the wild-type and mutant Runx2 proteins were tested in a reconstitution assay of Runx2-/- cell line. Cells were plated in 6-well dishes, and upon confluency infected with Runx2 adenovirus for 4 h. Cells were then washed and cultured for indicated days in osteogenic media (αMEM supplemented with 10% fetal bovine serum, 10 mm β-glycerol phosphate, and 50 μg/ml ascorbic acid). Control and Runx2-infected cells were harvested for RNA isolation at indicated days. Real-time reverse transcription-PCR analyses were performed to assess expression of the cell cycle, RUNX2 target, and other transcription factor genes during osteoblastic differentiation. Data are presented as relative transcript normalized to glyceraldehyde-3-phosphate dehydrogenase levels in the same sample. All experiments were performed in duplicates, and mean normalized values with standard deviation are shown.
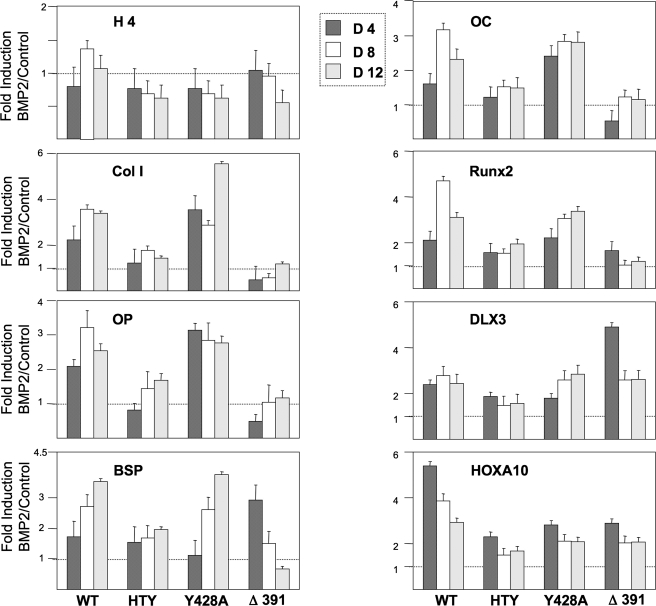
To provide evidence that the RUNX2-SMAD interaction is essential for incorporating a regulatory signal from the BMP2/TGFβ pathway for osteoblast differentiation, expression of the osteogenic marker genes was examined in response to BMP2 (Fig. 8). A modest anti-proliferative effect (<0.35-fold decreased histone H4 expression) was observed in the presence of BMP2 for the HTY, Y428A, and Δ391 mutant proteins; thus this BMP2 function is not dependent on RUNX2-SMAD interactions. The -fold induction values (BMP2/Control) showed generally similar BMP responsiveness between WT and Y428A for bone matrix-related RUNX2 target genes. However, BMP2 responsiveness is compromised by Runx2 HTY mutant proteins for bone matrix proteins, RUNX2, DLX3, and HOXA10, the three transcription factors whose expression in mesenchymal cells is induced by BMP2 (35, 36), further emphasizing the significance of RUNX2-SMAD-mediated BMP2 signaling. BMP2 effects of enhanced osteogenic differentiation for target gene expression are clearly integrated by wild-type and Y428A mutant RUNX2 proteins, which retain SMAD transduction signaling. BMP2 mediated stimulation (2-5 fold) is seen for collagen type I, osteopontin, BSP, osteocalcin, and Runx2. We note that for Y428A on day 12, the BMP2 -fold stimulation for Col-1 was highest in this representative study. Integration of the BMP2 signal for all marker genes was completely lost or significantly reduced in HTY and Δ391 RUNX2 mutants where SMAD interactions are disrupted. The anomaly of a 3-fold increased expression of BSP by Δ391 on day 4 was related to extremely low transcript levels as shown in Fig. 7.
FIGURE 8.
Integration of BMP2 osteogenic signal by RUNX2 during differentiation requires RUNX2-SMAD interaction. Runx2-/- cells were cultured in 6-well plates and 3 days later infected with the pre-determined m.o.i. of indicated Runx2 adenoviruses. Cells were fed every other day with osteogenic media in the presence or absence of 100 ng/ml BMP2. Cells were harvested at the indicated days, and RNA was isolated. Real time-PCR for selective marker genes were carried out in duplicate, and values were normalized to internal glyceraldehyde-3-phosphate dehydrogenase levels in the same sample. Data are presented as -fold induction (virus-infected BMP2-treated/virus-infected control).
