Abstract
STAT1 is an essential transcription factor for macrophage activation by IFN-γ and requires phosphorylation of the C-terminal Ser727 for transcriptional activity. In macrophages, Ser727 phosphorylation in response to bacterial lipopolysaccharide (LPS), UV irradiation, or TNF-α occurred through a signaling path sensitive to the p38 mitogen-activated protein kinase (p38 MAPK) inhibitor SB203580 whereas IFN-γ-mediated Ser727 phosphorylation was not inhibited by the drug. Consistently, SB203580 did not affect IFN-γ-mediated, Stat1-dependent transcription but inhibited its enhancement by LPS. Furthermore, LPS, UV irradiation, and TNF-α caused activation of p38 MAPK whereas IFN-γ did not. An essential role for p38 MAPK activity in STAT1 Ser727 phosphorylation was confirmed by using cells expressing an SB203580-resistant p38 MAPK. In such cells, STAT1 Ser727 phosphorylation in response to UV irradiation was found to be SB203580 insensitive. Targeted disruption of the mapkap-k2 gene, encoding a kinase downstream of p38 MAPK with a key role in LPS-stimulated TNF-α production and stress-induced heat shock protein 25 phosphorylation, was without a significant effect on UV-mediated Ser727 phosphorylation. The recombinant Stat1 C terminus was phosphorylated in vitro by p38MAPKα and β but not by MAPK-activated protein kinase 2. Janus kinase 2 activity, previously reported to be required for IFN-γ-mediated Ser727 phosphorylation, was not needed for LPS-mediated Ser727 phosphorylation, and activation of Janus kinase 2 did not cause the appearance of STAT1 Ser727 kinase activity. Our data suggest that STAT1 is phosphorylated at Ser727 by a stress-activated signaling pathway either through p38 MAPK directly or through an unidentified kinase downstream of p38MAPK.
Signal transducers and activators of transcription (STATs) cause rapid transcriptional responses to cytokines (1, 2). The prototype of this protein family, STAT1, lies at the heart of the immediate response to interferons (IFN). Targeted disruption of the STAT1 gene obstructs the IFN “system” and causes a loss of natural immunity to microbial pathogens (3, 4). In the case of bacteria, this is largely attributable to an impairment of macrophage activation by the Th1 cytokine IFN-γ. In a normal situation, IFN-γ activates STAT1 by triggering phosphorylation at two distinct sites. IFN-γ receptor-associated Janus kinases 1 and 2 (JAK1 and JAK2) phosphorylate Tyr701 and induce SH2 domain-mediated STAT1 dimerization, followed by nuclear translocation and binding to γ-interferon activation site DNA sequences (1, 5). The C-terminal Ser727 within a potential mitogen-activated protein kinase (MAPK) consensus PMSP motif is phosphorylated by (a) hitherto unknown kinase(s). This event strongly increases the transcription factor activity of STAT1 (6). In fact, STAT1 mutated to alanine at position 727 is unable to mediate interferon responses (7, 8).
Relevant to the biology of macrophages, S727 can serve to feed IFN-γ-independent signals into STAT1. For example, macrophage-activating components of bacterial cell walls like lipopolysaccharide (LPS) strongly activate a STAT1 Ser727 kinase independently of Tyr701 phosphorylation. Concomitantly, the presence of IFN-γ and LPS increases the efficiency of Ser727 phosphorylation over that of IFN-γ alone and thus produces a large pool of STAT1 molecules phosphorylated on both Tyr701 and Ser727 (9). The presence of bacterial signals thus enhances the synthesis of an arsenal of antimicrobial proteins that occurs upon IFN-γ-mediated macrophage activation because it promotes transcription of STAT1 target genes over that induced by IFN-γ alone. STAT1 Ser727 phosphorylation therefore contributes to the antibacterial strategies of the innate immune system. An open question remains whether LPS-derived signals target the same STAT1 Ser727 kinase that is also stimulated by IFN-γ.
The treatment of macrophages with LPS causes activation of all three major groups of MAPKs (10, 11). These are the extracellular signal-regulated kinases (ERK1 and ERK2), as well as the c-Jun kinases (JNKs) and the p38 MAPK isoenzymes. Although ERKs are usually referred to as growth factor-stimulated MAPKs, JNKs and p38 MAPK are classical stress-activated kinases, stimulated by a plethora of signals such as proinflammatory cytokines like TNF-α or IL-1, UV irradiation, microbial infection, osmotic stress, and others (12). Proteins such as transcription factors, translation initiation factors, or heat shock proteins that are targets of MAPK pathways and orchestrate a cellular growth factor or stress response may be direct substrates of ERKs, JNKs, or p38 MAPK, or alternatively may be phosphorylated by kinases that are their substrates. Several protein kinase substrates of MAPKs have been identified and are in vivo targets of the ERK1/2 pathway {MAPK-activated kinase [MAPKAP-K1/ribosomal S6 kinase (RSK)] (13, 14)}, the p38 MAPK pathway [MAPKAP-K2/3 (15–18), p38-regulated/activated kinase (PRAK) (19)], or both the ERK1/2 and p38 MAPK pathways [MAPK-interacting kinase 1/2 (MNK) (20, 21), mitogen- and stress-activated protein kinase 1 (MSK) (22)]. ERK1/2-dependent and p38 MAPK-dependent responses can be probed with specific inhibitors. The drug PD098059 inhibits activation of MAPK/ERK kinase (MEK) and therefore blocks the activation of ERK1/2 whereas p38 MAPK α and β can be inhibited specifically with the pyridinyl imidazole compound SB203580 (23, 24).
