Abstract
Makorin-2 belongs to the makorin RING zinc finger gene family, which encodes putative ribonucleoproteins. Here we cloned the Xenopus makorin-2 (mkrn2) and characterized its function in Xenopus neurogenesis. Forced overexpression of mkrn2 produced tadpoles with dorso-posterior deficiencies and small-head/short-tail phenotype, whereas knockdown of mkrn2 by morpholino antisense oligonucleotides induced double axis in tadpoles. In Xenopus animal cap explant assay, mkrn2 inhibited activin, and retinoic acid induced animal cap neuralization, as evident from the suppression of a pan neural marker, neural cell adhesion molecule. Surprisingly, the anti-neurogenic activity of mkrn2 is independent of the two major neurogenesis signaling cascades, BMP-4 and Wnt8 pathways. Instead, mkrn2 works specifically through the phosphatidylinositol 3-kinase (PI3K) and Akt-mediated neurogenesis pathway. Overexpression of mkrn2 completely abrogated constitutively active PI3K- and Akt-induced, but not dominant negative glycogen synthase kinase-3β (GSK-3β)-induced, neural cell adhesion molecule expression, indicating that mkrn2 acts downstream of PI3K and Akt and upstream of GSK-3β. Moreover, mkrn2 up-regulated the mRNA and protein levels of GSK-3β. These results revealed for the first time the important role of mkrn2 as a new player in PI3K/Akt-mediated neurogenesis during Xenopus embryonic development.
Makorin RING zinc finger (MKRN)3 gene family encodes putative ribonucleoproteins with a distinctive array of zinc finger domains. To date, nine MKRN family loci scattered throughout the human genome have been identified (1). The most studied member of this family is makorin-1 gene (MKRN1), which was recently documented to modulate telomere length homeostasis (2) and RNA polymerase II-dependent transcription (3). MKRN1 is widely transcribed in mammals, particularly in murine embryonic nervous system and adult testis (1). Such expression pattern suggests that members of this gene family may play an important role in embryonic development and neurogenesis.
We had previously identified a new gene makorin-2 (MKRN2), formerly designated as HSPC070, from human CD34+ hematopoietic stem/progenitor cells (4). The nucleotide sequence of this gene shares 105 bp in the 3′-untranslated region with an oncogene c-RAF in reversed transcription orientation (4, 5). However, the function of MKRN2 remains elusive.
Here we report the cloning of the Xenopus ortholog of human MKRN2 (mkrn2) and the characterization of its functions in Xenopus neurogenesis. In both gain-of-function and loss-of-function studies, we showed that dysregulation of mkrn2 produced morphological defects in neural development. To elucidate the underlying mechanism, we employed animal cap (AC) explant culture system to examine the effect of mkrn2 on the expression of a key neural marker, neural cell adhesion molecule (NCAM) (6). Xenopus AC is a unique model system for studying neural and mesodermal induction as it is capable of giving rise to either neural or mesodermal tissues in the presence of suitable inducers. We showed that forced expression of mkrn2 could inhibit activin- and retinoic acid-induced AC neuralization and NCAM expression. In contrast, knockdown of mkrn2 by morpholino antisense oligonucleotide led to enhanced neuralization and NCAM induction. To our surprise, mkrn2 is not involved in the two major neurogenesis signaling cascades, BMP-4 and Wnt8. Instead, it works specifically and exclusively in the phosphatidylinositol 3-kinase (PI3K)/Akt/GSK-3β-mediated neurogenesis pathway we previously reported (7).
EXPERIMENTAL PROCEDURES
Cloning of mkrn2—The cDNA encoding mkrn2 was cloned by reverse transcription-PCR (RT-PCR) using the primer pair: 5′-ATG AGT CCC AAG CAG GTG ACG TGC A-3′ (forward) and 5′-AGG AAC TCC CTG CAC CCT TTA ACT G-3′ (reverse). Total RNA isolated from Xenopus oocytes was used as template. The PCR product was ligated into PCR 3.1 vector, double-digested with restriction endonucleases (SmaI and StuI), and then subcloned into pSP64TS plasmid. The cDNA sequence of mkrn2 was deposited in GenBank™ (accession number EF626804).
Design of Morpholino Antisense Oligonucleotides—Two separate 25-mer morpholinos (Gene Tools LLC, Philomath, OR) were designed based on the cDNA sequence of mkrn2: MOa, 5′-TGC ACG TCA CCT GCT TGG GAC TCA T-3′, and MOb, 5′-CTG CAC GTC ACC TGC TTG GGA CTC A-3′.
Embryo Microinjection and Animal Cap Assay—Xenopus embryos obtained by artificial insemination were cultured as described previously (7). Embryos were staged according to Nieuwkoop and Faber (8). Embryos at two-cell stage were injected with various mRNAs and morpholinos in the animal poles. For microinjection at four-cell stage, mkrn2 sense RNA and morpholinos were injected into the two blastomeres at the dorsal marginal zone (DMZ) and ventral marginal zone (VMZ), respectively. Injected embryos were maintained up to stage 33 in 30% Marc's modified Ringer's solution for morphological observations.
In case of AC assay, ACs were dissected at stages 8.5–9 from two-cell stage injected embryos and cultured at 22 °C in 67% Leibovitz's L-15 medium (Invitrogen) with 7 mm Tris-HCI (pH 7.5) and gentamicin (50 μg/ml), in the presence or absence of retinoic acid (10-5 m) and activin (10 ng/ml). For each experiment, eight ACs were pooled at stage 22 for RT-PCR or Western blot analysis.