Taken together these findings demonstrate that three residues (HTY) in RUNX2 define a critical motif for formation of RUNX2-SMAD regulatory complex, which is essential for activation of gene networks that drive osteoblast differentiation. In summary our results clearly establish RUNX2 as a molecular end point that is required for execution and completion of TGFβ/BMP2 signaling in osteoblasts.
DISCUSSION
Our principal finding is the demonstration of a critical requirement for BMP-induced RUNX2-SMAD interaction to transduce the osteogenic signal that leads to osteoblast differentiation. Although RUNX2 has been characterized as an essential transcription factor for bone formation and BMP2, -4, and -7 have been appreciated as potent osteogenic morphogens (37-40), the interdependency of these two factors for promoting osteoblast differentiation is now established from our studies. We have demonstrated a definitive mechanism by which BMP signaling is transduced to an osteogenic signal through the formation of RUNX2-SMAD transcriptional regulatory complexes. This mechanism was characterized by site-directed mutagenesis of Runx2, identifying a requirement for mutation of three contiguous residues to abrogate the RUNX2-SMAD physical and functional interaction. This HTY mutant RUNX2 was incompetent to restore osteoblast differentiation of Runx2 null cells.
The consequences of the HTY RUNX2 mutation on osteoblast differentiation upon reconstitution of the Runx2 null cell line are observed to be nearly as severe as that of the RUNX2 Δ391 mutant with deletion of the entire C terminus. The comparison of these two mutants in this cell model supports an interpretation that the Runx2ΔC knock-in mouse phenotype lacking a mineralized skeleton can be attributed largely to the loss of SMAD interaction with RUNX2 (17). The HTY mutant RUNX2 mediates a 50-75% reduced activation of target gene (ALP, COL-1, OP, BSP, and OC) expression in a cellular context, although the mutants exhibit normal DNA binding, and equally activate a multimerized promoter. Yet, BMP2 addition fails to enhance transcriptional activity of HTY RUNX2.
A second key finding of our studies is that RUNX2 can function in the absence of exogenously added BMP2; however, BMP2 alone is insufficient for complete differentiation to the osteoblast phenotype in the absence of RUNX2. This novel finding could only have been established in an early embryonic osteogenic lineage cell devoid of the Runx2 gene. The establishment of a cell line from calvarial tissue of the E17.5 Runx2 null mice provided us with a powerful and unbiased biological assay system (27). Complete differentiation of our pre-osteogenic immortalized Runx2 null cell line by BMP2 is dependent on Runx2. These findings are similar to primary osteoblastic cells isolated from the Runx2-/- mouse (15, 26). However, one recent report has described a fibroblastic cell line immortalized from the Runx2 null mouse that can respond to BMP2 signal independent of Runx2 (41). Our cell line has allowed us to perform reconstitution studies with wild-type and mutant RUNX2 proteins to determine the independent and convergent nature of the RUNX2 and BMP osteogenic signals. It is interesting to note that cellular levels of other osteogenic factors (BMP, C/EBP, ATF4, steroid receptors, homeodomain, and Hox transcription factors) that may be present in these pre-osteogenic cells are not sufficient to mediate the BMP2 response for the complete program of osteoblast differentiation.
Significantly, the RUNX2 HTY mutation not only disrupts formation of the RUNX2-SMAD co-regulatory complex but is linked to the organization of RUNX2-SMAD complexes necessary for transduction of the BMP signal. RUNX2 is associated with the nuclear scaffold by a nuclear matrix targeting signal. Although SMADs enter the nucleus in response to a BMP ligand, there is a requirement for further trafficking and recruitment to RUNX2 foci to transduce the BMP2 signal to activation of RUNX2 target genes (24). Formation of tissue-specific transcriptional regulatory complexes involving their organization in subnuclear specific domains is a fundamental level of control for achieving either developmental or tissue-specific gene regulation. Although several important RUNX2 co-regulatory proteins have now been identified that interact in the C-terminal region (9, 10), the present studies firmly establish that the osteogenic effects of BMP2 are accounted for by RUNX2-SMAD interactions and suggest that these interactions contribute to the in vivo bone phenotypes in human and mouse.