Ser727 lies within a potential MAPK phosphorylation motif, and we have investigated the contribution of the different MAPK pathways to the serine phosphorylation of STAT1. Our data show that, unlike IFN-γ, stress signals cause STAT1 Ser727 phosphorylation through a pathway requiring p38 MAPK but not a downstream substrate, MAPK-activated protein kinase 2 (MAPKAP-2).
Materials and Methods
Cells, Cytokines, Drugs.
Bac1.2F5 macrophages (25), or the Bac1.2F5-derived clonal line C11 carrying a Stat-dependent luciferase reporter gene (9), were grown in DMEM with 10% FCS and 20% L-cell conditioned medium as a source of colony-stimulating factor 1 (CSF-1). 293 human embryonic kidney fibroblasts (293 cells) were grown in DMEM with 10% FCS. Derivatives of this line conditionally expressing wild-type (WT)-p38MAPK or an SB203580-resistant mutant (DR-p38) were recently described (26). The cells were cultured in DMEM + 10% FCS and the antibiotics Zeocin (Stratagene, 400 μg/ml) and G418 (GIBCO/BRL, 500 μg/ml). p38 MAPK expression was under the control of an Ecdysone-inducible promoter and was stimulated by treatment with 10 μM Ponasterone A (Invitrogen) for 24 h. Immortalized fibroblasts from MAPKAP-K2-deficient mice (27) or from WT mice were cultured in DMEM with 10% FCS. The cells were deprived of FCS for 16 h before treatment with IFN-γ or UV. Murine recombinant IFN-γ was kindly provided by G. Adolf (Boehringer Ingelheim) and was used at a concentration of 5 ng/ml. Human recombinant TNF-α was kindly provided by H. Kocher (Novartis Preclinical Research, Basel) and was used at 20 ng/ml. Murine granulocyte-macrophage CSF (GM-CSF) was a 1:500 dilution of supernatant from X6310 cells, transfected with a GM-CSF expression construct. LPS (Sigma) was either from salmonella or Escherichia coli and was used at a concentration of 1 μg/ml. The p38MAPK inhibitor SB203580 was kindly provided by Ken Murray (SmithKline Beecham) and was used at 10 μM unless otherwise indicated. The drug was added 30–60 min before further treatment. UVC irradiation (254 nm) was with either 40 J/m2 in the case of macrophages and immortalized fibroblasts or 200 J/m2 in the case of 293 embryonic kidney fibroblasts.
Antibodies.
Rabbit antiserum to phosphoserine727-Stat1 (anti-pS727 (9) was used in Western blots at a dilution of 1:5,000. Rabbit antiserum to the Stat1α C terminus (9) was used for immunoprecipitation at a dilution of 1:100 and in Western blots at a dilution of 1:5,000. Rabbit antiserum to Y701-phosphorylated STAT1 was purchased from New England Biolabs and was used at a dilution of 1:1,000. Rabbit antiserum to the JAK2 kinase was a kind gift from Andrew Zimiecki (Laboratory for Clinical and Experimental Cancer Research, Bern, Switzerland). It was used at 1:100 dilution for immunoprecipitation and 1:1,000 dilution for Western blots. Monoclonal antibody Py20, directed against phosphotyrosine, was purchased from Transduction Laboratories (Lexington, KY) and was used for Western blots at 2 μg/ml. Antibodies to phosphorylated p38MAPK and to p38MAPK were purchased from New England Biolabs and were used for Western Blots at a dilution of 1:1,000.
Cellular Extracts.
For immunoprecipitation and/or Western blot analysis, cells were lysed in lysis buffer containing 10 mM Tris⋅HCl (pH 7.05), 50 mM NaCl, 30 mM NaPPi, 50 mM NaF, 2 mM EDTA, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 0.1 mM sodium vanadate, 1 μg/ml pepstatin, 0.5 μg/ml leupeptin, and 3 μg/ml aprotinin. Extracts were cleared by centrifugation at 15,000 rpm (20,000 × g), 4°C, 8 min.
In Vitro Kinase Assay for p38MAPK and MAPKAP-K2 Activity.
A portion (10 μg) glutathione S-transferase (GST)–STAT1 (AA711–750) or the otherwise identical GST-STAT Ser727Ala was incubated for 30 min at 30°C with either activated GST-p38MAPKα (10 units/ml; one unit of p38MAPK was that amount that catalyzed the phosphorylation of 1 nmol of myelin basic protein, 0.33 mg/ml, in the assay described below) or activated GST-p38MAPKβ2 (10 units/ml) in the presence of 50 mM Tris⋅HCl (pH 7.4), 0.1 mM EGTA, 0.1 mM sodium orthovanadate, 0.1 (wt/vol) 2-mercaptoethanol, 0.1 mg/ml BSAe, 10 mM magnesium acetate, and 0.1 mM [32P] ATP. The reactions were terminated with 2% (wt/vol) SDS, and, after electrophoresis on a 10% SDS/polyacrylamide gel, phosphorylated proteins were detected by autoradiography for 3 h at −80°C. GST p38MAPKα and GST p38MAPKβ2 were activated by prior incubation with Mg-ATP and a constitutively active DD MAPK kinase 6 (28). Kinase activity of constitutively active MAPKAP-K2 (Δ3BΔPC) was determined as described (29). GST-STAT1 (AA 711–750), GST-STAT1Ser727Ala and heat shock protein 25 were used as substrates. The reaction was terminated by addition of 8 μl of 4× SDS sample buffer (1× sample buffer is 50 mM Tris⋅HCl (pH 6.8), 0.1 mM EDTA, 5% (vol/vol) glycerol, and 5% (vol/vol) β-mercaptoethanol). Proteins were separated by SDS/PAGE, and 32P-labeled proteins were detected by Bio Imaging using the Cyclone Imaging System and the optiquant software package (Packard).
Immunoprecipitation and Western Blot Analysis.
