Abstract
The concentrative nucleoside transporter (CNT) protein family in humans is represented by three members, hCNT1, hCNT2, and hCNT3. Belonging to a CNT subfamily phylogenetically distinct from hCNT1/2, hCNT3 mediates transport of a broad range of purine and pyrimidine nucleosides and nucleoside drugs, whereas hCNT1 and hCNT2 are pyrimidine and purine nucleoside-selective, respectively. All three hCNTs are Na+-coupled. Unlike hCNT1/2, however, hCNT3 is also capable of H+-mediated nucleoside cotransport. Using site-directed mutagenesis in combination with heterologous expression in Xenopus oocytes, we have identified a C-terminal intramembranous cysteine residue of hCNT3 (Cys-561) that reversibly binds the hydrophilic thiol-reactive reagent p-chloromercuribenzene sulfonate (PCMBS). Access of this membrane-impermeant probe to Cys-561, as determined by inhibition of hCNT3 transport activity, required H+, but not Na+, and was blocked by extracellular uridine. Although this cysteine residue is also present in hCNT1 and hCNT2, neither transporter was affected by PCMBS. We conclude that Cys-561 is located in the translocation pore in a mobile region within or closely adjacent to the nucleoside binding pocket and that access of PCMBS to this residue reports a specific H+-induced conformational state of the protein.
Nucleosides are hydrophilic molecules that require specialized nucleoside transporter proteins (NTs)4 to cross cellular membranes (1–3). NT-mediated transport is necessary for salvage of nucleotide precursors for nucleic acid biosynthesis and is a critical determinant of the therapeutic efficacy of antineoplastic and antiviral nucleoside drugs (3, 4). By regulating adenosine availability to cell-surface purinoreceptors, NTs also profoundly affect neurotransmission, vascular tone, and other physiological processes (5, 6). Two different NT protein families are present in human and other mammalian cells and tissues; that is, the SLC28 concentrative nucleoside transporter (CNT) family and the structurally unrelated SLC29 equilibrative nucleoside transporter family (3, 6–8). Equilibrative nucleoside transporters are present in most, possibly all, cell types (8). In contrast, CNTs are found predominantly in intestinal and renal epithelia and other specialized cells where they have important roles in absorption, secretion, distribution, and elimination of nucleosides and nucleoside drugs (1–4, 6, 7).
In humans (h), the CNT protein family is represented by three members, hCNT1, hCNT2, and hCNT3. Similar to their orthologs in other mammalian species, hCNT1 and hCNT2 are pyrimidine nucleoside-selective and purine nucleoside-selective, respectively, whereas hCNT3 transports both pyrimidine and purine nucleosides (9–11). Non-mammalian members of the CNT protein family that have been characterized functionally include hfCNT from an ancient marine prevertebrate, the Pacific hagfish Eptatretus stouti (12), CaCNT from the pathogenic yeast Candida albicans (13), CeCNT3 from the nematode Caenorhabditis elegans (14), and NupC from the bacterium Escherichia coli (15).
hCNT1 and hCNT2 function exclusively as Na+-coupled nucleoside transporters, whereas hCNT3 can utilize electrochemical gradients of either Na+ or H+ to accumulate nucleosides within cells (16–18). hfCNT is Na+-coupled (12), whereas CaCNT, CeCNT3, and NupC are H+-coupled (13–15). Na+: nucleoside coupling stoichiometries are 1:1 for hCNT1 and hCNT2 and 2:1 for hCNT3 and hfCNT (12, 16–18). H+:nucleoside coupling ratios for hCNT3 and CaCNT are both 1:1 (13, 17). In the presence of both Na+ and H+, charge/uptake experiments suggest that hCNT3 binds one Na+ and one H+ (17). Na+ and H+ activate CNTs through mechanisms to increase nucleoside apparent binding affinity (16, 17). Consistent with their different energetics and cation specificities, Na+- and H+-coupled hCNT3 and Na+-coupled hCNT1/2 belong to separate CNT subfamilies (11, 18).
hCNT1–3 and other eukaryote CNT family members are predicted to have a 13 (or possibly 15) transmembrane helix (TM) architecture (19), and multiple alignments reveal strong sequence similarities within the C-terminal halves of the proteins (11). Prokaryote CNTs lack the first three TMs of their eukaryote counterparts, and functional expression of N-terminal-truncated human and rat CNT1 in Xenopus oocytes has established that the first three TMs are not required for Na+-dependent uridine transport activity (19). Consistent with these findings, chimeric studies between hCNT1 and hfCNT (12) and between hCNT1 and hCNT3 (17) have demonstrated that residues involved in nucleoside selectivity and Na+- and H+-coupling reside within the C-terminal half of the protein. In hCNT1, mutagenesis studies have identified serine, glutamine, and leucine residues in TMs 7 and 8 with roles in nucleoside selectivity and apparent binding affinity for cations (20, 21). Glutamate residues in hCNT1 TM7 and in the region between TMs 11 and 12 have also been shown to function in Na+-mediated nucleoside cotransport (22).
Whereas Na+-coupled hCNT3 transports a broad range of physiological purine and pyrimidine nucleosides as well as anticancer and antiviral nucleoside drugs, H+-coupled hCNT3 is unable to transport guanosine, 3′-azido-3′-deoxythymidine, or 2′,3′-dideoxycytidine (11, 17). This suggests that Na+- and H+-bound versions of hCNT3 have significantly different conformations of the nucleoside binding pocket and/or translocation channel. Here, we describe a conformationally sensitive pore-lining cysteine residue in TM12 of hCNT3 whose accessibility to the hydrophilic membrane-impermeant thiol reagent p-chloromercuribenzene sulfonate (PCMBS) reports a specific H+-activated state of the transporter.
EXPERIMENTAL PROCEDURES
Site-directed Mutagenesis and DNA Sequencing—hCNT3 cDNA (GenBank™ accession number AF305210) in the Xenopus expression vector pGEM-HE (23) provided the template for construction of hCNT3 mutants by the oligonucleotide-directed technique (24) using reagents from the QuikChange® site-directed mutagenesis kit (Stratagene) according to the manufacturer's directions. Constructs were sequenced in both directions by Taq dideoxy-terminator cycle sequencing to ensure that only the correct mutation had been introduced. hCNT3 cDNA also provided the template for the construction of a cysteine-free version of hCNT3 (hCNT3C-) in which all 14 endogenous cysteine residues of wild-type hCNT3 were converted to serine (25). Also in pGEM-HE, hCNT3C- was then used as the template for the construction of hCNT3C- mutants.
Production of Wild-type and Mutant hCNT3 Proteins in Xenopus Oocytes—hCNT3 cDNAs were transcribed with T7 polymerase using the mMESSAGE mMACHINE™ (Ambion) transcription system and produced in oocytes of Xenopus laevis by standard procedures (26). Healthy defolliculated stage VI oocytes were microinjected with 20 nl of water or 20 nl of water containing RNA transcript (20 ng) and incubated in modified Barth's medium (changed daily) at 18 °C for 72 h before the assay of transport activity. Oocytes producing recombinant wild-type hCNT1 and hCNT2 (GenBank™ accession numbers U62968 and AF036109, respectively) were prepared identically.
Flux Assays—Transport was measured as described previously (21, 26, 27). Groups of 12 oocytes were incubated at room temperature (20 °C) in 200 μl of transport medium containing either 100 mm NaCl or choline chloride (ChCl) and 2 mm KCl, 1 mm CaCl2, 1 mm MgCl2, and 10 mm HEPES, pH 7.5 and 8.5, or MES, pH 5.5. The uridine concentration was 20 μm unless otherwise stated. Uptake was traced with 14C- or 3H-labeled uridine (1 or 2 μCi/ml, respectively) (GE Healthcare) using a 1-min uptake interval to measure initial rates of transport (influx). In transport assays with adenosine, 1 μm deoxycoformycin was also included to inhibit adenosine breakdown by adenosine deaminase. At the end of the incubation period, extracellular label was removed by six rapid washes in ice-cold Na+-free 100 mm ChCl transport medium, pH 7.5, and individual oocytes were dissolved in 1% (w/v) SDS for quantitation of oocyte-associated radioactivity by liquid scintillation counting (LS 6000 IC; Beckman). In PCMBS inhibition studies oocytes were pretreated with PCMBS on ice for 10 min and then washed 5 times with ice-cold transport medium to remove excess organomercurial before the assay of transport activity. Control experiments established that a 10-min exposure to PCMBS resulted in maximum inhibition of hCNT3 transport activity. Corresponding pretreatment with the methanethiosulfonate (MTS) reagents MTSEA, MTSES, and MTSET was performed at room temperature (20 °C) for 5 min. In substrate protection experiments, unlabeled uridine (20 mm) was included along with PCMBS (28). To demonstrate reversal of inhibition, PCMBS-treated oocytes were subjected to a second preincubation with 5 mm dithiothreitol (DTT) at room temperature for 1 min (28). The flux values shown represent mediated transport, corrected for basal uridine uptake measured in control water-injected oocytes, and are the means ± S.E. of 10–12 oocytes. IC50 values (±S.E.) were calculated using ENZFITTER software (Elsevier-Biosoft). Each experiment was performed at least twice on different batches of cells.