In summary, our studies provide direct evidence that RUNX2 does not require BMP2 for expression of osteoblast genes, but BMP2 requires RUNX2 for induction of osteoblast differentiation. The loss of SMAD function by the HTY mutation coupled with the inability of this mutant to be organized with co-regulatory proteins in subnuclear domains, establishes a critical structural requirement for RUNX2-SMAD interactions in RUNX2-specific subnuclear domains, as necessary for mediating BMP/TGFβ signaling related to the control of osteoblast differentiation. BMP2, -4, and -7 have established roles in skeletogenesis (37). We propose that the RUNX2-SMAD interaction is a mechanism for specification of the osteogenic activity of the multi-functional BMP morphogens that can affect the development of other cell phenotypes and tissues (muscle, nerve, and skin) (42-45). Development of skeletal lineage cells requires coordinated activity of both RUNX2 and BMP2, and, in the absence of the RUNX2 platform, osteogenic signaling of BMP pathways is blunted.
This work is supported by National Institutes of Health Grants P01 AR48818 and DE012528. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: BMP, bone morphogenetic protein; TGFβ, transforming growth factor-β; Runx2, runt-related transcription factor 2; RHD, runt homology domain; SMID, SMAD interacting domain; ALP, alkaline phosphatase; NMTS, nuclear matrix targeting signal; αMEM, α minimal essential medium; HA, hemagglutinin; m.o.i., multiplicity of infection; WT, wild type.
References
- 1.Liu, F., Hata, A., Baker, J. C., Doody, J., Carcamo, J., Harland, R. M., and Massague, J. (1996) Nature 381 620-623 [DOI] [PubMed] [Google Scholar]
- 2.Massague, J., Seoane, J., and Wotton, D. (2005) Genes Dev. 19 2783-2810 [DOI] [PubMed] [Google Scholar]
- 3.Massague, J. (2003) Clin. Adv. Hematol. Oncol. 1 576-577 [PubMed] [Google Scholar]
- 4.Li, X., Nie, S., Chang, C., Qiu, T., and Cao, X. (2006) Exp. Cell Res. 312 854-864 [DOI] [PubMed] [Google Scholar]
- 5.Haag, J., and Aigner, T. (2006) Arthritis Rheum. 54 3878-3884 [DOI] [PubMed] [Google Scholar]
- 6.Hendy, G. N., Kaji, H., Sowa, H., Lebrun, J. J., and Canaff, L. (2005) Horm. Metab Res. 37 375-379 [DOI] [PubMed] [Google Scholar]
- 7.Suzuki, A., Raya, A., Kawakami, Y., Morita, M., Matsui, T., Nakashima, K., Gage, F. H., Rodriguez-Esteban, C., and Izpisua Belmonte, J. C. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 10294-10299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Itoh, S., and ten Dijke, P. (2007) Curr. Opin. Cell Biol. 19 176-184 [DOI] [PubMed] [Google Scholar]
- 9.Lian, J. B., Javed, A., Zaidi, S. K., Lengner, C., Montecino, M., van Wijnen, A. J., Stein, J. L., and Stein, G. S. (2004) Crit. Rev. Eukaryot. Gene Expr. 14 1-41 [PubMed] [Google Scholar]
- 10.Schroeder, T. M., Jensen, E. D., and Westendorf, J. J. (2005) Birth Defects Res. C. Embryo. Today 75 213-225 [DOI] [PubMed] [Google Scholar]
- 11.Yang, X., and Karsenty, G. (2002) Trends Mol. Med. 8 340-345 [DOI] [PubMed] [Google Scholar]
- 12.Franceschi, R. T., and Xiao, G. (2003) J. Cell. Biochem. 88 446-454 [DOI] [PubMed] [Google Scholar]