Antibodies and Immunoblotting—A peptide composed of 20 amino acids at the C-terminal end of mkrn2 (EVGKLGELFMHLSGADRGTP) was used to generate polyclonal antibodies in rabbits (Boster Co., Wuhan, China). For Western blot analysis, embryos were homogenized in solution of 10 mm HEPES (pH 7.9), 10 mm KCl, 1.5 mm MgCl2, 0.5 mm dithiothreitol, 0.5 mm phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, and 10 μg/ml leupeptin. Protein extracts were then subjected to SDS-PAGE and transferred to polyvinylidene difluoride membrane followed by immunoblotting analysis.
RT-PCR—Total RNA was extracted with TRIzol reagent (Invitrogen) according to the manufacturer's instructions. RT-PCR amplification was performed using SuperScript first-strand synthesis system (Invitrogen). Primer sets and PCR conditions for elongation factor 1α (EF-1α) (27 cycles), NCAM (29 cycles), OTX2 (29 cycles), HoxB9 (30 cycles), and En2 (28 cycles) have already been described (9, 10). For each RT-PCR primer pair, a pilot experiment was done to determine the linear amplification range. Then the optimal cycle number was chosen as the approximate midpoint of the linear range. The forward and reverse primer pairs for mkrn2, Krox20, and GSK-3β were as follows: mkrn2, 5′-AAG AGA TGT CGC CCG TAC TCA G-3′ and 5′-GGC AGT AGG TGT GGT TAC AGT TGG-3′; Krox20, 5′-CTA CCT CCA GAA GAT GAG ACT C-3′ and 5′-GGA GCC ACT GCC TTC GTA ATC G-3′; GSK-3β, 5′-AGA AAT GAA CCC CAA CTA CAC-3′ and 5′-GAT GAG AAT TGA GGA GAG AGA A-3′. PCR conditions for mkrn2, Krox20, and GSK-3β were as follows: 15 min at 95 °C followed by 1 min at 94 °C, 1 min at 55 °C (or 60 °C for mkrn2), and 1 min at 72 °C for 29 cycles (or 30 cycles for Krox20). PCR products were subjected to electrophoresis on 1% ethidium bromide-stained agarose gel. The band intensity was quantitated by ImageQuant software (GE Healthcare) and summarized as mean value with standard error.
Statistical Analysis—All RT-PCR and Western blot results were confirmed by three independent experiments, in which only the representative bands were shown. Differences, as compared with control, were considered to be statistically significant at p < 0.05 in two-tailed Student's t test (GraphPad Prism 3.0, GraphPad Software, San Diego, CA). We also performed analysis of variance to determine the statistical significance between the mRNA levels of different treatment groups. Differences were established at 95% confidence interval.
RESULTS
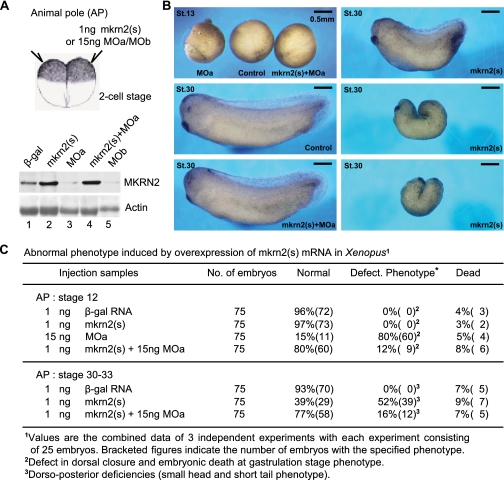
Ectopic Overexpression of mkrn2 in Xenopus Embryos Produced Tadpoles with Dorso-posterior Deficiencies—To explore the role of mkrn2 during Xenopus embryonic development, we conducted both gain-of-function and loss-of-function studies by microinjecting into the two animal poles of two-cell stage embryos with mkrn2 sense RNA (mkrn2(s)) and two morpholino antisense oligonucleotides (MOa and MOb) that specifically suppressed the translation of endogenous mkrn2. By Western blotting analysis using polyclonal antibody targeting mkrn2, it was confirmed that microinjection of MOa or MOb significantly reduced, whereas mkrn2(s) enhanced, the protein level of mkrn2 in embryos (Fig. 1A). Furthermore, we showed that mkrn2 is required for early development as knockdown of mkrn2 caused embryonic death, possibly due to the defect in dorsal closure (Fig. 1B, top left panel, and Fig. 1C, 60/75, 80%). In contrast, microinjection of mkrn2(s) did not cause death at early stages but produced tadpoles with different degrees of dorso-posterior deficiencies and small head-short tail phenotype at stages 30–33 (Fig. 1B, right panels, and Fig. 1C, 39/75, 52%). However, these phenotypes could be rescued by the co-injection of MOa, suggesting that their effects are mkrn2-specific (Fig. 1B, bottom left panel, and Fig. 1C). These observations suggested that mkrn2 may have anti-neurogenic activity in Xenopus embryonic development.