Electrophysiology—Steady-state and presteady-state currents in hCNT3-producing oocytes were measured at room temperature (20 °C) using the whole-cell, two-electrode voltage clamp (GeneClamp 500B, Molecular Devices Corp.) as previously described (16–18). The GeneClamp 500B was interfaced to an IBM-compatible PC via a Digidata 1322A A/D converter and controlled by pCLAMP software (Version 9.0, Molecular Devices Corp.). The microelectrodes were filled with 3 m KCl and had resistances ranging from 0.5 to 1.5 megaohms. After microelectrode penetration, resting membrane potential was measured over a 10-min period before the start of the experiment. Oocytes exhibiting an unstable membrane potential or a potential more positive than -30 mV were discarded. Individual oocytes with good resting membrane potentials were clamped at -50 mV. Steady-state currents were measured using a uridine concentration of 100 μm. Presteady-state (transient) currents were studied using a voltage pulse protocol. Membrane voltage was stepped from the holding potential (Vh) of -50 mV to a range of test potentials (Vt) from -130 to +30 mV in 20 mV increments. The voltage rise time of the clamp was adjusted by use of an oscilloscope such that it varied between 200 and 500 μs. Current measurements were sampled before and after incubation with 500 μm PCMBS (100 mm NaCl, pH 5.5, 10 min) in the appropriate transport medium of the same composition used in radioisotope transport assays (100 mm NaCl or ChCl, pH 8.5). Current signals were filtered at 2 kHz (fourpole Bessel filter) at a sampling interval of 200 μs/point (corresponding to a sampling frequency of 5 kHz). For data presentation, the current at each test potential was averaged from 5 sweeps and further filtered at 0.75 kHz by pCLAMP 9.0 software (Molecular Devices Corp.).
RESULTS
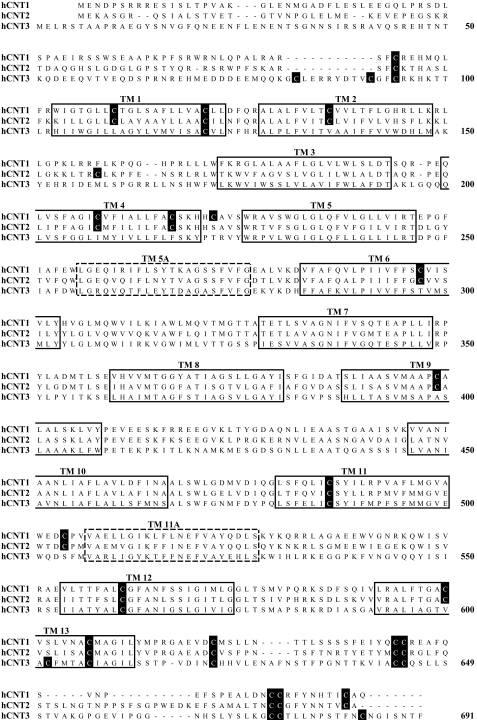
In contrast to Na+-specific hCNT1 and hCNT2, hCNT3 is able to couple uphill nucleoside transport to both Na+ and H+ electrochemical gradients (9–11, 16–18). hCNT3 has 14 endogenous cysteine residues compared with 20 each for hCNT1 and hCNT2, of which 10 are common to all three hCNTs (Fig. 1). Five of the hCNT3 cysteine residues are located in putative TMs (TMs 1, 11, 12, and 13), and the remainder reside within the extramembranous N- and C-terminal regions of the protein. In this report we have used site-directed mutagenesis and heterologous expression of hCNT1–3 in Xenopus oocytes to reveal a unique H+-activated conformational state of hCNT3 in which one of the conserved cysteine residues becomes reactive to PCMBS.
FIGURE 1.
Cysteine residues in hCNT1, hCNT2, and hCNT3. An alignment of the amino acid sequences of hCNT1, hCNT2, and hCNT3 (GenBank™ accession numbers AAB53839, AAB88539, and AAG22551, respectively). The positions of 13 putative TMs are indicated by solid boxes. Two additional TMs present in an alternative 15 TM model of hCNT membrane architecture are indicated by dashed boxes. Cysteine residues are shown on black squares. Numbers refer to hCNT3 residue positions.
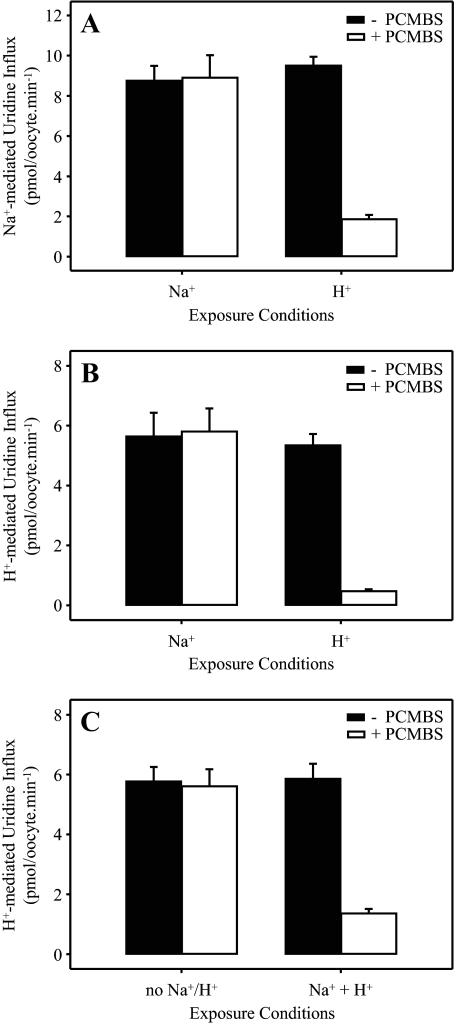
PCMBS Inhibition of hCNT3—Despite having multiple endogenous cysteine residues, we have previously established that wild-type hCNT1 is not inhibited by PCMBS (21). In Fig. 2, we show the effects of PCMBS (500 μm) on wild-type hCNT3. Because hCNT3 is both Na+- and H+-coupled (11, 16–18), the protein was exposed to PCMBS either in the presence of Na+ or H+ (100 mm NaCl, pH 8.5, and 100 mm ChCl, pH 5.5, respectively). Exposure to PCMBS was performed on ice to minimize diffusion of organomercurial across the lipid bilayer (21, 28). By employing Na+-containing medium buffered at pH 8.5, we avoided the small but significant amount of hCNT3 H+ activation that occurs at pH 7.5 (17, 18). We have undertaken control experiments to verify that Na+-coupled uridine transport by hCNT3 at pH 8.5 is kinetically indistinguishable from that at
FIGURE 2.
PCMBS inhibition of hCNT3. hCNT3-mediated influx of 20 μm [14C]uridine in Na+-containing (A) or H+-containing (B and C) medium (100 mm NaCl, pH 8.5, or 100 mm ChCl, pH 5.5, respectively; 1 min at 20 °C) was measured after 10 min of incubation on ice in the absence (solid bars) or presence (open bars) of 500 μm PCMBS in media containing Na+ but not H+ (100 mm NaCl, pH 8.5), H+ but not Na+ (100 mm ChCl, pH 5.5), lacking both Na+ and H+ (100 mm ChCl, pH 8.5), or containing both Na+ and H+ (100 mm NaCl, pH 5.5) as indicated. Values are corrected for basal non-mediated uptake in control water-injected oocytes and are the means ± S.E. of 10–12 oocytes.
pH 7.5.5 After treatment with PCMBS, the same two media were used in assays of uridine transport activity.
Similar to the previous findings with hCNT1 (21) and independent of whether uridine transport activity was subsequently determined in Na+- or H+-containing medium (Figs. 2, A and B, respectively), hCNT3-mediated uridine influx was unaffected by PCMBS when exposed in H+-reduced medium containing Na+ (100 mm NaCl, pH 8.5). In contrast, however, Fig. 2, A and B, also reveal marked inhibition (80–90%) of both Na+- and H+-coupled uridine influx when hCNT3 was reacted with PCMBS under acidified, Na+-free conditions (100 mm ChCl, pH 5.5). As described under “Experimental Procedures,” the flux values shown in Figs. 2, A and B, depict mediated transport activity, defined as the difference in uptake between RNA transcript-injected and control water-injected oocytes. In this and subsequent experiments, uridine uptake in water-injected oocytes was <0.02 pmol/oocyte·min-1 under all conditions tested (data not shown).