Cellular extracts were normalized on the basis of protein determinations. Normalized volumes of extracts were incubated for 2–4 h at 4°C with specific antibodies. Immunecomplexes were collected after incubation (2–12 h at 4°C) with Protein A-Sepharose beads. Immunoprecipitates were washed 4 times with lysis buffer. The beads were eluted by boiling in Laemmli sample buffer. Proteins were resolved by 10% SDS/PAGE and were electrotransferred to nitrocellulose. Membranes were blocked with 2% bovine serum albumin in PBST (PBS including 0.1% Tween 20) and were incubated with the appropriate concentration of specific antibody, diluted in PBST + 2% BSA. After washing, the blots were incubated with horseradish peroxidase-conjugated second antibody and were developed by using an ECL system (Amersham Pharmacia) according to the manufacturer’s instructions.
Luciferase Assays.
Extracts from C11 macrophages after appropriate stimulation were assayed for luciferase activity according to standard protocols (30). Inducibility was calculated by dividing cpm light emission of stimulated cells by cpm light emission of unstimulated cells.
Results
Phosphorylation of Stat1 S727 by Signals of Inflammation and Stress Is Blocked by SB203580.
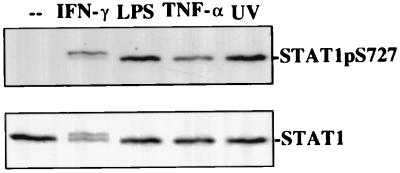
In macrophages, phosphorylation of STAT1 Ser727 occurs in response to IFN-γ and bacterial LPS, both of which cause or promote inflammation responses (9). We sought to determine whether other stimuli of inflammation or stress caused STAT1 Ser727 phosphorylation as well. Macrophages were treated for 25 min with the proinflammatory cytokine TNF-α or were subjected to UVC irradiation. STAT1 Ser727 phosphorylation occurred in both cases, with the stronger signal resulting from UV irradiation (Fig. 1). Thus, STAT1 Ser727 phosphorylation appears to be one of the hallmarks of the macrophages’ inflammatory and stress responses. In marked contrast to IFN-γ, neither UV irradiation nor TNF-α treatment caused tyrosine phosphorylation of Stat1, as indicated by the absence of the typical mobility shift in SDS/PAGE in Fig. 1 (compare the lane marked IFN-γ with all other lanes). This finding was further confirmed by electrophoretic mobility shift assays (data not shown). Therefore, like LPS, TNF-α and UV irradiation target Stat1 without concomitant activation of a JAK-Stat1 pathway.
Figure 1.
Stat1 serine phosphorylation in response to signals of inflammation and stress. Bac1.2F5 macrophages were treated for 25 min with IFN-γ, LPS, or TNF-α or were irradiated with UV (40 J/m2). Cellular extracts were assayed in Western blots for STAT Ser727 phosphorylation and Stat1 expression.
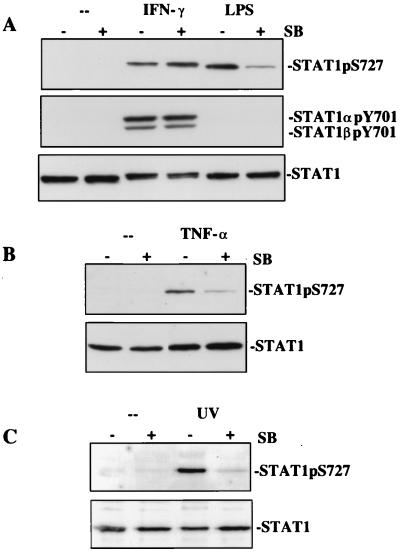
LPS, UV irradiation, and TNF-α activate signaling pathways targeting the stress-activated kinases of the JNK and p38 MAPK families (31, 32). In the case of LPS, the ERK pathway is activated as well (9). We treated macrophages with the pathway-specific inhibitors PD98059 and SB203580. The MAPK/ERK kinase (MEK) inhibitor did not affect LPS-mediated phosphorylation of Stat1 (data not shown). By contrast, SB203580 inhibited STAT1 Ser727 phosphorylation in response to LPS, TNF-α, and UV irradiation by >80% (Fig. 2 A–C). This suggests an involvement of p38 MAPKα, because macrophages predominantly express this isoform, but hardly any β isoform (32). SB203580 may only cause a reduction of ≈80% first because of incomplete inhibition of P38MAPKα by the drug or second because another stress-activated protein kinase that is insensitive to SB203580 phosphorylates this site. One candidate kinase is p38MAPKδ, which is expressed in macrophages (32). IFN-γ-mediated STAT1 Ser727 phosphorylation was not affected by SB203580 (Fig. 2), indicating that the cytokine uses a pathway for STAT1 Ser727 phosphorylation that differs from that used in response to LPS, UV irradiation, or TNF-α. Control experiments showed that SB203580 did not influence JAK-Stat signal transduction (Fig. 2A Middle). In the presence of the drug, IFN-γ-mediated Stat1 Tyr701 phosphorylation was unaffected, and SB203580 did not cause aberrant activation of Stat1 in untreated or LPS-treated macrophages.
Figure 2.
SB203580 inhibits STAT1 Ser727 phosphorylation by LPS, TNF-α, and UV irradiation but not IFN-γ. Bac1.2F5 macrophages were stimulated with IFN-γ or LPS (A) or TNF-α (B) or were irradiated with UV (C) as described for Fig. 1. Where indicated, 10 μM SB203580 was added 30 min before subsequent treatment. Stat1 phosphorylation and expression were determined in Western blots with appropriate antibodies.
SB203580 does not affect the stress-induced phosphorylation of c-Jun, a downstream substrate of the JNKs (33). Consistently, STAT1 Ser727 phosphorylation in response to LPS or IFN-γ in primary bone marrow macrophages from jnk1−/− mice was normal (data not shown).
p38 MAPK Activation and STAT1 Ser727 Phosphorylation Are Consecutive Events in Stress Responses.