FIGURE 1.
mkrn2 is required for early Xenopus embryonic development. A, Western blotting of embryo lysates isolated from stage 12 embryos microinjected into the animal pole at two-cell stage with β-galactosidase (β-gal) (control), two different morpholinos targeting mkrn2 (MOa and MOb), or mkrn2 sense RNA (mkrn2(s)). Actin was shown as control. B, embryos at the two-cell stage were injected into the animal pole area with mkrn2(s) or MOa or both, as indicated. At stage 13 (St.13), embryos injected with MOa showed defect in dorsal closure. At stage 30, embryos injected with mkrn2(s) showed dorso-posterior deficiencies with small head and short tail phenotype. Co-injection of mkrn2(s) and MOa resulted in normal phenotype similar to control. C, the percentage (and number) of normal, defective, and dead embryos of each animal pole injection group.
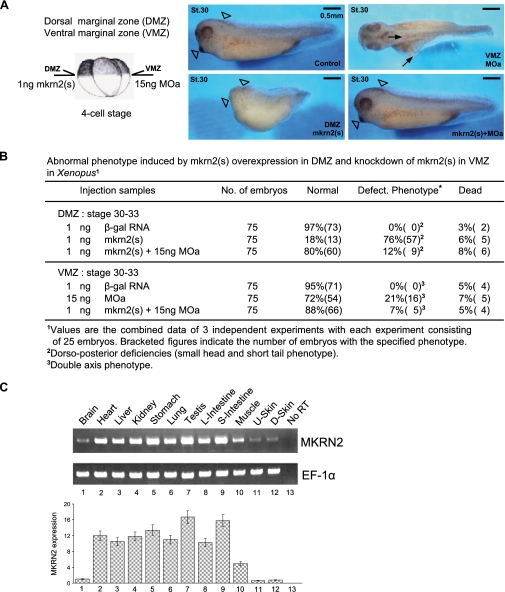
mkrn2 Regulated Neurogenesis and Axis Formation in Xenopus Embryos—We further validated the anti-neurogenic activity of mkrn2 by microinjecting its sense RNA into the DMZ of four-cell stage embryos. DMZ is a key embryonic region involved in body axis organization and neural induction. When compared with β-galactosidase-injected control embryos at stage 30, overexpression of mkrn2 at DMZ resulted in tadpoles with diminished head structure and short tail phenotype (Fig. 2A, bottom left panel, and Fig. 2B, 57/75, 76%), and the phenotype could be rescued by co-injection of MOa (Fig. 2B). Moreover, knockdown of mkrn2 by microinjection of MOa at VMZ did not cause embryonic death; however, the surviving tadpoles displayed axis duplication (Fig. 2A, top right panel, the arrows indicate the double axis, and Fig. 2B, 16/75, 21%), and the phenotype could be rescued by co-injection of mkrn2(s) (Fig. 2B). Taken together, these results strongly suggested that mkrn2 plays a negative role in neurogenesis and axis formation during Xenopus embryonic development.
FIGURE 2.
mkrn2 affects neural development in Xenopus embryos. A, mkrn2(s) and MOa were injected into the DMZ and VMZ of four-cell stage Xenopus embryos, respectively. The embryos were cultured until the equivalent of stage 30 (St.30) for photography. Embryos injected with mkrn2(s) at DMZ showed dorso-posterior deficiencies with small head and short tail phenotype, whereas those injected with MOa at VMZ showed double axis phenotype. Co-injection of mkrn2(s) and MOa at either DMZ or VMZ rescued the defect phenotype. B, the percentage (and number) of normal, defective (Defect.), and dead embryos of each treatment group. β-gal, β-galactosidase. C, RT-PCR analysis of mkrn2 expression in different tissues of adult Xenopus. Elongation factor 1α (EF-1α) expression was used as a loading control. Reaction without the addition of reverse transcriptase (No RT) served as a negative control to confirm the absence of contaminating genomic DNA. L-Intestine, large intestine; S-Intestine, small intestine; U-Skin, upper skin; D-Skin, dermal skin.
mkrn2 Expression Was Comparatively Low in Adult Xenopus Brain and Skin Tissues—By RT-PCR analysis, we showed that high levels of mkrn2 mRNA could be detected in testis, small intestine, stomach, heart, liver, kidney, lung, large intestine, and muscle of adult male Xenopus (Fig. 2C). Consistent with its potential role as a neurogenesis inhibitor, mkrn2 mRNA levels were significantly lower in brain and skin.
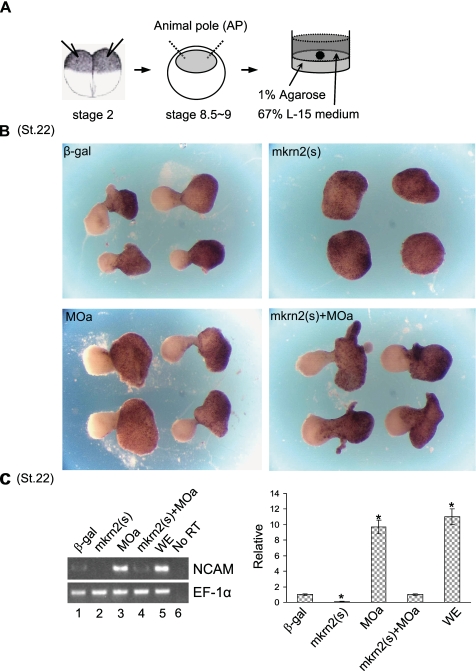
Overexpression of mkrn2 Inhibited Animal Cap Neuralization and NCAM Expression—Using the Xenopus AC explant system (Fig. 3A), we examined the effect of microinjection of mkrn2(s) or morpholino antisense oligonucleotide on the morphology of AC explant at stage 22 equivalent. As expected, in the presence of a neural-inducing factor retinoic acid and mesoderm-inducing factor activin, microinjection of control β-galactosidase mRNA was sufficient to induce neural-like extension in AC (Fig. 3B, β-gal). We found that overexpression of mkrn2 abrogated the retinoic acid plus activin induced AC neuralization, as the resulting AC exhibited round shaped epidermal-like phenotype (Fig. 3B, mkrn2(s)). Furthermore, knockdown of mkrn2 by MOa further enhanced the AC elongation and neuralization phenotype (Fig. 3B, MOa), and the co-injection of mkrn2(s) fully restored the normal AC phenotype (Fig. 3B, mkrn2(s)+MOa).