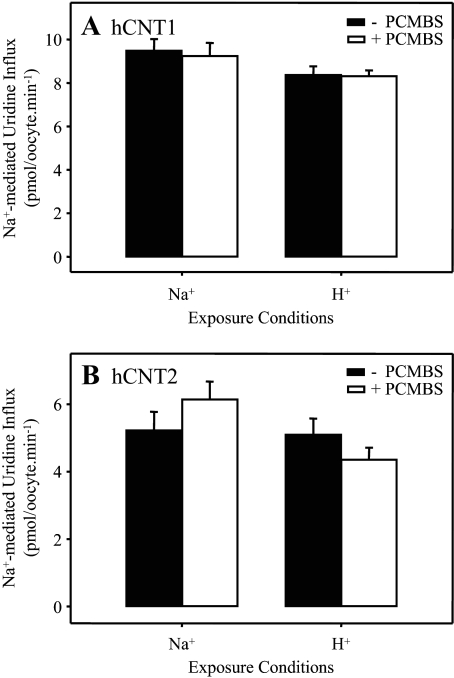
As demonstrated in Fig. 2C, hCNT3-mediated uridine influx was unaffected by PCMBS (500 μm) when incubated in H+-reduced medium also lacking Na+ (100 mm ChCl, pH 8.5). Inhibition was evident, however, when exposure to PCMBS (500 μm) occurred in acidified, Na+-containing medium (100 mm NaCl, pH 5.5) (Fig. 2C). These findings eliminate the possibility that Na+ exerts a protective effect against PCMBS binding and suggest instead that inhibition of hCNT3 by PCMBS results from a specific H+-dependent exofacial conformational shift which exposes the PCMBS-sensitive residue(s) to the extracellular medium. The H+-dependent effect of PCMBS on hCNT3 was not secondary to enhanced chemical reactivity of PCMBS with cysteinyl sulfhydryl groups under acidic conditions or to nonspecific pH-induced changes in protein conformation, because control experiments confirmed that uridine transport by Na+-specific hCNT1 was unchanged by exposure to PCMBS (500 μm) either in the presence of Na+ or H+ (100 mm NaCl, pH 8.5, and 100 mm ChCl, pH 5.5, respectively) (Fig. 3A). Fig. 3B extends this finding of PCMBS insensitivity to Na+-specific hCNT2.
FIGURE 3.
PCMBS insensitivity of hCNT1 and hCNT2. Influx of 20 μm [14C]uridine in the presence of Na+ (100 mm NaCl, pH 7.5; 1 min at 20 °C) was measured in oocytes producing hCNT1 (A) or hCNT2 (B) after 10 min of incubation on ice in the absence (solid bars) or presence (open bars) of 500 μm PCMBS in media containing either Na+ but not H+ (100 mm NaCl, pH 8.5) or H+ but not Na+ (100 mm ChCl, pH 5.5) as indicated. Values are corrected for basal non-mediated uptake in control water-injected oocytes and are the means ± S.E. of 10–12 oocytes.
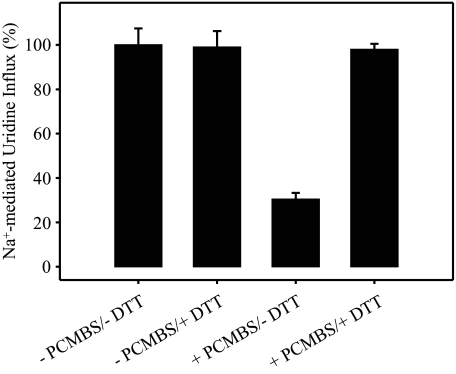
H+-induced PCMBS inhibition of hCNT3 was also evident in influx assays employing other physiological nucleosides. In a representative experiment measuring Na+-coupled influx of a panel of 20 μm 14C-labeled nucleosides after incubation in the presence and absence of 500 μm PCMBS (100 mm ChCl, pH 5.5), the percentage inhibition of influx of uridine, thymidine, cytidine, adenosine, inosine, and guanosine was 89 ± 1, 91 ± 2, 90 ± 1, 87 ± 1, 89 ± 1, and 89 ± 1%, respectively. Therefore, PCMBS binding interferes with translocation of both purine and pyrimidine nucleosides. PCMBS-inhibited hCNT3 nucleoside transport activity was restored by incubation with 5 mm DTT, verifying a specific and reversible interaction of PCMBS with hCNT3 cysteine residue(s) (Fig. 4). Because PCMBS is both hydrophilic and membrane-impermeant (28, 29), the targeted residue(s) must be accessible from the external medium and is most likely located within an outward-facing conformation of the hCNT3 translocation pore.
FIGURE 4.
Reversal of PCMBS inhibition of hCNT3-mediated uridine uptake by DTT. hCNT3-expressing oocytes were incubated in the absence or presence of 500 μm PCMBS (100 mm ChCl, pH 5.5; 10 min on ice) followed by a second incubation in the absence or presence of 5 mm DTT (100 mm ChCl, pH 7.5; 1 min at 20 °C) before measuring uptake of 20 μm [14C]uridine in Na+-containing transport medium (100 mm NaCl, pH 7.5; 1 min at 20 °C). Data are presented as mediated transport, calculated as uptake in RNA-injected oocytes minus uptake in water-injected oocytes and are normalized to the respective influx of uridine in the absence of PCMBS and DTT (12.8 ± 1.0 pmol/oocyte·min-1). Each value is the mean ± S.E. of 10–12 oocytes.
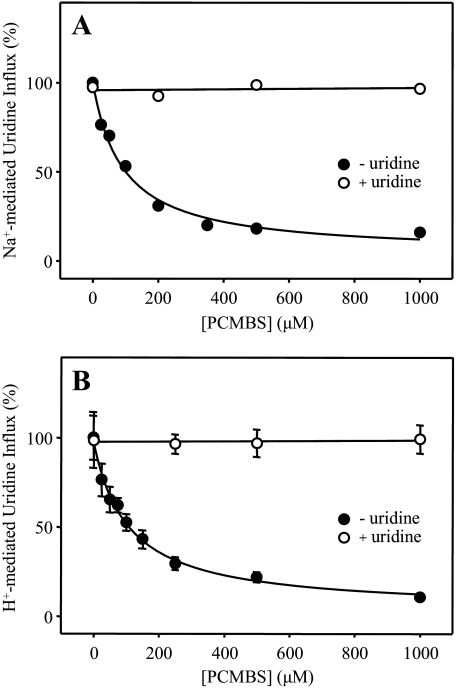
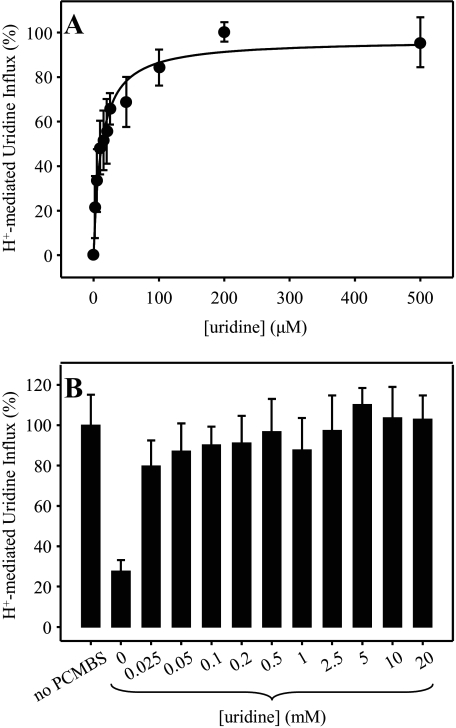
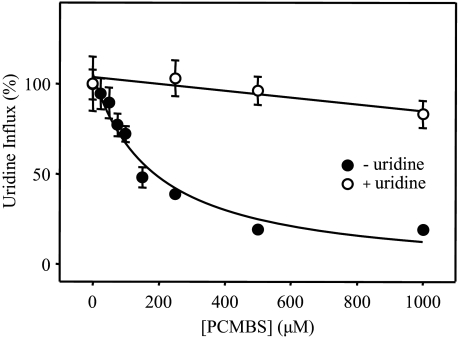
Concentration Dependence and Uridine Protection of PCMBS Inhibition—Dose-response curves for PCMBS inhibition of hCNT3 in the concentration range 25 μm to 1 mm are presented in Fig. 5. Quantitatively, both Na+- and H+-mediated modes of uridine influx were equally affected, with IC50 values of 130 ± 20 μm (Fig. 5A) and 93 ± 18 μm (Fig. 5B), respectively. Figs. 5, A and B, also demonstrate the ability of extracellular uridine (20 mm) to fully protect the transporter against this inhibition. As shown in Figs. 6, A and B, the uridine concentration required for half-maximal protection against PCMBS was 12 ± 2 μm. This compares favorably with a previously determined apparent Km value of 110 μm for H+-coupled uridine influx (17), especially if an anticipated increase in substrate apparent affinity at low temperature is taken into consideration (the uridine protection was performed on ice). Therefore, the PCMBS binding residue(s) is likely located in a position within or closely adjacent to the nucleoside binding pocket.
FIGURE 5.
PCMBS inhibition of hCNT3: concentration dependence and uridine protection. Influx of 20 μm [14C]uridine in both Na+- and H+-containing media (A and B, respectively) was measured after hCNT3-producing oocytes were incubated with various concentrations of PCMBS under acidic conditions either in the absence (solid circles) or in the presence (open circles) of 20 mm extracellular uridine as described in Fig. 2. Data are presented as mediated transport, calculated as uptake in RNA-injected oocytes minus uptake in water-injected oocytes, and are normalized to the respective influx of uridine in the absence of inhibitor (8.7 ± 0.5 (A) and 5.3 ± 0.7 (B) pmol/oocyte·min-1). Each value is the mean ± S.E. of 10–12 oocytes. Error bars are not shown where values were smaller than that represented by the symbols.