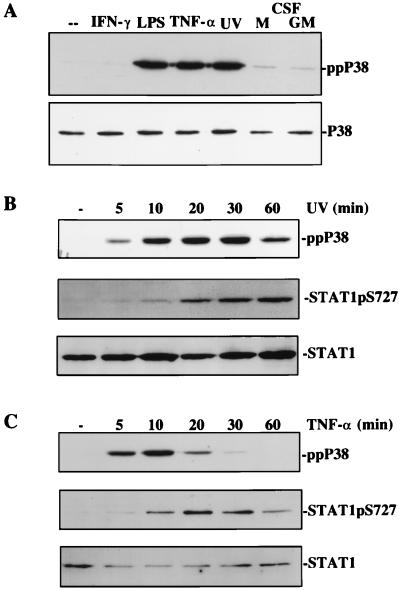
In further experiments, we assayed p38 MAPK activation in macrophages and its correlation to the phosphorylation of STAT1 Ser727. As expected, LPS, TNF-α, and UV irradiation treatments caused a strong phosphorylation of p38 MAPK (Fig. 3A). In marked contrast and consistent with the results obtained after SB203580 treatment, IFN-γ was without effect on p38 phosphorylation. This finding was confirmed at 5, 10, 20, 30, and 60 min after IFN-γ treatment (data not shown). M-CSF and GM-CSF, strong activators of the ERK1/2 pathway, caused minor increases in p38 MAPK activation and no detectable phosphorylation of STAT1 Ser727 (Fig. 5; ref. 9).
Figure 3.
p38MAPK activation and its correlation to STAT1 Ser727 phosphorylation. (A) BAC1.2F5 macrophages were treated with the indicated cytokines or were irradiated with UV, and p38MAPK activation was determined by assaying its phosphorylation status with the use of phosphospecific antibodies. (B and C) UV irradiation (B) or TNF-α treatment (C) of Bac1.2F5 macrophages was carried out for different time periods, and p38 MAPK or STAT1 Ser727 phosphorylation were determined in Western blots with phosphospecific antisera.
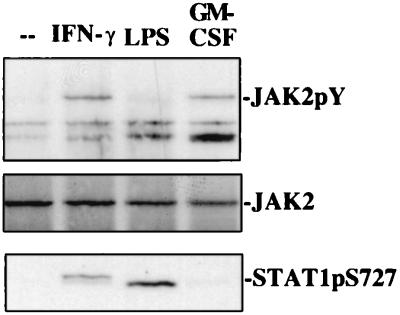
Figure 5.
Correlation between JAK2 activity and phosphorylation of STAT1 Ser727. BAC1.2F5 cells were treated with IFN-γ, LPS, or GM-CSF for 20 min. JAK2 activation was determined after immunoprecipitation from cellular extracts and Western blots with phosphotyrosine-specific monoclonal antibody Py20. Phosphorylation of STAT1 Ser727 was determined in the same extracts by Western blotting.
We wanted to determine the kinetic relationship between p38 MAPK activation and STAT1 Ser727 phosphorylation. UV irradiation (Fig. 3B) and LPS (data not shown) caused p38 MAPK activation with maximal levels reached already after ≈10 min. Maximal levels of STAT1 Ser727 phosphorylation lagged behind p38 MAPK by ≈10 min, and the onset of STAT1 S727 phosphorylation was delayed with respect to that of p38MAPK. A similar lag in maximal phosphorylation of the two molecules was obtained with TNF-α. Notably, the more transient activation of p38 MAPK in this response was paralleled by a more transient activation of STAT1 Ser727 (Fig. 3C). Together, these results are consistent with a role of p38 MAPK; however, the lag between p38 phosphorylation and the phosphorylation of STAT1 Ser727 may indicate the execution of intermediate steps.
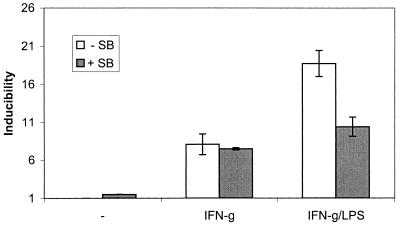
SB203580 Reverses the LPS Enhancement of Stat1-Dependent Transcription.
LPS causes STAT1 Ser727 phosphorylation and thereby increases IFN-γ-induced, Stat1-dependent transcriptional responses. From the results shown above, one would expect SB203580 to inhibit the augmenting effect of LPS but to be without a significant effect on transcriptional induction by IFN-γ alone. To test this assumption, macrophages stably transfected with a Stat-dependent luciferase gene were used (9). Treatment of the cells with IFN-γ resulted in luciferase gene expression, and this remained unaffected in the presence of SB203580 (Fig. 4). Consistent with our previous reports, a brief pretreatment with LPS doubled IFN-γ-dependent luciferase gene expression. SB203580 reduced the LPS/IFN-γ level of luciferase expression back to the level of IFN-γ alone. These results confirm our hypotheses and place the SB203580-sensitive kinase in a LPS-induced but not an IFN-γ-induced pathway.
Figure 4.
Effect of SB203580 on synergistic induction of STAT1-dependent transcription by IFN-γ and LPS. C11 cells, a clonal line derived from Bac1.2F5 macrophages and stably transfected with a Stat-dependent luciferase gene, were left untreated or were treated with either IFN-γ alone or a 30-min pretreatment with LPS followed by IFN-γ and LPS. Where indicated, SB203580 was added 30 min before subsequent treatment. After 2 h of IFN-γ or IFN-γ/LPS treatment, luciferase activity was determined in cellular extracts. The data represent mean values +/− standard deviations of triplicate measurements from three independent experiments.