FIGURE 3.
mkrn2 inhibits neural tissue induction in animal cap. A, schematic illustration of AC assay as described under “Experimental Procedures.” Dissected AC explants were cultured until the equivalent of stage 22 for photography or RT-PCR analysis. B, morphology of AC treated with β-galactosidase (β-gal) mRNA (control), mkrn2(s), MOa, or mkrn2(s) plus MOa, in the presence of retinoic acid and activin. St.22, stage 22. C, RT-PCR analysis of the expression of pan-neural marker NCAM. NCAM expression in whole embryo (WE) was used as positive control. No RT, reaction without the addition of reverse transcriptase.
To further verify the neuroinhibitory activity of mkrn2, we measured its effect on the expression of a key pan-neural marker NCAM in AC explant by RT-PCR. We showed that overexpression of mkrn2 suppressed, whereas knockdown of mkrn2 by MOa significantly stimulated, NCAM expression (Fig. 3C, lanes 2 and 3). Moreover, co-injection of mkrn2(s) fully abrogated MOa-induced NCAM expression (Fig. 3C, lane 4). These results are consistent with observations in AC morphology assay, further suggesting that mkrn2 is a neurogenesis inhibitor.
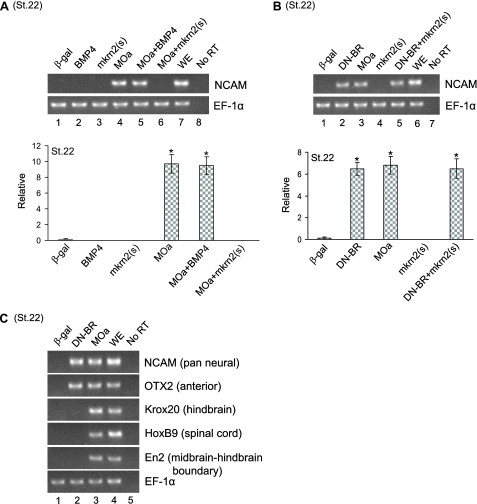
mkrn2 Did Not Affect DN-BR-mediated Neurogenesis—It is well documented that blocking BMP-4 signaling by the dominant negative mutant of BMP-4 receptor (DN-BR) induces neural differentiation in Xenopus ectoderm (11–13). We tested whether mkrn2 is involved in this pathway. We first confirmed by RT-PCR that knockdown of mkrn2 by MOa was sufficient to induce NCAM expression even in the absence of retinoic acid and activin (Fig. 4A, lane 4). BMP-4 could not abolish MOa-induced NCAM expression, whereas mkrn2 effectively abolished MOa-induced NCAM expression (Fig. 4A, lanes 5 and 6). Furthermore, we showed that suppression of BMP-4 signaling by DN-BR induced NCAM expression (Fig. 4B, lane 2), and mkrn2 failed to suppress DN-BR-induced NCAM expression (Fig. 4B, lane 5). These results indicated that the anti-neurogenic activity of mkrn2 is independent of DN-BR signaling pathway.
FIGURE 4.
Effect of mkrn2 on DN-BR-mediated neurogenesis. AC assay with the injection of mkrn2(s), MOa, mRNAs encoding β-galactosidase (β-gal) (control), BMP-4, DN-BR, or combinations were performed. A, mkrn2(s), but not BMP-4, abolished MOa-induced NCAM expression. St.22, stage 22; WE, whole embryo; No RT, reaction without the addition of reverse transcriptase. B, mkrn2(s) did not abolish DN-BR-induced NCAM expression. C, differential neural markers NCAM, OTX2, Krox20, HoxB9, and En2 expression induced by DN-BR and MOa.
It has been reported that DN-BR mainly induces the expressions of NCAM and anterior neural marker OTX2 in AC explant. We then compared the ability of MOa and DN-BR to induce different neural markers. We found that knockdown of mkrn2 by MOa not only induced the expressions of NCAM and OTX2 but also induced the expressions of hindbrain marker Krox20, spinal cord marker HoxB9, and midbrain-hindbrain boundary marker En2 (Fig. 4C, lane 3).