FIGURE 6.
PCMBS inhibition of hCNT3: concentration dependence of uridine protection. Influx of 20 μm [3H]uridine in H+-containing medium was measured in oocytes producing hCNT3 after incubation with 500 μm PCMBS under acidic conditions in the presence of 0–500 μm (A) or 0–20 mm (B) extracellular uridine as described in Fig. 2. Data are presented as mediated transport, calculated as uptake in RNA-injected oocytes minus uptake in water-injected oocytes, and normalized to the respective influx of uridine in the absence of inhibitor (8.3 ± 1.3 (A) and 7.3 ± 0.6 (B) pmol/oocyte·min-1). Each value is the mean ± S.E. of 10–12 oocytes. Error bars are not shown where values were smaller than that represented by the symbols (A).
Electrophysiology of PCMBS Inhibition—Steady-state electrophysiological experiments confirmed (i) that PCMBS inhibition of wild-type hCNT3 required exposure under acidified conditions and (ii) that uridine-induced Na+ and H+ inward currents were equally affected by bound PCMBS (data not shown).
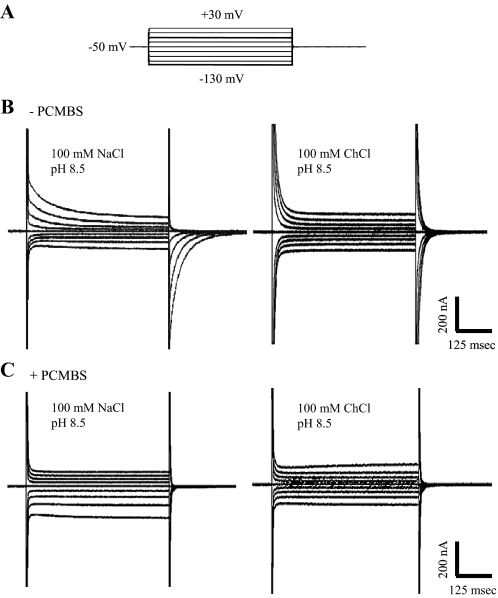
In parallel electrophysiological experiments performed under presteady-state conditions and in the absence of uridine (16, 18), hCNT3-producing oocytes were voltage-clamped at a holding potential (Vh) of -50 mV, and presteady-state currents were activated by voltage steps to the series of test potentials (Vt) outlined in the voltage pulse protocol shown in Fig. 7A. Current recordings in a representative hCNT3-producing oocyte are shown in Fig. 7B in both the presence (left) and absence (right) of Na+ (100 mm NaCl and ChCl, pH 8.5, respectively). As reported previously for both hCNT1 (16, 22) and hCNT3 (30), current relaxations persisting for tens of ms after the time required to charge the membrane capacitance were apparent in both the ON response (when Vh was stepped to Vt) and in the OFF response (when Vt was returned to Vh). These presteady-state currents were reduced, but not eliminated, upon removal of external Na+. Representing both Na+- and carrier-associated charge movements within the membrane, these presteady-state currents were absent in control water-injected oocytes (data not shown). As demonstrated in Fig. 7C for the same oocyte, exposure to 500 μm PCMBS in H+-containing medium (100 mm NaCl, pH 5.5) abolished presteady-state currents in both the presence and absence of Na+ (Fig. 7C, left and right, respectively). Confirming the specificity of PCMBS for hCNT3 and as anticipated from the lack of effect on transport seen in Fig. 3A, hCNT1 presteady-state currents were unaffected by incubation with PCMBS (data not shown).
FIGURE 7.
Time courses of presteady-state currents measured in an hCNT3-expressing oocyte elicited by voltage pulses before and after treatment with PCMBS. A, voltage pulse protocol; the oocyte membrane was held at a holding potential (Vh) of -50 mV and stepped to a range of test potentials (Vt). Shown are Vt from -130 to +30 mV (20-mV increments). B, representative total membrane current records. An hCNT3-producing oocyte displays slow current relaxations in the presence (100 mm NaCl, pH 8.5; left current record) and absence (100 mm ChCl, pH 8.5; right current record) of Na+ in response to voltage pulses before incubation with PCMBS (-PCMBS). C, presteady-state currents were measured in the same hCNT3-expressing oocyte after incubation with PCMBS (500 μm; 10 min). Currents were measured in the presence (100 mm NaCl, pH 8.5; left current record) and absence (100 mm ChCl, pH 8.5; right current record) of Na+.
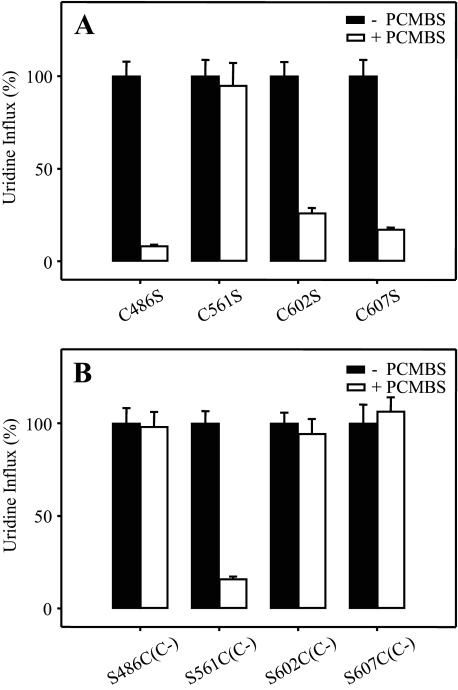
PCMBS Inhibition of hCNT3 Mutants—hCNT3 contains 14 endogenous cysteine residues, of which five lie within predicated TMs (TMs 1, 11, 12, and 13) (Fig. 1). Because TMs 1–3 of mammalian CNTs are not required for transport activity (19) and because we have also determined that the C-terminal half of CNTs comprises the functional domain for cation-coupling (12, 17), we identified cysteine residues 486 (TM11), 561 (TM12), 602 (TM13), and 607 (TM13) as potential candidate residues responsible for H+-induced PCMBS binding. Of these, Cys-486, Cys-561, and Cys-607 are conserved in all three human CNTs (Fig. 1). Using site-directed mutagenesis, the four hCNT3 cysteine residues were individually mutated to serine, generating hCNT3 mutants C486S, C561S, C602S, and C607S. All four constructs were functional when produced in Xenopus oocytes (Fig. 8A). Similar to wild-type hCNT3 (Fig. 2B), inhibition of uridine transport after incubation with 500 μm PCMBS in H+-containing medium was evident for C486S, C602S, and C607S, but there was no effect on mutant C561S (Fig. 8A). None of the mutants were affected by exposure to PCMBS in medium containing 100 mm NaCl, pH 8.5 (data not shown).
FIGURE 8.
Effects of PCMBS on hCNT3 and hCNT3C- mutants. Influx of 20 μm [14C]uridine in H+-containing medium was measured after oocytes producing hCNT3 mutants C486S, C561S, C602S, and C607S (A) and hCNT3C- mutants S486C(C-), S561C(C-), S602C(C-), and S607C(C-) (B) were incubated with or without 500 μm PCMBS (open and solid bars, respectively) under acidic conditions as described for wild-type hCNT3 in Fig. 2. Data are presented as mediated transport, calculated as uptake in RNA-injected oocytes minus uptake in water-injected oocytes, and normalized to the respective values of mediated uridine influx in the absence of inhibitor (7.4 ± 0.6, 3.7 ± 0.3, 7.2 ± 0.5, and 6.7 ± 0.6 pmol/oocyte·min-1 for hCNT3 mutants C486S, C561S, C602S, and C607S, respectively, and 5.1 ± 0.4, 7.9 ± 0.5, 4.2 ± 0.2, and 6.3 ± 0.6 pmol/oocyte·min-1 for hCNT3C- mutants S486C(C-), S561C(C-), S602C(C-), and S607C(C-), respectively). Each value is the mean ± S.E. of 10–12 oocytes.
PCMBS Inhibition of hCNT3C- Mutants—In subsequent experiments, site-directed mutagenesis was also used to generate revertant mutants in the cysteine-free background of hCNT3C- (25). As shown in Fig. 8B, each of the four revertant mutants (S486C(C-), S561C(C-), S602C(C-), and S607C(C-)) were functional when produced in oocytes. Complementary to the results presented for the corresponding hCNT3 mutants (Fig. 8A), only S561C(C-) showed inhibition of uridine influx after incubation with 500 μm PCMBS in H+-containing medium (Fig. 8B). The extent of inhibition was similar to that observed with wild-type hCNT3 (Fig. 2B). None of the mutants was affected by exposure to PCMBS in medium containing 100 mm NaCl, pH 8.5 (data not shown).