JAK2 Activity Is Not Required for LPS-Mediated STAT1 Ser727 Kinase Activation.
STAT1 Ser727 phosphorylation in response to IFN-γ does not occur in JAK2-deficient cells (34). We asked whether JAK2 activity might be required for other signaling paths targeting the STAT1 Ser727 kinase. First, we tested JAK2 tyrosine phosphorylation, a necessary requirement for kinase activity, after LPS treatment of macrophages. No evidence for JAK2 tyrosine phosphorylation was found (Fig. 5). Next, we asked whether activated JAK2 necessarily causes the appearance of STAT1 Ser727 kinase activity. Treatment of macrophages with GM-CSF resulted in JAK2 tyrosine phosphorylation as expected but did not cause STAT1 Ser727 phosphorylation. Together, these results indicate that in the context of an LPS response JAK2 activity is not necessary for the phosphorylation of STAT1 Ser727 and that activated JAK2 alone is not sufficient for Stat1 serine phosphorylation.
SB203580 Resistance of p38 MAPK Causes SB203580 Resistance of STAT1 Ser727 Phosphorylation.
The findings reported so far provide pharmacological and correlative evidence for an involvement of p38MAPK in Stat1 phosphorylation. However, although SB203580 inhibition of STAT1 Ser727 phosphorylation is a strong indication for p38MAPK involvement, other SB203580 sensitive enzymes are known (26). Among these is the c-Raf kinase, which is activated in response to LPS (35).
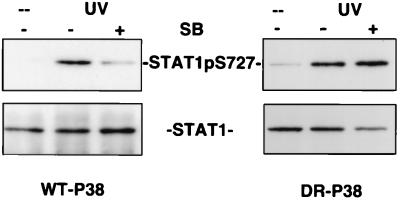
The crucial amino acid for SB203580 binding to p38 MAPK is Thr106, and mutation of Thr106 to an amino acid with a “bulkier” side chain causes a loss of SB203580 sensitivity (28, 36, 37). This finding was recently exploited to engineer 293 fibroblasts that conditionally express either a WT-p38 MAPK or a Thr106Met, His107Pro, Leu108Phe triple mutant of p38 MAPK (DR-p38 MAPK), which is insensitive to SB203580 (26). This cellular system allows one to unequivocally assign biological responses to a p38 MAPK-dependent pathway by testing whether DR-p38 MAPK reverses the sensitivity to SB203580.
293 cells with WT-p38 MAPK or DR-p38 MAPK transgenes were treated with Ponasterone A to induce expression of the enzymes. Both lines were subsequently irradiated with UV in the presence or absence of 1 μM SB203580. This treatment caused a strong phosphorylation of STAT1 Ser727 in both cell lines (Fig. 6). Although SB203580 inhibited the UV effect in cells expressing WT-p38 MAPK, no significant effect of the drug on Stat1 Ser727 phosphorylation was observed in cells expressing DR-p38 MAPK. Control experiments demonstrated that the phosphorylation of STAT1 Ser727 in WT-p38MAPK and DR-p38MAPK expressing cells was closely paralleled by the phosphorylation and activation of the p38MAPK substrate MAPKAP-K2 (data not shown). These results establish beyond doubt that the phosphorylation of STAT1 Ser727 occurs by a stress-induced signaling path involving p38 MAPK.
Figure 6.
SB203580 resistance of p38MAPK causes SB203580 resistance of STAT1 Ser727 phosphorylation. 293 human embryonic kidney fibroblasts harboring either WT-p38MAPK or SB203580-resistant p38MAPK (DR-p38) under control of the ecdysone receptor were treated for 24 h with Ponasterone A to induce expression of the kinase transgenes. Subsequently, the cells were stimulated by UV irradiation (200 J/m2), and STAT1 expression and Ser727 phosphorylation were determined with appropriate antisera. Where indicated, 1 μM SB203580 was added 1 h before UV irradiation.
Stress-Induced STAT1 Ser727 Phosphorylation Occurs in the Absence of MAPKAP-K2.
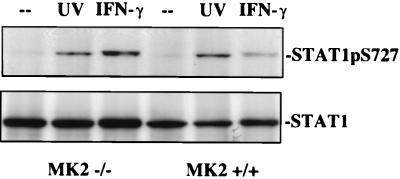
As shown above, the kinetics of p38 MAPK activation and STAT1 Ser727 phosphorylation can be separated by ≈10 min. For these reasons, we considered the possibility that one of the known p38 MAPK-dependent kinases might be a Stat1 kinase. Particularly interesting in this regard is MAPKAP-K2 because it mediates in vivo effects of LPS (27). Moreover, p38 MAPK phosphorylates MAPKAP-K2 and causes it to relocate to the cytoplasm, the compartment in which STAT1 Ser727 phosphorylation most likely occurs (38).
Immortalized fibroblasts derived from MAPKAP-K2-deficient mice (27) were irradiated with UV and, as a control, were treated with IFN-γ. Stat1 Ser727 phosphorylation in response to UV irradiation occurred with similar intensity in WT and MAPKAP-K2-deficient cells (Fig. 7). IFN-γ-mediated STAT1 Ser727 phosphorylation was somewhat enhanced; we do not as yet understand the underlying mechanism for this effect.
Figure 7.
UV irradiation-induced STAT1 Ser727 phosphorylation in cells with a disrupted mapkap-k2 gene. Immortalized fibroblasts derived from WT mice or from mapkap-k2-deficient mice were starved from serum for 16 h and then were stimulated for 25 min with IFN-γ or UV irradiation (40 J/m2). STAT1 Ser727 phosphorylation was determined in a Western blot with a phosphospecific antiserum.
In Vitro Phosphorylation of Recombinant Stat1 by p38MAPK but not by MAPKAP-K2.