- 13.Zhang, Y. W., Yasui, N., Ito, K., Huang, G., Fujii, M., Hanai, J., Nogami, H., Ochi, T., Miyazono, K., and Ito, Y. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 10549-10554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Otto, F., Kanegane, H., and Mundlos, S. (2002) Hum. Mutat. 19 209-216 [DOI] [PubMed] [Google Scholar]
- 15.Komori, T., Yagi, H., Nomura, S., Yamaguchi, A., Sasaki, K., Deguchi, K., Shimizu, Y., Bronson, R. T., Gao, Y.-H., Inada, M., Sato, M., Okamoto, R., Kitamura, Y., Yoshiki, S., and Kishimoto, T. (1997) Cell 89 755-764 [DOI] [PubMed] [Google Scholar]
- 16.Otto, F., Thornell, A. P., Crompton, T., Denzel, A., Gilmour, K. C., Rosewell, I. R., Stamp, G. W. H., Beddington, R. S. P., Mundlos, S., Olsen, B. R., Selby, P. B., and Owen, M. J. (1997) Cell 89 765-771 [DOI] [PubMed] [Google Scholar]
- 17.Choi, J.-Y., Pratap, J., Javed, A., Zaidi, S. K., Xing, L., Balint, E., Dalamangas, S., Boyce, B., van Wijnen, A. J., Lian, J. B., Stein, J. L., Jones, S. N., and Stein, G. S. (2001) Proc. Natl. Acad. Sci., U. S. A. 98 8650-8655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang, L., Guo, B., Javed, A., Choi, J.-Y., Hiebert, S., Lian, J. B., van Wijnen, A. J., Stein, J. L., Stein, G. S., and Zhou, G. W. (1999) J. Biol. Chem. 274 33580-33586 [DOI] [PubMed] [Google Scholar]
- 19.Javed, A., Guo, B., Hiebert, S., Choi, J.-Y., Green, J., Zhao, S.-C., Osborne, M. A., Stifani, S., Stein, J. L., Lian, J. B., van Wijnen, A. J., and Stein, G. S. (2000) J. Cell Sci. 113 2221-2231 [DOI] [PubMed] [Google Scholar]
- 20.Zaidi, S. K., Javed, A., Pratap, J., Schroeder, T. M., Westendorf, J., Lian, J. B., van Wijnen, A. J., Stein, G. S., and Stein, J. L. (2006) J. Cell. Physiol. 209 935-942 [DOI] [PubMed] [Google Scholar]
- 21.Young, D. W., Hassan, M. Q., Pratap, J., Galindo, M., Zaidi, S. K., Lee, S., Yang, X., Xie, R., Underwood, J., Furcinitti, P., Imbalzano, A. N., Penman, S., Nickerson, J. A., Montecino, M. A., Lian, J. B., Stein, J. L., van Wijnen, A. J., and Stein, G. S. (2007) Nature 445 442-446 [DOI] [PubMed] [Google Scholar]
- 22.Javed, A., Barnes, G. L., Jassanya, B. O., Stein, J. L., Gerstenfeld, L., Lian, J. B., and Stein, G. S. (2001) Mol. Cell. Biol. 21 2891-2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Afzal, F., Pratap, J., Ito, K., Ito, Y., Stein, J. L., van Wijnen, A. J., Stein, G. S., Lian, J. B., and Javed, A. (2005) J. Cell. Physiol. 204 63-72 [DOI] [PubMed] [Google Scholar]
- 24.Zaidi, S. K., Sullivan, A. J., van Wijnen, A. J., Stein, J. L., Stein, G. S., and Lian, J. B. (2002) Proc. Natl. Acad. Sci., U. S. A. 99 8048-8053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, K. S., Kim, H. J., Li, Q. L., Chi, X. Z., Ueta, C., Komori, T., Wozney, J. M., Kim, E. G., Choi, J. Y., Ryoo, H. M., and Bae, S. C. (2000) Mol. Cell. Biol. 20 8783-8792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Young, D. W., Pratap, J., Javed, A., Weiner, B., Ohkawa, Y., van Wijnen, A., Montecino, M., Stein, G. S., Stein, J. L., Imbalzano, A. N., and Lian, J. B. (2005) J. Cell. Biochem. 94 720-730 [DOI] [PubMed] [Google Scholar]