PCMBS Inhibition of Mutant S561C(C-): Concentration Dependence and Uridine Protection—Dose-response and uridine protection experiments were undertaken to confirm the identity of Cys-561 in TM12 as the residue responsible for PCMBS binding. Similar to wild-type hCNT3 (Fig. 5), exposure of S561C(C-) to PCMBS in H+-containing medium produced inhibition of uridine transport activity with an IC50 value of 190 ± 60 μm (Fig. 9). Furthermore, extracellular uridine (20 mm) protected the mutant transporter against this inhibition (Fig. 9).
FIGURE 9.
PCMBS inhibition of hCNT3C- mutant S561C(C-); concentration dependence and uridine protection. Influx of 20 μm [14C]uridine in H+-containing medium was measured after S561C(C-)-producing oocytes were incubated with various concentrations of PCMBS under acidic conditions either in the absence (solid circles) or presence (open circles) of 20 mm extracellular uridine as described in Fig. 2. Data are presented as mediated transport, calculated as uptake in RNA-injected oocytes minus uptake in water-injected oocytes, and normalized to the influx of uridine in the absence of inhibitor (7.5 ± 1.1 pmol/oocyte·min-1). Each value is the mean ± S.E. of 10–12 oocytes. Error bars are not shown where values were smaller than that represented by the symbols.
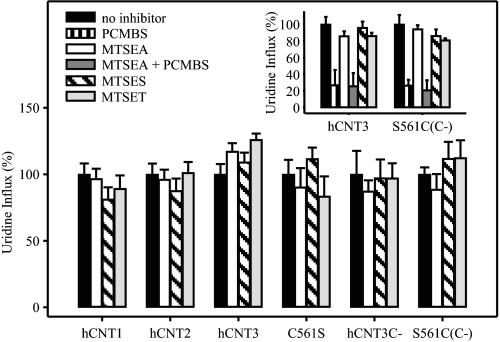
Effects of MTS Reagents—In addition to PCMBS, we also tested the inhibitory effects of three MTS derivatives, MTSEA, MTSES, and MTSET. Included in the analysis were wild-type hCNT1, hCNT2, and hCNT3, hCNT3 mutant C561S, cysteine-free hCNT3C-, and hCNT3C- mutant S561C(C-). Reflecting their different reactivities toward thiol groups, MTSEA, MTSES, and MTSET were used at concentrations of 2.5, 10, and 1 mm, respectively. Similar to PCMBS, oocytes producing each of the constructs were exposed to MTS reagents in acidified 100 mm ChCl transport medium, pH 5.5, then assayed for radiolabeled uridine transport activity either in 100 mm NaCl, pH 7.5 (hCNT1 and hCNT2), or 100 mm ChCl, pH 5.5 (hCNT3, C561S, hCNT3C-, and S561C(C-)). In no case was transport activity affected (Fig. 10). Similarly, and in agreement with previously published studies for hCNT1 (22) and hCNT3C- mutant S561C(C-) (25) under non-acidified conditions, there was also no inhibition of uridine uptake when constructs were incubated with the three MTS reagents in Na+-containing medium at pH 8.5 (data not shown).
FIGURE 10.
Effects of MTS reagents on hCNT1, hCNT2, hCNT3, and mutants. Oocytes producing hCNT1, hCNT2, hCNT3, C561S, hCNT3C-, or S561C(C-) were incubated under acidic conditions in the absence of inhibitor or in the presence of 500 μm PCMBS (inset only), 2.5 mm MTSEA, 2.5 mm MTSEA followed by 500 μm PCMBS (inset only), 10 mm MTSES or 1 mm MTSET. After incubation, 20 μm [3H]uridine influx was measured in the presence of 100 mm NaCl, pH 7.5 (hCNT1 and hCNT2), or 100 mm ChCl, pH 5.5 (hCNT3, C561S, hCNT3C- and S561C(C-)), as described in Fig. 2. Data are presented as mediated transport, calculated as uptake in RNA-injected oocytes minus uptake in water-injected oocytes, and normalized to the influx of uridine in the absence of inhibitor (7.0 ± 0.6, 6.5 ± 0.5, 6.6 ± 0.5, 3.6 ± 0.4, 1.0 ± 0.2, and 4.1 ± 0.2 pmol/oocyte·min-1 for hCNT1, hCNT2, hCNT3, C561S, hCNT3C-, and S561C(C-), respectively (large panel), and 8.2 ± 0.8 and 7.0 ± 0.8 pmol/oocyte·min-1 for hCNT3 and S561C(C-), respectively (inset). Each value is the mean ± S.E. of 10–12 oocytes.
To eliminate the possibility that MTS reagents bind to hCNT3 Cys-561 without affecting transport, oocytes producing wild-type hCNT3 or revertant hCNT3C- mutant S561C(C-) were incubated first in acidified medium (100 mm ChCl, pH 5.5) containing 2.5 mm MTSEA followed by a subsequent incubation under the same conditions with 500 μm PCMBS (Fig. 10, inset). Oocytes producing hCNT3 or S561C(C-) both showed the expected degree of uridine transport inhibition when treated with PCMBS alone (compare with Figs. 2B and 7B, respectively), and the extent of this inhibition was unaffected by pre-exposure to MTSEA. Therefore, MTSEA, the smallest of the MTS reagents examined, was unable to access Cys-561.
Substituted Cysteine Accessibility Methodology (SCAM) Analysis of TM12—To explore the relationship between Cys-561 and other TM12 residues, the cysteine-free background of hCNT3C- was used in conjunction with SCAM to systematically screen all 21 putative positions in the helix for PCMBS sensitivity. In this analysis a series of hCNT3C- mutants with individual TM12 residues mutated to cysteine were produced in Xenopus oocytes. Functional mutants together with hCNT3C- as control were investigated for inhibition by 200 μm PCMBS both in the presence of Na+ or H+ and, where inhibition was obtained, uridine protection (Table 1).
TABLE 1.
Effects of PCMBS on uridine uptake in Xenopus oocytes expressing hCNT3C – and single-cysteine mutants
Influx of 10 μm [3H]uridine was measured in both Na+- and H+-containing medium (100 mm NaCl, pH 8.5, or 100 mm ChCl, pH 5.5, respectively; 1 min; 20 °C) after 10 min of incubation on ice in the absence or presence of 200 μm PCMBS or 200 μm PCMBS + 20 mm uridine in media of the same composition (i.e. Na+- or H+-containing, as indicated). Values are corrected for basal non-mediated uptake in control water-injected oocytes and are presented as a percentage of mediated uridine influx in the absence of inhibitor for each individual mutant. Each value is the mean ± S.E. of 10–12 oocytes. ND, not determined because of low functional activity.