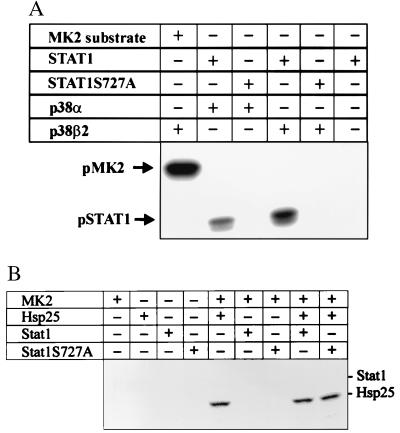
To investigate the possibility of a direct enzyme-substrate relationship between p38MAPK and Stat1 kinase, we performed in vitro assays with highly activated kinase. Fig. 8A demonstrates that a GST fusion protein of the STAT1 C terminus (AA711–750) was phosphorylated by the α and β2 isoforms of p38MAPK, albeit weaker than a MAPKAP-K2 control substrate (lane 1). Compared with a MAPKAP-K2 substrate, the calculated rate of incorporation of 32P into STAT1 was lower by a factor of >10 (data not shown). Phosphorylation occurred specifically at Ser727, as shown by an otherwise identical STAT1 substrate with the Ser727Ala mutation.
Figure 8.
The STAT1 C terminus can be phosphorylated by p38MAPK but not by MAPKAP-K2. (A) GST-STAT1 (AA711–750) and GST-STAT1Ser727Ala (AA711–750) were incubated with activated P38MAPKα and p38MAPKβ2 as described in Materials and Methods. In the first lane, MAPKAP-K2 (MK2) substrate is shown as a positive control. Phosphorylation was visualized by autoradiography. (B) Kinase assays were carried out by using constitutively active MAPKAP-K2 (MK2) and either GST-STAT1 C terminus, GST-STAT1 C terminus (S727A), or heat shock protein 25. Phosphorylation products were separated by SDS/PAGE, and the amount of phosphorylation was determined by phosphoimaging.
In marked contrast to p38MAPK, constitutively active MAPKAP-K2 did not phosphorylate Stat1 (Fig. 8B) while readily phosphorylating its in vivo substrate heat shock protein 25. This result is consistent with Fig. 7 showing that MAPKAP-K2 is not required for STAT1 Ser727 phosphorylation in cells.
Discussion
In this paper, we report that (i) the phosphorylation of STAT1 Ser727 and thus a potential enhancement of Stat1-mediated transcription occurs whenever cells encounter signals of stress and (ii) in accordance with pharmacological evidence as well as experiments with the SB203580-resistant p38 MAPK and with cells from mapkap-k2 knock-out mice, activation of the Stat1 kinase is relayed through a stress-activated signaling pathway involving p38 MAPK but not one of its substrate kinases, MAPKAP-K2. Consistently, p38MAPK phosphorylated STAT1 Ser727 in vitro whereas MAPKAP-K2 did not. These findings with regard to p38MAPK are consistent with a previous report that SB203580 blocks Stat1 serine phosphorylation in response to a combination of IL-12 and IL-2 (39). Importantly, IFN-γ signaling to the Stat1 S727 kinase does not include p38 MAPK, indicating different Stat1 kinases and/or different signaling paths in stress and IFN-γ responses. In further contrast to the IFN-γ response (34), JAK2 activity is not needed in the context of stress-activated STAT1 Ser727 phosphorylation.
An obvious question remaining to be answered is the nature of the Stat1 serine kinase in stress responses. Here we report that p38 MAPK can phosphorylate STAT1 Ser727 in vitro with high specificity. However, the low rate of phosphorylation compared with the MAPKAP-K2 substrate may indicate that p38MAPK in vitro mimics the action of an unknown, equally specific kinase with a much higher rate of turning over STAT1 substrates in vivo. Alternatively, our assay conditions may not faithfully reproduce the interaction between p38MAPK and STAT1 in vivo. Although the latter question can be addressed by offering different STAT1 portions for phosphorylation in vitro, additional studies will be needed to ascertain an in vivo role of p38MAPK. Assuming p38MAPK is not a direct STAT1 kinase, the question arises whether IFN-γ and stress signals target the same molecule via p38MAPK-dependent and p38MAPK-independent pathways. Clearly, some of the p38MAPK substrate kinases can also be targeted by ERKs that have previously been implicated in IFN-γ-signaling (40). However, for reasons discussed below, we argue that the known substrate kinases for both ERKs and p38MAPK do not qualify as STAT1 Ser727 kinases.
The more rapid kinetics of p38 MAPK activation compared with STAT1 Ser727 phosphorylation could potentially be explained by a need for p38 MAPK to relocate to the cytoplasm; an alternative explanation is that of intermediate signals between p38 MAPK and the Stat1 kinase.