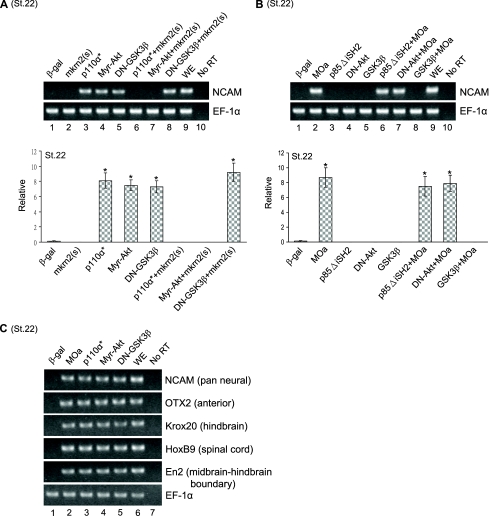
mkrn2 Acted Downstream of PI3K and Akt and Upstream of GSK-3β—We previously reported a novel role of PI3K/Akt signaling pathway in axis formation and neural development during Xenopus embryogenesis (7). To determine whether and how mkrn2 modulates the PI3K signal, we interfered with the PI3K/Akt cascade to investigate the effect of mkrn2 on this signaling pathway. We first co-expressed mkrn2(s) with PI3K signal inducers, including the active form of the PI3K catalytic subunit (p110α*), the active form of Akt (Myr-Akt), and the dominant negative mutant of GSK-3β (DN-GSK3β) and examined their effects on NCAM expression in an AC explant assay. We showed that mkrn2 completely inhibited p110α*- and Myr-Akt-induced, but not DN-GSK3β-induced, NCAM expression (Fig. 5A, lanes 6–8).
FIGURE 5.
Role of mkrn2 in PI3K/Akt signaling pathway. AC assay was performed with the injection of mkrn2(s), MOa, mRNAs encoding β-galactosidase (β-gal) (control), the active form of PI3K catalytic subunit (p110α*) and truncated mutant of PI3K (p85ΔiSH2), the active form of Akt (Myr-Akt) and its dominant negative mutant (DN-Akt), GSK-3β and its dominant negative mutant (DN-GSK3β), or combinations. A, mkrn2(s) inhibited p110α* and Myr-Akt-induced, but not DN-GSK3β-induced, NCAM expression. WE, whole embryo; No RT, reaction without the addition of reverse transcriptase. B, MOa-induced NCAM induction was inhibited by co-expression of GSK-3β; however, co-expression of p85ΔiSH2 or DN-Akt could not abolish MOa-induced NCAM expression. C, induction of specific neural markers expression by MOa and PI3K signal inducers, including p110α*, Myr-Akt, and DN-GSK3β.
In contrast, we employed a truncated form of the p85 regulatory subunit of PI3K (p85ΔiSH2) and dominant negative mutant of Akt (DN-Akt) to suppress the activities of endogenous PI3K and Akt, respectively. Ectopic expression of p85ΔiSH2 or DN-Akt could not inhibit MOa-induced NCAM expression, whereas MOa-induced NCAM expression was suppressed by GSK-3β (Fig. 5B, lanes 6–8). Taken together, these results indicated that mkrn2 acts downstream of PI3K and Akt and upstream of GSK-3β.
In addition, we examined the ability of PI3K signals to induce different neural markers. Surprisingly, the neural marker expression patterns of p110α*, Myr-Akt, and DN-GSK3β (Fig. 5C, lanes 3–5) were essentially the same as that triggered by MOa (Fig. 5C, lane 2). These results further support the hypothesis that mkrn2 inhibits neurogenesis through modulating the PI3K/Akt signaling pathway.
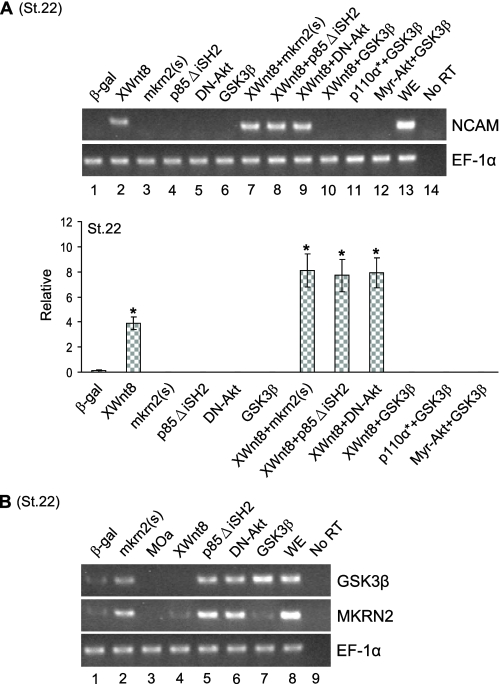
Cross-talk between PI3K and Wnt8 Signaling Pathways at GSK-3β but Not mkrn2—As Wnt8 signaling is one of the major pathways for neural development in Xenopus embryos (14) and GSK-3β is a common downstream target of both PI3K and Wnt8 signaling, we tested whether mkrn2 plays a role in the cross-talk between Wnt8- and PI3K-induced NCAM expression. We showed that XWnt8-induced NCAM expression (Fig. 6A, lane 2) could not be abolished by co-expression of mkrn2(s), p85ΔiSH2, or DN-Akt (Fig. 6A, lanes 7–9). In contrast, GSK-3β abolished XWnt8-mediated NCAM induction (Fig. 6A, lane 12) and XWnt8 did not affect mkrn2 expression (Fig. 6B, lane 4), indicating that the cross-talk between PI3K and Wnt signaling pathways is mediated by GSK-3β, but not mkrn2.
FIGURE 6.
mkrn2 is not involved in the cross-talk of PI3K and Wnt8 signaling. A, GSK-3β, but not mkrn2, abolished XWnt8-induced NCAM expression in AC explant.β-gal, β-galactosidase; WE, whole embryo; No RT, reaction without the addition of reverse transcriptase; ST.22, stage 22. B, XWnt8 did not affect the mRNA expression of mkrn2.