|
Na+a
|
H+b
|
|||
|---|---|---|---|---|
| +PCMBS | +PCMBS and uridine | +PCMBS | +PCMBS and uridine | |
| hCNT3C– | 109 ± 9 | – | 103 ± 13 | – |
| I554C(C–) | 98 ± 16 | – | 52 ± 6 | 67 ± 9 |
| I555C(C–) | 73 ± 8 | – | 66 ± 7 | – |
| A556C(C–) | 97 ± 13 | – | 114 ± 21 | – |
| T557C(C–) | 7 ± 1 | 16 ± 2 | 22 ± 2 | 33 ± 2 |
| Y558C(C–) | 92 ± 15 | – | 19 ± 3 | 71 ± 11 |
| A559C(C–) | 106 ± 13 | – | 117 ± 14 | – |
| L560C(C–) | 101 ± 14 | – | 106 ± 14 | – |
| S561C(C–) | 102 ± 19 | – | 54 ± 5 | 94 ± 8 |
| G562C(C–) | 104 ± 17 | – | 92 ± 18 | – |
| F563C(C–) | ND | – | ND | – |
| A564C(C–) | 110 ± 31 | – | 88 ± 13 | – |
| N565C(C–) | 37 ± 5 | 109 ± 8 | 46 ± 9 | 106 ± 16 |
| I566C(C–) | 85 ± 19 | – | 71 ± 6 | – |
| G567C(C–) | 17 ± 4 | 97 ± 19 | 14 ± 5 | 94 ± 15 |
| S568C(C–) | ND | – | ND | – |
| L569C(C–) | 112 ± 17 | – | 113 ± 12 | – |
| G570C(C–) | 107 ± 15 | – | 93 ± 12 | – |
| I571C(C–) | 44 ± 8 | 85 ± 9 | 30 ± 3 | 90 ± 8 |
| V572C(C–) | 89 ± 11 | – | 104 ± 9 | – |
| I573C(C–) | 92 ± 11 | – | 109 ± 15 | – |
| G574C(C–) | 92 ± 23 | – | 95 ± 16 | – |
Values for mediated uridine influx in 100 mm NaCl, pH 8.5 (pmol/oocyte·min–1), in the absence of inhibitor are: hCNT3C– (1.5 ± 0.2); I554C(C–) (2.0 ± 0.3); I555C(C–) (1.9 ± 0.2); A556C(C–) (1.5 ± 0.3); T557C(C–) (2.7 ± 0.3); Y558C(C–) (0.11 ± 0.01); A559C(C–) (0.78 ± 0.05); L560C(C–) (2.1 ± 0.3); S561C(C–) (2.4 ± 0.4); G562C(C–) (0.75 ± 0.2); A564C(C–) (0.50 ± 0.1); N565C(C–) (2.6 ± 0.4); I566C(C–) (0.91 ± 0.2); G567C(C–) (1.4 ± 0.3); L569C(C–) (1.8 ± 0.3); G570C(C–) (1.1 ± 0.2); I571C(C–) (3.2 ± 0.4); V572C(C–) (3.3 ± 0.3); I573C(C–) (3.8 ± 0.5); G574C(C–) (0.18 ± 0.04)
Values for mediated uridine influx in 100 mm ChCl, pH 5.5 (pmol/oocyte·min–1), in the absence of inhibitor are: hCNT3C– (1.5 ± 0.3); I554C(C–) (1.5 ± 0.2); I555C(C–) (2.1 ± 0.2); A556C(C–) (1.5 ± 0.1); T557C(C–) (1.9 ± 0.1); Y558C(C–) (0.50 ± 0.04); A559C(C–) (1.0 ± 0.1); L560C(C–) (2.1 ± 0.3); S561C(C–) (3.6 ± 0.2); G562C(C–) (0.51 ± 0.1); A564C(C–) (0.10 ± 0.01); N565C(C–) (0.79 ± 0.1); I566C(C–) (0.35 ± 0.1); G567C(C–) (0.9 ± 0.1); L569C(C–) (1.5 ± 0.2); G570C(C–) (0.69 ± 0.1); I571C(C–) (1.4 ± 0.1); V572C(C–) (2.0 ± 0.2); I573C(C–) (1.8 ± 0.2); G574C(C–) (0.13 ± 0.03)
Of the 21 mutants investigated, only two (F563C(C-) and S568C(C-)) had functional activity too low to characterize further (10 μm uridine influx <0.1 pmol/oocyte·min-1). F563C(C-) is also nonfunctional when produced in yeast, whereas S568C(C-) exhibits very low transport activity (25). Because both mutant proteins are localized to the yeast plasma membrane in amounts similar to hCNT3 and hCNT3C- (25), it is likely that residues Phe-563 and Ser-568 are structurally and/or functionally important for hCNT3 transport activity. Both residues are highly conserved within the CNT protein family (Fig. 11). Of the remaining 19 cysteine substitutions, seven resulted in substantial (>40%) inhibition by 200 μm PCMBS (I554C(C-), T557C(C-), Y558C(C-), S561C(C-), N565C(C-), G567C(C-), and I571C(C-)). Of these, two resembled S561C(C-) and were selectively inhibited by PCMBS in the presence of H+ but not Na+ (I554C(C-) and Y558C(C-)), whereas four were approximately equally PCMBS-sensitive in either Na+- or H+-containing media (T557C(C-), N565C(C-), G567C(C-), and I571C(C-)). The presence of uridine (20 mm) during exposure to PCMBS resulted in partial protection of Y558C(C-) and essentially complete protection of S561C(C-), N565C(C-), G567C(C-), and I571C(C-). No uridine protection was seen for I554C(C-) or T557C(C-). Two additional mutants that were more weakly inhibited by PCMBS (I555C(C-) and I566C(C-)) in both Na+- and H+-containing media were not tested for uridine protection.
FIGURE 11.
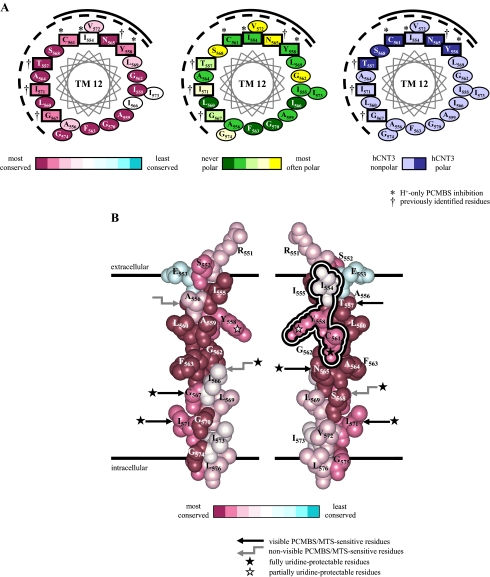
Molecular modeling of hCNT3 TM12. Analysis of residue conservation in the region corresponding to residues 551–576 of hCNT3 and its homologs was performed by the ConSeq method (40) on the aligned sequences of 126 eukaryote and prokaryote CNT family members. A presents three identical α-helical wheel projections of hCNT3 TM12 viewed from the extracellular side of the membrane and colored either to indicate degrees of residue conservation (left), polarity based on analysis of the multiple sequence alignment (middle), or polarity of hCNT3 residues (right). Residue positions in hCNT3 sensitive to inhibition by PCMBS are boxed. Those reactive with PCMBS in H+-containing medium only are indicated by an asterisk (*) Four residue positions previously shown to be reactive toward MTS reagents (25) are indicated by †. The same four residues are also characterized by PCMBS inhibition in the presence of both Na+ and H+. B shows corresponding views of an α-helical space-filling model of the region. The view on the left differs from that on the right by a 180° rotation. To permit comparison with the left-hand helical wheel projection in A, the views are colored to indicate degrees of residue conservation. The conformationally mobile cluster of three residues specifically reactive with PCMBS only in H+-containing medium are outlined in the schematic on the right. Other PCMBS/MTS-sensitive residues are indicated by black straight arrows where visible or gray elbow arrows where present on the non-visible, opposite face of the helix. PCMBS/MTS-sensitive positions that are fully and partially uridine-protected and, therefore, likely to be within or closely adjacent to the nucleoside binding pocket are indicated by black and white stars, respectively.
DISCUSSION
As shown in Fig. 1, current models of hCNT topology have 13 putative TMs (19). Computer algorithms also weakly predict two additional potential transmembrane regions, designated in Fig. 1 as 5A and 11A (19). Consistent with both a 13 and 15 TM membrane architecture, the loop linking TMs 4 and 5 has been shown to be cytoplasmic, whereas the N terminus and glycosylated C terminus are intracellular and extracellular, respectively (19). Initial SCAM analyses of TMs 11, 12, and 13 of hCNT3C- using MTS reagents (25) as well as other previously published structure/function studies (e.g. Refs. 20 and 21) are also consistent with both models. Recent investigations of hCNT1 glutamate residues (22), however, favor a 15 TM membrane architecture.
Previously, TMs 7 and 8 (Fig. 1) were identified to contain residues of functional importance and are predicted to be pore-lining (20–22). The presently revealed H+-activated reactivity of wild-type hCNT3 Cys-561 to PCMBS, as determined by inhibition of uridine transport activity, now establishes that TM12 is also pore-lining. Pore-lining status has also been ascribed to this TM on the basis of an initial series of SCAM analyses of cysteine-free hCNT3C- (25). Performed at neutral pH, this study found partial sensitivity of TM12 mutants T557C(C-), N565C(C-), G567C(C-) and I571C(C-) to MTS reagents and variable protection against that inhibition by uridine (25). S561C(C-) was unreactive to MTS reagents in that analysis, a finding confirmed here in experiments performed with MTSEA, MTSES, and MTSET under both acidified and H+-reduced conditions and with Cys-561 present in either a cysteine-free (hCNT3C-) or wild-type (hCNT3) background.
PCMBS (31, 32) and MTS (33, 34) reagents both react preferentially with the ionized thiolate form of cysteine (-S-) rather than with the uncharged thiol form (-SH) (35). PCMBS and the MTS derivatives MTSEA, MTSES, and MTSET differ, however, in charge (PCMBS and MTSES are negatively charged, MTSEA and MTSET are positively charged) and membrane permeability (PCMBS, MTSES, and MTSET are membrane-impermeable, MTSEA is membrane-permeable). They also differ in size (PCMBS < MTSEA < MTSES < MTSET). It is possible, therefore, that steric and/or electrostatic factors contributed to the specificity of the interaction of Cys-561 with PCMBS. MTSEA, the smallest of the MTS reagents tested, failed to block addition of PCMBS to hCNT3 Cys-561, confirming the inability of MTS reagents to react with this residue.
hCNT3 Cys-561 is, at least transiently, pore-lining because (i) only cysteines on the water-accessible surface of the protein will ionize to a significant extent, and (ii) hydrophilic negatively charged PCMBS is unlikely to enter hydrophobic regions in the lipid bilayer or protein interior. Because PCMBS was added extracellularly, the aqueous pathway that it traverses to reach Cys-561 must be contiguous with the external medium and, therefore, part of the outward-facing aspect of the hCNT3 translocation pore. The exofacial pore-lining status of Cys-561 is confirmed by the ability of micromolar concentrations of uridine in the extracellular medium to protect the transporter against PCMBS inhibition. Opposite to the predicted effect of pH on thiol group chemical reactivity toward PCMBS, inhibition of hCNT3 only occurred under acidified conditions. Na+ had no influence on PCMBS inhibition of the transporter.