Therefore, a sensible assumption was that of a role for p38 MAPK kinase substrates in Stat1 phosphorylation. Particularly, MAPKAP-K2 was an attractive candidate, because of its involvement in inflammatory responses (27) and its p38 MAPK-dependent shuttling between nucleus and cytoplasm (38). However, MAPKAP-K2 could be ruled out by using cells with a deleted mapkap-k2 gene and by in vitro kinase assays. The significance of these results extends to MAPKAP-K3, which has an identical substrate specificity (18), and probably also to other members belonging to the family of MAPK-activated protein kinases. Unlike the MAPK, these are not proline-directed kinases. In fact, mutation of an SV motif within a synthetic peptide substrate to SP strongly reduced the kinase activity of MAPKAP-K2/3 (18). The PMSP motif of Stat1 suggests phosphorylation by a proline-directed kinase. Therefore, in light of our results with MAPKAP-K2 and judging from known substrate phosphorylation sites, neither the MAPK-interacting kinases, mitogen- and stress-activated protein kinases, nor p38-regulated/activated kinases (MNK, MSK, or PRAK) are likely candidates for a Stat1 kinase. A further reason for considering MAPK-interacting or mitogen- and stress-activated protein kinases (MNK or MSK) as unlikely candidates is that these kinases are activated by ERK1/2 as well as p38 MAPK (20–22). Therefore, in a situation in which both pathways are active, as in the case of LPS treatment, STAT1 Ser727 phosphorylation would be expected to be at least partially sensitive to the MAPK/ERK kinase inhibitor PD98056. Our unpublished results document that this is clearly not the case. Moreover, growth factor stimulation of the ERK pathway, as in the case of GM-CSF or M-CSF (CSF-1), which also activate MAPK-interacting and mitogen- and stress-activated protein kinases (MNK and MSK), does not induce significant levels of STAT1 Ser727 phosphorylation (Fig. 5 and (9). In conclusion, although p38 MAPK is a more attractive candidate as a STAT1 kinase than its known substrate kinases, a correct assessment of its function in vivo will require extensive work in the future.
Acknowledgments
We thank Gustav Ammerer for valuable discussion and Manuela Baccarini for critical reading of the manuscript. Gifts of JAK antisera from Andrew Zimiecki (Laboratory for Clinical and Experimental Cancer Research, Bern, Switzerland), murecIFN-γ from Guenther Adolf (Boehringer Ingelheim), and SB203580 from Ken Murray (SmithKline Beecham) are gratefully acknowledged. This work was funded by the Austrian Research Foundation (Fonds zur Förderung der Wissenschaftlichen Forschung) through Grant P13640-GEN to T.D.
Abbreviations
- CSF
colony-stimulating factor
- ERK
extracellular signal-regulated kinase
- GM-CSF
granulocyte-macrophage CSF
- GST
glutathione S-transferase
- JAK
Janus kinase
- JNK
c-Jun NH2-terminal kinase
- LPS
lipopolysaccharide
- MAPK
mitogen-activated protein kinase
- MAPKAP-K
MAPK-activated protein kinase
- MEK
MAPK/ERK kinase
- STAT
signal transducer and activator of transcription
- WT
wild-type
Note Added in Proof
Shortly after the acceptance of our manuscript for publication, a paper was published by Goh et al. describing the involvement of p38MAPK in IFN-γ signaling, particularly in the phosphorylation of STAT1 at Ser727 (41). In very similar experiments to the ones performed in our study, Goh et al. report an ≈2-fold activation of p38MAPK by IFN-γ in HeLa cells 5 min after stimulation and a repression below basal activity after 10 min. Addition of 10 μM SB203580 inhibited STAT1 Ser727 phosphorylation to some extent Moreover, an overexpressed, dominant-negative allele of p38MAPK inhibited Stat1 serine phosphorylation in response to IFN-γ. We think the latter result could potentially be explained by a lack of specificity of the overexpressed dominant-negative kinase. p38MAPK activation 5 min after IFN-γ treatment and an inhibition of Stat1 Ser727 phosphorylation by SB203580, even if not very robust, are clearly not in agreement with our results. We have tested the effect of SB203580 on the IFN-γ-mediated STAT1 Ser727 phosphorylation in macrophages in many independent experiments with negative results. We have tested p38MAPK activation by IFN-γ in macrophages, 293 human embryonic kidney fibroblasts, and human HepG2 hepatoma cells, also with negative results. However, it is still possible that HeLa cells use an IFN-γ-responsive signalling path distinct from that used by the cell types used in our studies.
References
- 1.Darnell J E, Jr, Kerr I M, Stark G R. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 2.Leonard W J, O’Shea J J. Annu Rev Immunol. 1998;16:293–322. doi: 10.1146/annurev.immunol.16.1.293. [DOI] [PubMed] [Google Scholar]
- 3.Durbin J E, Hackenmiller R, Simon M C, Levy D E. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 4.Meraz M A, White J M, Sheehan K C, Bach E A, Rodig S J, Dighe A S, Kaplan D H, Riley J K, Greenlund A C, Campbell D, et al. Cell. 1996;84:431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- 5.Decker T, Kovarik P, Meinke A. J Interferon Cytokine Res. 1997;17:121–134. doi: 10.1089/jir.1997.17.121. [DOI] [PubMed] [Google Scholar]