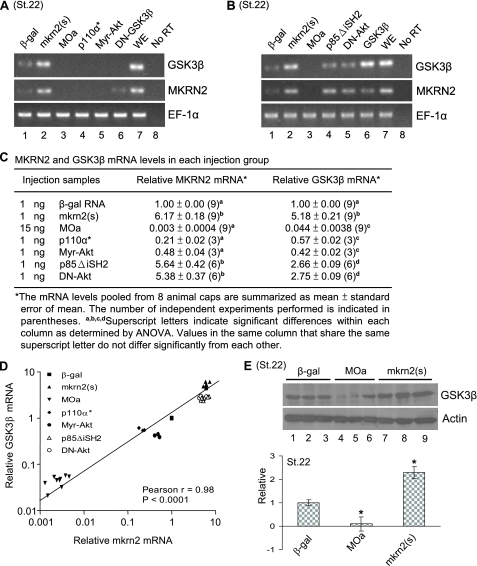
mkrn2 Positively Regulated GSK-3β mRNA Expression—As mkrn2 had been showed to work upstream of GSK-3β, we investigated how mkrn2 regulates GSK-3β. We found by RT-PCR that overexpression of mkrn2 induced GSK-3β expression, whereas knockdown of mkrn2 suppressed the expression of GSK-3β (Fig. 7A, lanes 2 and 3). We also noticed that PI3K signal inducers (p110α* and Myr-Akt) inhibited mkrn2 and GSK-3β expression (Fig. 7A, lanes 4 and 5). In contrast, the mRNA levels of mkrn and GSK-3β significantly increased upon the suppression of PI3K and Akt (Fig. 7B, lanes 4 and 5). These results suggested that upstream PI3K/Akt signal inhibits both GSK-3β and mkrn2 at mRNA level, and mkrn2 positively regulates GSK-3β.
FIGURE 7.
mkrn2 induces GSK-3β mRNA and protein expression. A, mkrn2 induced GSK-3β mRNA expression, whereas p110α* and Myr-Akt suppressed mkrn2 expression. ST.22, stage 22; β-gal, β-galactosidase; WE, whole embryo; No RT, reaction without the addition of reverse transcriptase. B, knockdown of mkrn2 suppressed GSK-3β mRNA expression. C, summary of the mean mRNA levels of mkrn2 and GSK-3β in different injection groups. ANOVA, analysis of variance. D, correlation analysis of mkrn2 and GSK-3β mRNA levels. The Kolmogorov-Smirnov test shows that the data are normally distributed after logarithmic transformation. Both axes are in log10 scale to represent the transformation. E, Western blot analysis of GSK-3β protein level in response to overexpression or knockdown of mkrn2.
To study the correlation between mkrn2 and GSK-3β, we measured mkrn2 and GSK-3β mRNA levels in different injection groups (Fig. 7C). We found a significant positive correlation between the mRNA levels of mkrn2 and GSK-3β (Fig. 7D). Consistent with the result from RT-PCR, Western blot analysis showed that mkrn2 also up-regulated the protein level of GSK-3β (Fig. 7E). These results suggested that mkrn2 preferentially up-regulates the expression of GSK-3β mRNA and subsequently leads to an increased level of GSK-3β protein.
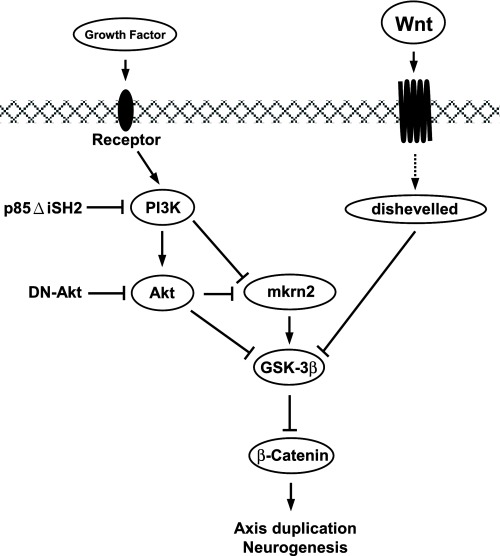
In summary, we uncovered a novel role of mkrn2 in inhibiting axis formation and neurogenesis during Xenopus embryonic development. Based on our previous report that the signal of PI3K activation is transmitted through its downstream target Akt, which in turn inhibits the activity of GSK-3β and regulates neural development (7), we provided evidence that mkrn2 achieves its anti-neurogenic effect by acting downstream of PI3K/Akt to up-regulate the expression of GSK-3β (Fig. 8).
FIGURE 8.
Inhibition of neurogenesis and axis duplication by mkrn2 via PI3K/Akt signaling pathway.
DISCUSSION
MKRN2 is the latest member in the makorin gene family. This gene is highly conserved throughout evolution, and its ancestral origin can be traced back to 450 million years ago, possibly arising from gene duplication of MKRN1 (5). We have performed sequence analysis and found that human MKRN1 and MKRN2 possess the same set of functional protein domains, suggesting that they may share similar functions or regulatory mechanisms (supplemental Fig. 1).
Since MKRN2 is rarely studied and its biological function has never been reported, we employed Xenopus laevis as a model system to characterize the role of mkrn2 in Xenopus neurogenesis. We demonstrated that misexpression of mkrn2 resulted in tadpoles with dorsal-posterior deficiencies (Fig. 1), whereas knockdown by microinjection of morpholino was sufficient to induce axis duplication (Fig. 2). In AC explants, mkrn2 overexpression inhibited AC neuralization and neural marker NCAM expression (Fig. 3). These results provided strong indication that mkrn2 inhibits neurogenesis and is essential for proper neural development in Xenopus embryos. In fact, mkrn2 is highly expressed in adult Xenopus tissues that do not require extensive neural development, and only a low level of mkrn2 expression is observed in brain and skin (Fig. 2C). This tissue expression pattern can be rationally explained by our proposed anti-neurogenic activity of mkrn2.