- 27.Bae, J. S., Gutierrez, S., Narla, R., Pratap, J., Devados, R., van Wijnen, A. J., Stein, J. L., Stein, G. S., Lian, J. B., and Javed, A. (2007) J. Cell. Biochem. 100 434-449 [DOI] [PubMed] [Google Scholar]
- 28.Miyazono, K., Maeda, S., and Imamura, T. (2005) Cytokine Growth Factor Rev. 16 251-263 [DOI] [PubMed] [Google Scholar]
- 29.Leboy, P., Grasso-Knight, G., D'Angelo, M., Volk, S. W., Lian, J. V., Drissi, H., Stein, G. S., and Adams, S. L. (2001) J. Bone Joint Surg. Am. 83 Suppl. 1, S15-S22 [PubMed] [Google Scholar]
- 30.Lee, K. S., Hong, S. H., and Bae, S. C. (2002) Oncogene 21 7156-7163 [DOI] [PubMed] [Google Scholar]
- 31.Xiao, G., Gopalkrishnan, R. V., Jiang, D., Reith, E., Benson, M. D., and Franceschi, R. (2002) J. Bone Miner. Res. 17 101-110 [DOI] [PubMed] [Google Scholar]
- 32.Phimphilai, M., Zhao, Z., Boules, H., Roca, H., and Franceschi, R. T. (2006) J. Bone Miner. Res. 21 637-646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hassan, M. Q., Tare, R. S., Lee, S., Mandeville, M., Morasso, M. I., Javed, A., van Wijnen, A. J., Stein, J. L., Stein, G. S., and Lian, J. B. (2006) J. Biol. Chem. 281 40515-40526 [DOI] [PubMed] [Google Scholar]
- 34.Dobreva, G., Chahrour, M., Dautzenberg, M., Chirivella, L., Kanzler, B., Farinas, I., Karsenty, G., and Grosschedl, R. (2006) Cell 125 971-986 [DOI] [PubMed] [Google Scholar]
- 35.Holleville, N., Mateos, S., Bontoux, M., Bollerot, K., and Monsoro-Burq, A. H. (2007) Dev. Biol. 304 860-874 [DOI] [PubMed] [Google Scholar]
- 36.Hassan, M. Q., Tare, R. S., Lee, S., Mandeville, M., Weiner, B., Montecino, M., van Wijnen, A. J., Stein, J. L., Stein, G. S., and Lian, J. B. (2007) Mol. Cell. Biol. 27 3337-3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bandyopadhyay, A., Tsuji, K., Cox, K., Harfe, B. D., Rosen, V., and Tabin, C. J. (2006) PLoS. Genet. 2 e216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanzler, B., Foreman, R. K., Labosky, P. A., and Mallo, M. (2000) Development 127 1095-1104 [DOI] [PubMed] [Google Scholar]
- 39.Lee, M. H., Kim, Y. J., Kim, H. J., Park, H. D., Kang, A. R., Kyung, H. M., Sung, J. H., Wozney, J. M., Kim, H. J., and Ryoo, H. M. (2003) J. Biol. Chem. 278 34387-34394 [DOI] [PubMed] [Google Scholar]
- 40.Zhu, W., Rawlins, B. A., Boachie-Adjei, O., Myers, E. R., Arimizu, J., Choi, E., Lieberman, J. R., Crystal, R. G., and Hidaka, C. (2004) J. Bone Miner. Res. 19 2021-2032 [DOI] [PubMed] [Google Scholar]
- 41.Liu, T., Gao, Y., Sakamoto, K., Minamizato, T., Furukawa, K., Tsukazaki, T., Shibata, Y., Bessho, K., Komori, T., and Yamaguchi, A. (2007) J. Cell. Physiol. 211 728-735 [DOI] [PubMed] [Google Scholar]
- 42.Zhao, G. Q. (2003) Genesis 35 43-56 [DOI] [PubMed] [Google Scholar]
- 43.Knockaert, M., Sapkota, G., Alarcon, C., Massague, J., and Brivanlou, A. H. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 11940-11945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fukuda, S., Abematsu, M., Mori, H., Yanagisawa, M., Kagawa, T., Nakashima, K., Yoshimura, A., and Taga, T. (2007) Mol. Cell. Biol. 27 4931-4937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xia, Y., Yu, P. B., Sidis, Y., Beppu, H., Bloch, K. D., Schneyer, A. L., and Lin, H. Y. (2007) J. Biol. Chem. 282 18129-18140 [DOI] [PubMed] [Google Scholar]