- 6.Wen Z, Zhong Z, Darnell J E., Jr Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 7.Bromberg J F, Horvath C M, Wen Z, Schreiber R D, Darnell J E., Jr Proc Natl Acad Sci USA. 1996;93:7673–7678. doi: 10.1073/pnas.93.15.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horvath C M, Darnell J E., Jr J Virol. 1996;70:647–650. doi: 10.1128/jvi.70.1.647-650.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kovarik P, Stoiber D, Novy M, Decker T. EMBO J. 1998;17:3660–3668. doi: 10.1093/emboj/17.13.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seger R, Krebs E G. FASEB J. 1995;9:726–735. [PubMed] [Google Scholar]
- 11.Whitmarsh A J, Davis R J. J Mol Med. 1996;74:589–607. doi: 10.1007/s001090050063. [DOI] [PubMed] [Google Scholar]
- 12.Kyriakis J M, Avruch J. J Biol Chem. 1996;271:24313–24316. doi: 10.1074/jbc.271.40.24313. [DOI] [PubMed] [Google Scholar]
- 13.Sturgill T W, Ray L B, Erikson E, Maller J L. Nature (London) 1988;334:715–718. doi: 10.1038/334715a0. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Y, Bjorbaek C, Weremowicz S, Morton C C, Moller D E. Mol Cell Biol. 1995;15:4353–4363. doi: 10.1128/mcb.15.8.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rouse J, Cohen P, Trigon S, Morange M, Alonso-Llamazares A, Zamanillo D, Hunt T, Nebreda A R. Cell. 1994;78:1027–1037. doi: 10.1016/0092-8674(94)90277-1. [DOI] [PubMed] [Google Scholar]
- 16.Freshney N W, Rawlinson L, Guesdon F, Jones E, Cowley S, Hsuan J, Saklatvala J. Cell. 1994;78:1039–1049. doi: 10.1016/0092-8674(94)90278-x. [DOI] [PubMed] [Google Scholar]
- 17.McLaughlin M M, Kumar S, McDonnell P C, Van Horn S, Lee J C, Livi G P, Young P R. J Biol Chem. 1996;271:8488–8492. doi: 10.1074/jbc.271.14.8488. [DOI] [PubMed] [Google Scholar]
- 18.Clifton A D, Young P R, Cohen P. FEBS Lett. 1996;392:209–214. doi: 10.1016/0014-5793(96)00816-2. [DOI] [PubMed] [Google Scholar]
- 19.New L, Jiang Y, Zhao M, Liu K, Zhu W, Flood L J, Kato Y, Parry G C, Han J. EMBO J. 1998;17:3372–3384. doi: 10.1093/emboj/17.12.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waskiewicz A J, Flynn A, Proud C G, Cooper J A. EMBO J. 1997;16:1909–1920. doi: 10.1093/emboj/16.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fukunaga R, Hunter T. EMBO J. 1997;16:1921–1933. doi: 10.1093/emboj/16.8.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deak M, Clifton A D, Lucocq L M, Alessi D R. EMBO J. 1998;17:4426–4441. doi: 10.1093/emboj/17.15.4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alessi D R, Cuenda A, Cohen P, Dudley D T, Saltiel A R. J Biol Chem. 1995;270:27489–27494. doi: 10.1074/jbc.270.46.27489. [DOI] [PubMed] [Google Scholar]
- 24.Cuenda A, Rouse J, Doza Y N, Meier R, Cohen P, Gallagher T F, Young P R, Lee J C. FEBS Lett. 1995;364:229–233. doi: 10.1016/0014-5793(95)00357-f. [DOI] [PubMed] [Google Scholar]
- 25.Morgan C, Pollard J W, Stanley E R. J Cell Physiol. 1987;130:420–427. doi: 10.1002/jcp.1041300316. [DOI] [PubMed] [Google Scholar]
- 26.Eyers P A, van den, van den IJssel P, Quinlan R A, Goedert M, Cohen P. FEBS Lett. 1999;451:191–196. doi: 10.1016/s0014-5793(99)00552-9. [DOI] [PubMed] [Google Scholar]
- 27.Kotlyarov A, Neininger A, Schubert C, Eckert R, Birchmaier C, Volk H-D, Gaestel M. Nat Cell Biol. 1999;1:94–97. doi: 10.1038/10061. [DOI] [PubMed] [Google Scholar]
- 28.Eyers P A, Craxton M, Morrice N, Cohen P, Goedert M. Chem Biol. 1998;5:321–328. doi: 10.1016/s1074-5521(98)90170-3. [DOI] [PubMed] [Google Scholar]
- 29.Engel K, Schultz H, Martin F, Kotlyarov A, Plath K, Hahn M, Heinemann U, Gaestel M. J Biol Chem. 1995;270:27213–27221. doi: 10.1074/jbc.270.45.27213. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 31.Hambleton J, Weinstein S L, Lem L, DeFranco A L. Proc Natl Acad Sci USA. 1996;93:2774–2778. doi: 10.1073/pnas.93.7.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hale K K, Trollinger D, Rihanek M, Manthey C L. J Immunol. 1999;162:4246–4252. [PubMed] [Google Scholar]
- 33.Hazzalin C A, Cano E, Cuenda A, Barratt M J, Cohen P, Mahadevan L C. Curr Biol. 1996;6:1028–1031. doi: 10.1016/s0960-9822(02)00649-8. [DOI] [PubMed] [Google Scholar]
- 34.Zhu X J, Wen Z L, Xu L Z, Darnell J E. Mol Cell Biol. 1997;17:6618–6623. doi: 10.1128/mcb.17.11.6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reimann T, Buscher D, Hipskind R A, Krautwald S, Lohmann Matthes M L, Baccarini M. J Immunol. 1994;153:5740–5749. [PubMed] [Google Scholar]
- 36.Wilson K P, McCaffrey P G, Hsiao K, Pazhanisamy S, Galullo V, Bemis G W, Fitzgibbon M J, Caron P R, Murcko M A, Su M S. Chem Biol. 1997;4:423–431. doi: 10.1016/s1074-5521(97)90194-0. [DOI] [PubMed] [Google Scholar]
- 37.Tong L, Pav S, White D M, Rogers S, Crane K M, Cywin C L, Brown M L, Pargellis C A. Nat Struct Biol. 1997;4:311–316. doi: 10.1038/nsb0497-311. [DOI] [PubMed] [Google Scholar]
- 38.Engel K, Kotlyarov A, Gaestel M. EMBO J. 1998;17:3363–3371. doi: 10.1093/emboj/17.12.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gollob J A, Schnipper C P, Murphy E A, Ritz J, Frank D A. J Immunol. 1999;162:4472–4481. [PubMed] [Google Scholar]
- 40.Takaoka A, Tanaka N, Mitani Y, Miyazaki T, Fujii H, Sato M, Kovarik P, Decker T, Schlessinger J, Taniguchi T. EMBO J. 1999;18:2480–2488. doi: 10.1093/emboj/18.9.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goh K C, Haque S J, Williams B R G. EMBO J. 1999;18:5601–5608. doi: 10.1093/emboj/18.20.5601. [DOI] [PMC free article] [PubMed] [Google Scholar]