BMP-4 inhibition and activation of canonical Wnt signaling cascades are the two major mechanisms for neural induction (13–15). Using an AC explant culture system, we found that mkrn2 is involved in neither BMP-4 nor XWnt8 signaling pathway (Figs. 4 and 6A), but alternatively, participated in PI3K-mediated neurogenesis that we previously reported (Fig. 5) (7). PI3K is involved, through its downstream target Akt, in a wide range of cellular processes, such as cell survival (16), myogenic differentiation (17, 18), protein synthesis (19), angiogenesis (20), and transformation (21). Activated PI3K transmits survival signals from growth factors to Akt (22). Akt then phosphorylates GSK-3β at residue Ser-9 and thereby inhibits its activity (23). Inactivation of GSK-3β leads to decreased suppression of β-catenin and thus subsequently promotes axis formation and neurogenesis. Here we showed that mkrn2 abolished p110α*- and Myr-Akt-, but not DN-GSK3β-induced, NCAM expression (Fig. 5A). In addition, mkrn2 up-regulated the expression of GSK-3β at both mRNA and protein levels (Fig. 7). These results suggested that mkrn2 acts between Akt and GSK-3β in the PI3K pathway to antagonize the inactivation of GSK-3β (Fig. 8).
The molecular mechanism involved in Akt/mkrn2/GSK-3β signaling is still far from clear at present. Since Akt is a serine threonine kinase, it is possible that Akt phosphorylates and inactivates mkrn2 by a mechanism similar to that of GSK-3β suppression. In silico kinase-specific phosphorylation prediction using Scansite (24) and NetPhosK (25) revealed that mkrn2 has a potential phosphorylation site at Ser-252.
mkrn2 possesses four C3H zinc fingers that may be responsible for RNA binding (26). With these RNA-binding domains, mkrn2 may up-regulate the expression of GSK-3β mRNA at post-transcriptional level, rather than increasing the transcription of the GSK-3β gene. mkrn2 may be essential for the stabilization of GSK-3β mRNA or the transport of GSK-3β mRNA from nucleus to cytoplasm for translation via RNA binding activity (27, 28). The tandem repeat of C3H zinc fingers may provide high specificity for RNA binding (26). Alternatively, as mkrn2 presumptively possesses ubiquitin-protein isopeptide ligase (E3) activity intimated by the presence of a highly conserved C3HC4 RING zinc finger domain (2, 29), it is tempting to speculate that mkrn2 may favor the expression of GSK-3β by degrading yet unknown GSK-3β inhibitors via ubiquitin-proteasome pathway.
Dysregulation of PI3K/Akt pathway has been implicated in a wide range of human cancers (30, 31). Data from the Gene Expression Omnibus (GEO), a public repository for microarray gene expression data, indicated that human MKRN2 is overexpressed in various cancer cell lines. For example, MKRN2 was significantly up-regulated by more than 4-fold in metastatic colon cancer cell line SW620, as compared with its primary tumor counterpart SW840 (GEO accession number GDS756). Moreover, MKRN2 was found to be up-regulated by more than 2-fold in breast cancer cell line MDA-MB-436, as compared with normal mammary epithelial cells (GEO accession numbers GDS820 and GDS823). The overexpression of mammalian MKRN2 in cancer cell lines raises the possibility that MKRN2 may involve in PI3K/Akt pathway for tumor progression.
In conclusion, our results illustrate for the first time a functional role of mkrn2 in negatively regulating neurogenesis via PI3K/Akt signaling in Xenopus embryos. The detailed molecular mechanisms and the potential functions of mammalian MKRN2 remain to be studied.
Supplementary Material
Acknowledgments
We thank Oscar G. W. Wong and Vanessa W. S. Leung for critical review and thoughtful advice on this manuscript.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s) EF626804.
This work was supported by Research Grants Council of the Hong Kong, China Grants N_CUHK721/03 and CUHK7328/04M (to H.-F. K.), Foundation of Guangzhou Science and Technology Bureau Grant 2005Z1-E0131 (to H.-F. K.), and National Natural Science Fund of China Grants 30570589 (to Y. P.) and 30318001 and 30200112 (to Z. C. and Q.-H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
Footnotes
The abbreviations used are: MKRN, makorin; mkrn2, Xenopus makorin-2; AC, animal cap; NCAM, neural cell adhesion molecule; PI3K, phosphatidylinositol 3-kinase; BMP, bone morphogenetic protein; DN, dominant-negative; DN-BR, DN mutant of BMP-4 receptor; GSK-3β, glycogen synthase kinase 3β; RT-PCR, reverse transcription-PCR; DMZ, dorsal marginal zone; VMZ, ventral marginal zone; MO, morpholino.