Conserved in all three human CNTs, accessibility of Cys-561 to PCMBS was unique to hCNT3. hCNT3 differs from Na+-specific hCNT1 and hCNT2 by being able to couple uphill nucleoside transport to both Na+ and H+ electrochemical gradients (9–11, 16–18). It is proposed, therefore, that PCMBS reactivity with hCNT3 Cys-561 reports a specific H+-activated conformational state of the protein. Because exofacial uridine occludes this residue and blocks access to PCMBS, hCNT3 Cys-561 is likely located within, or closely adjacent to, the nucleoside binding pocket of the transporter. H+-induced changes in hCNT3 nucleoside and nucleoside drug selectivity (11, 17) are also strongly indicative of a H+-specific conformation of the nucleoside binding pocket and/or translocation pore. H+-coupled hCNT3 is also distinguished by a cation:nucleoside stoichiometry of 1:1, compared with 2:1 for Na+ (17, 18). In acidified Na+-containing transport medium, when both cations are present together, charge/uptake experiments suggest that the transporter binds one Na+ and one H+ (17).
We interpret hCNT1–3 kinetics and cation coupling in terms of a conformational equilibrium model of secondary active transport (36, 37). Developed by Krupka, this modified ordered binding model of secondary active transport alleviates the stringent sequential carrier states of earlier models and instead allows for flexible cation interactions such as those observed for Na+- and H+-coupling of hCNT3. In the model, binding of cation (Na+ and/or H+ in the case of hCNT3) shifts the equilibrium between two carrier states to “unlock” or open the nucleoside binding site, thereby promoting active transport. The reactivity of hCNT3 Cys-561 to PCMBS senses unique characteristics of the H+-bound transporter.
As part of a larger, more comprehensive analysis encompassing the entire C-terminal half of hCNT3C-,5 SCAM experiments have been undertaken utilizing PCMBS to investigate the molecular and functional properties of all 21 putative residues of TM12 (Table 1). In addition to S561C(C-), these studies have identified two further cysteine-substituted constructs in TM12, I554C(C-) and Y558C(C-), which also exhibit H+-activated inhibition by PCMBS. Consistent with their anticipated relative depth within the membrane (and likely proximity to the nucleoside binding pocket) and as demonstrated in Fig. 9 for S561C(C-), the constructs exhibited no protection (I554C(C-)), partial protection (Y558C(C-)) and full protection by extracellular uridine (S561C(C-)). More than Cys-561 alone, therefore, three adjacent pore-lining residues of hCNT3 (Ile-554, Tyr-558, and Cys-561) combine to delineate a conformationally sensitive exofacial pore-lining region of TM12 specifically responsive to H+ binding. In addition, each of the four residue positions in TM12 previously shown to be sensitive to MTS reagents at neutral pH (Thr-557, Asn-565, Gly-567, and Ile-571) were also found to display inhibition by PCMBS in the presence of both Na+ and H+. All, except Thr-557 (the most exofacial) were uridine-protectable. None of the remaining 14 residue positions in TM12 were strongly PCMBS-sensitive.
H+-activation, therefore, distinguishes two discrete classes of PCMBS-sensitive residues within TM12 of hCNT3. To provide additional information on these residues and the possible nature of the H+-induced conformational shift, Fig. 11 presents α-helical wheel projections (Fig. 11A) and space-filling models of hCNT3 TM12 (Fig. 11B), although it is appreciated that the true structure of this region of the protein may differ appreciably from the perfect α-helix illustrated. Based on the aligned sequences of 126 eukaryote and prokaryote CNT family members, individual residues are color-coded to indicate patterns of residue conservation and polarity within the helix. Residues Ile-554, Ser-558, and Cys-561 and Thr-557, Asn-565, Gly-567, and Ile-571 are highlighted. TM12 has the membrane orientation shown in Fig. 11 irrespective of whether hCNT3 has a 13 or 15 TM membrane architecture (Fig. 1).
All 21 residue positions in TM12 exhibited restricted variability, showing a high degree of conservation among CNT family members (Fig. 11A). This suggests involvement either in maintaining the structures of the transporters or in cation and nucleoside binding and translocation, these being features of CNT family members that are held in common. Thus, TM12 residues are likely either to face another helix or to line the translocation pore.
The helical wheel projections in Fig. 11A establish that residues insensitive to PCMBS (and MTS reagent) inhibition are localized to one-half of the helix surface. Consistent with a role in helix-helix packing, this half of TM12 contains six residue positions exhibiting the highest level of sequence conservation (Ile-555, Ala-559, Gly-562, Phe-563, Gly-570, and Gly-574). In contrast, the other side of TM12 is mostly PCMBS/MTS-reactive and likely, therefore, to be pore-lining. The uridine protection observed with many of these residues and the demonstration that polar residues localize predominantly to this face of the helix support this conclusion. The four residue positions (Thr-557, Asn-565, Gly-567, and Ile-571), which show inhibition by PCMBS in the presence of both Na+ and H+, are distributed throughout the PCMBS-sensitive face of the helix, whereas Ser-561 and the two adjacent residue positions (Ile-554 and Tyr-558), which exhibit H+-activated PCMBS inhibition, come together in one quadrant of that surface.
Fig. 11B spatially differentiates within the plane of the membrane the H+-dependent class of PCMBS-sensitive residues from those that are inhibited in the presence of both Na+ and H+. Residues that are inhibited by PCMBS only in the presence of H+ cluster together in a small exofacial aspect of the helix specific to the H+-bound conformation of hCNT3, whereas those which exhibit inhibition by PCMBS in the presence of both Na+ and H+ have more endofacial locations. Additionally, Fig. 11B reveals that the five TM12 residue positions that are both PCMBS-sensitive and uridine-protectable (Tyr-558, Cys-561, Asn-565, Gly-567, and Ile-571) are grouped centrally within the putative pore-lining face of TM12 in a position that likely delineates the location of the uridine binding pocket. Presteady-state current measurements of hCNT1 (16) and hCNT35 reveal that the site(s) of Na+ binding also reside approximately halfway across the membrane.
Within the plane of the membrane, uridine-protectable Cys-561, the residue identified and characterized in the present study, is located at the interface between those residues sensitive to inhibition by PCMBS in H+-containing medium only and those where inhibition occurs in the presence of both Na+ and H+. Among other possibilities, we hypothesize that the surface domain of hCNT3 TM12 represented by residues Ile-554, Tyr-558, and Cys-561 may be masked to PCMBS in the H+-unbound state, and upon H+ binding, movement of the helix occurs such that these residues now become accessible to the aqueous translocation pore. hCNT3, unlike hCNT1/2, has two cation binding sites. Our previous findings suggest that one site is Na+-specific, whereas the second site may functionally interact with both H+ and Na+. Our hypothesis is that hCNT3 Cys-561 senses conformational changes associated with H+ binding to the second of these sites.
hCNT3 presteady-state currents have components contributed by carrier-associated charge movements as well as by Na+ binding. PCMBS addition to Cys-561 will contribute a negative charge to the modified transporter. The total abolition of hCNT3 presteady-state currents that is seen after treatment with PCMBS is consistent with blockade of hCNT3 function by a mechanism involving loss of cation binding and locking of the transporter in a conformationally restricted state.
In addition to the pore-lining residues present in TMs 7 and 8 of hCNT1 (20–22), the present study identifies a conformationally sensitive pore-lining residue in TM12 of hCNT3. A glutamate residue in hCNT1 and hCNT3 with a critical role in cation binding has also been identified within the conserved motif (G/A)XKX3NEFVA(Y/M/F) of TM11A (22). In the 15 TM model of hCNT membrane architecture, TMs 7/8 and 11A/12 are separated by a large and likely flexible cytoplasmic loop, evident in Fig. 1 as the 37-residue linker region between TMs 9 and 10. It is possible that this loop enables TMs 7/8 and 11A/12 to come together in the translocation pore in a manner that facilitates conformational transitions within the cation/nucleoside translocation cycle.
An emerging theme of recently solved high resolution molecular structures of cation transporters such as LeuTAa (38) and GltPh (39) is close-proximity integration of cation/solute binding and transport within a common cation/permeant translocation pore. The present results for hCNT3 Cys-561 mirror this principle and reveal a residue centrally positioned within a mobile region of the cation/nucleoside translocation machinery. Further investigation of this conformationally sensitive residue is likely to provide mechanistic and structural insights into differences between hCNT3 and hCNT1/2.
This work was supported in part by the National Cancer Institute of Canada with funds from the Canadian Cancer Society and the Alberta Cancer Board. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: NT, nucleoside transporter; CNT, concentrative nucleoside transporter; TM, putative transmembrane helix; PCMBS, p-chloromercuribenzene sulfonate; MES, 2-(N-morpholino)ethanesulfonic acid; MTS, methanethiosulfonate; MTSEA, 2-aminoethyl methanethiosulfonate hydrobromide; MTSES, sodium (2-sulfonatoethyl) methanethiosulfonate; MTSET, [(triethylammonium)ethyl] methanethiosulfonate bromide; DTT, dithiothreitol; SCAM, substituted cysteine accessibility method; ChCl, choline chloride.
M. D. Slugoski, A. M. L. Ng, S. Y. M. Yao, K. M. Smith, C. C. Lin, J. Zhang, E. Karpinski, C. E. Cass, S. A. Baldwin, and J. D. Young, unpublished results.
References
- 1.Cass, C. E. (1995) in Drug Transport in Antimicrobial and Anticancer Chemotherapy (Georgopapadakou, N. H., ed) pp. 403-451, Marcel Dekker, Inc., New York
- 2.Griffith, D. A., and Jarvis, S. M. (1996) Biochim. Biophys. Acta 1286 153-181 [DOI] [PubMed] [Google Scholar]
- 3.Young, J. D., Cheeseman, C. I., Mackey, J. R., Cass, C. E., and Baldwin, S. A. (2000) in Current Topics in Membranes (Barrett, K. E., and Donowitz, M., eds) Vol. 50, pp. 329-378, Academic Press, Inc., San Diego [Google Scholar]
- 4.Damaraju, V. L., Damaraju, S., Young, J. D., Baldwin, S. A., Mackey, J., Sawyer, M. B., and Cass, C. E. (2003) Oncogene 22 7524-7536 [DOI] [PubMed] [Google Scholar]
- 5.Latini, S., and Pedata, F. (2001) J. Neurochem. 79 463-484 [DOI] [PubMed] [Google Scholar]
- 6.King, A. E., Ackley, M. A., Cass, C. E., Young, J. D., and Baldwin, S. A. (2006) Trends Pharmacol. Sci. 27 416-425 [DOI] [PubMed] [Google Scholar]
- 7.Gray, J. H., Owen, R. P., and Giacomini, K. M. (2004) Pfluegers Arch. Eur. J. Physiol. 447 728-734 [DOI] [PubMed] [Google Scholar]
- 8.Baldwin, S. A., Beal, P. R., Yao, S. Y. M., King, A. E., Cass, C. E., and Young, J. D. (2004) Pfluegers Arch. Eur. J. Physiol. 447 735-743 [DOI] [PubMed] [Google Scholar]
- 9.Ritzel, M. W. L., Yao, S. Y. M., Huang, M. Y., Elliott, J. F., Cass, C. E., and Young, J. D. (1997) Am. J. Physiol. 272 C707-C714 [DOI] [PubMed] [Google Scholar]
- 10.Ritzel, M. W. L., Yao, S. Y. M., Ng, A. M. L., Mackey, J. R., Cass, C. E., and Young, J. D. (1998) Mol. Membr. Biol. 15 203-211 [DOI] [PubMed] [Google Scholar]
- 11.Ritzel, M. W. L., Ng, A. M. L., Yao, S. Y. M., Graham, K., Loewen, S. K., Smith, K. M., Ritzel, R. G., Mowles, D. A., Carpenter, P., Chen, X.-Z., Karpinski, E., Hyde, R. J., Baldwin, S. A., Cass, C. E., and Young, J. D. (2001) J. Biol. Chem. 276 2914-2927 [DOI] [PubMed] [Google Scholar]
- 12.Yao, S. Y. M., Ng, A. M. L., Loewen, S. K., Cass, C. E., Baldwin, S. A., and Young, J. D. (2002) Am. J. Physiol. Cell Physiol. 283 155-168 [DOI] [PubMed] [Google Scholar]
- 13.Loewen, S. K., Ng, A. M. L., Mohabir, N. N., Baldwin, S. A., Cass, C. E., and Young, J. D. (2003) Yeast 20 661-675 [DOI] [PubMed] [Google Scholar]
- 14.Xiao, G., Wang, J., Tangen, T., and Giacomini, K. M. (2001) Mol. Pharmacol. 59 339-348 [DOI] [PubMed] [Google Scholar]
- 15.Loewen, S. K., Yao, S. Y. M., Slugoski, M. D., Mohabir, N. N., Turner, R. J., Mackey, J. R., Weiner, J. H., Gallagher, M. P., Henderson, P. J., Baldwin, S. A., Cass, C. E., and Young, J. D. (2004) Mol. Membr. Biol. 21 1-10 [DOI] [PubMed] [Google Scholar]
- 16.Smith, K. M., Ng, A. M. L., Yao, S. Y. M., Labedz, K. A., Knaus, E. E., Wiebe, L. I., Cass, C. E., Baldwin, S. A., Chen, X.-Z., Karpinski, E., and Young, J. D. (2004) J. Physiol. (Lond.) 558 807-823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith, K. M., Slugoski, M. D., Loewen, S. K., Ng, A. M. L., Yao, S. Y. M., Chen, X.-Z., Karpinski, E., Cass, C. E., Baldwin, S. A., and Young, J. D. (2005) J. Biol. Chem. 280 25436-25449 [DOI] [PubMed] [Google Scholar]
- 18.Smith, K. M., Slugoski, M. D., Cass, C. E., Baldwin, S. A., Karpinski, E., and Young, J. D. (2007) Mol. Membr. Biol. 24 53-64 [DOI] [PubMed] [Google Scholar]
- 19.Hamilton, S. R., Yao, S. Y. M., Ingram, J. C., Hadden, D. A., Ritzel, M. W. L., Gallagher, M. P., Henderson, P. J. F., Cass, C. E., Young, J. D., and Baldwin, S. A. (2001) J. Biol. Chem. 276 27981-27988 [DOI] [PubMed] [Google Scholar]
- 20.Loewen, S. K., Ng, A. M. L., Yao, S. Y. M., Cass, C. E., Baldwin, S. A., and Young, J. D. (1999) J. Biol. Chem. 274 24475-24484 [DOI] [PubMed] [Google Scholar]
- 21.Slugoski, M. D., Loewen, S. K., Ng, A. M. L., Smith, K. M., Yao, S. Y. M., Karpinski, E., Cass, C. E., Baldwin, S. A., and Young, J. D. (2007) Biochemistry 46 1684-1693 [DOI] [PubMed] [Google Scholar]
- 22.Yao, S. Y. M., Ng, A. M. L., Slugoski, M. D., Smith, K. M., Mulinta, R., Karpinski, E., Cass, C. E., Baldwin, S. A., and Young, J. D. (2007) J. Biol. Chem. 282 30607-30617 [DOI] [PubMed] [Google Scholar]
- 23.Liman, E. R., Tytgat, J., and Hess, P. (1992) Neuron 9 861-871 [DOI] [PubMed] [Google Scholar]
- 24.Kirsch, R. D., and Joly, E. (1998) Nucleic Acids Res. 26 1848-1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang, J., Tackaberry, T., Ritzel, M. W., Raborn, T., Barron, G., Baldwin, S. A., Young, J. D., and Cass, C. E. (2006) Biochem. J. 394 389-398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao, S. Y. M., Cass, C. E., and Young, J. D. (2000) in Membrane Transport. A Practical Approach (Baldwin, S. A., ed.) pp. 47-78, Oxford University Press, New York
- 27.Huang, Q. Q., Yao, S. Y. M., Ritzel, M. W., Paterson, A. R., Cass, C. E., and Young, J. D. (1994) J. Biol. Chem. 269 17757-17760 [PubMed] [Google Scholar]
- 28.Yao, S. Y. M., Sundaram, M., Chomey, E. G., Cass, C. E., Baldwin, S. A., and Young, J. D. (2001) Biochem. J. 353 387-393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rothstein, A. (1970) Curr. Top. Membr. Transp. 1 135-176 [Google Scholar]
- 30.Hu, H., Endres, C. J., Chang, C., Umapathy, N. S., Lee, E.-W., Fei, Y.-J., Itagaki, S., Swaan, P. W., Ganapathy, V., and Unadkat, J. D. (2006) Mol. Pharmacol. 69 1542-1553 [DOI] [PubMed] [Google Scholar]
- 31.Yan, R. T., and Maloney, P. C. (1993) Cell. 75 37-44 [PubMed] [Google Scholar]
- 32.Yan, R. T., and Maloney, P. C. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 5973-5976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akabas, M. H., Stauffer, D. A., Xu, M., and Karlin, A. (1992) Science 258 307-310 [DOI] [PubMed] [Google Scholar]
- 34.Stauffer, D. A., and Karlin, A. (1994) Biochemistry 33 6840-6849 [DOI] [PubMed] [Google Scholar]
- 35.Williams, D. B., and Akabas, M. H. (1999) Biophys. J. 77 2563-2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krupta, R. M. (1993) Biochim. Biophys. Acta 1183 105-113 [DOI] [PubMed] [Google Scholar]
- 37.Krupta, R. M. (1994) Biochim. Biophys. Acta 1193 165-178 [DOI] [PubMed] [Google Scholar]
- 38.Yamashita, A., Singh, S. K., Kawate, T., Jin, Y., and Gouaux, E. (2005) Nature 437 215-223 [DOI] [PubMed] [Google Scholar]
- 39.Boudker, O., Ryan, R. M., Yernool, D., Shimamoto, K., and Gouaux, E. (2007) Nature 445 387-393 [DOI] [PubMed] [Google Scholar]
- 40.Berezin, C., Glaser, F., Rosenberg, J., Paz, I., Pupko, T., Fariselli, P., Casadio, R., and Ben-Tal, N. (1994) Bioinformatics 20 1322-1324 [DOI] [PubMed] [Google Scholar]