References
- 1.Gray, T. A., Hernandez, L., Carey, A. H., Schaldach, M. A., Smithwick, M. J., Rus, K., Marshall Graves, J. A., Stewart, C. L., and Nicholls, R. D. (2000) Genomics 66 76-86 [DOI] [PubMed] [Google Scholar]
- 2.Kim, J. H., Park, S. M., Kang, M. R., Oh, S. Y., Lee, T. H., Muller, M. T., and Chung, I. K. (2005) Genes Dev. 19 776-781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Omwancha, J., Zhou, X. F., Chen, S. Y., Baslan, T., Fisher, C. J., Zheng, Z., Cai, C., and Shemshedini, L. (2006) Endocrine 29 363-373 [DOI] [PubMed] [Google Scholar]
- 4.Zhang, Q. H., Ye, M., Wu, X. Y., Ren, S. X., Zhao, M., Zhao, C. J., Fu, G., Shen, Y., Fan, H. Y., Lu, G., Zhong, M., Xu, X. R., Han, Z. G., Zhang, J. W., Tao, J., Huang, Q. H., Zhou, J., Hu, G. X., Gu, J., Chen, S. J., and Chen, Z. (2000) Genome Res. 10 1546-1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gray, T. A., Azama, K., Whitmore, K., Min, A., Abe, S., and Nicholls, R. D. (2001) Genomics 77 119-126 [DOI] [PubMed] [Google Scholar]
- 6.Kintner, C. R., and Melton, D. A. (1987) Development (Camb.) 99 311-325 [DOI] [PubMed] [Google Scholar]
- 7.Peng, Y., Jiang, B. H., Yang, P. H., Cao, Z., Shi, X., Lin, M. C., He, M. L., and Kung, H. F. (2004) J. Biol. Chem. 279 28509-28514 [DOI] [PubMed] [Google Scholar]
- 8.Nieuwkoop, P. D., and Faber, J. (1994) Normal Table of Xenopus laevis (Daudin) pp. 162-188, Garland Publishing, New York
- 9.Hemmati-Brivanlou, A., and Melton, D. A. (1994) Cell 77 273-281 [DOI] [PubMed] [Google Scholar]
- 10.Umbhauer, M., Penzo-Mendez, A., Clavilier, L., Boucaut, J., and Riou, J. (2000) J. Cell Sci. 113 2865-2875 [DOI] [PubMed] [Google Scholar]
- 11.Maeno, M., Ong, R. C., Suzuki, A., Ueno, N., and Kung, H. F. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 10260-10264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki, A., Thies, R. S., Yamaji, N., Song, J. J., Wozney, J. M., Murakami, K., and Ueno, N. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 10255-10259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu, R. H., Kim, J., Taira, M., Zhan, S., Sredni, D., and Kung, H. F. (1995) Biochem. Biophys. Res. Commun. 212 212-219 [DOI] [PubMed] [Google Scholar]
- 14.Baker, J. C., Beddington, R. S., and Harland, R. M. (1999) Genes Dev. 13 3149-3159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stern, C. D. (2005) Development (Camb.) 132 2007-2021 [DOI] [PubMed] [Google Scholar]
- 16.Khwaja, A., Rodriguez-Viciana, P., Wennstrom, S., Warne, P. H., and Downward, J. (1997) EMBO J. 16 2783-2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang, B. H., Aoki, M., Zheng, J. Z., Li, J., and Vogt, P. K. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 2077-2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang, B. H., Zheng, J. Z., and Vogt, P. K. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 14179-14183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vogt, P. K. (2001) Trends Mol. Med. 7 482-484 [DOI] [PubMed] [Google Scholar]
- 20.Jiang, B. H., Zheng, J. Z., Aoki, M., and Vogt, P. K. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 1749-1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang, H. W., Aoki, M., Fruman, D., Auger, K. R., Bellacosa, A., Tsichlis, P. N., Cantley, L. C., Roberts, T. M., and Vogt, P. K. (1997) Science 276 1848-1850 [DOI] [PubMed] [Google Scholar]
- 22.Duronio, V., Scheid, M. P., and Ettinger, S. (1998) Cell. Signal. 10 233-239 [DOI] [PubMed] [Google Scholar]
- 23.Cross, D. A., Alessi, D. R., Cohen, P., Andjelkovich, M., and Hemmings, B. A. (1995) Nature 378 785-789 [DOI] [PubMed] [Google Scholar]
- 24.Obenauer, J. C., Cantley, L. C., and Yaffe, M. B. (2003) Nucleic Acids Res. 31 3635-3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blom, N., Sicheritz-Ponten, T., Gupta, R., Gammeltoft, S., and Brunak, S. (2004) Proteomics 4 1633-1649 [DOI] [PubMed] [Google Scholar]
- 26.Hudson, B. P., Martinez-Yamout, M. A., Dyson, H. J., and Wright, P. E. (2004) Nat. Struct. Mol. Biol. 11 257-264 [DOI] [PubMed] [Google Scholar]
- 27.Lai, W. S., Carballo, E., Strum, J. R., Kennington, E. A., Phillips, R. S., and Blackshear, P. J. (1999) Mol. Cell. Biol. 19 4311-4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De, J., Lai, W. S., Thorn, J. M., Goldsworthy, S. M., Liu, X., Blackwell, T. K., and Blackshear, P. J. (1999) Gene (Amst.) 228 133-145 [DOI] [PubMed] [Google Scholar]
- 29.Joazeiro, C. A., and Weissman, A. M. (2000) Cell 102 549-552 [DOI] [PubMed] [Google Scholar]
- 30.Wymann, M. P., and Marone, R. (2005) Curr. Opin. Cell Biol. 17 141-149 [DOI] [PubMed] [Google Scholar]
- 31.Vivanco, I., and Sawyers, C. L. (2002) Nat. Rev. Cancer 2 489-501 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.