Abstract
ATRX is a member of the SNF2 family of helicase/ATPases that is thought to regulate gene expression via an effect on chromatin structure and/or function. Mutations in the hATRX gene cause severe syndromal mental retardation associated with α-thalassemia. Using indirect immunofluorescence and confocal microscopy we have shown that ATRX protein is associated with pericentromeric heterochromatin during interphase and mitosis. By coimmunofluorescence, ATRX localizes with a mouse homologue of the Drosophila heterochromatic protein HP1 in vivo, consistent with a previous two-hybrid screen identifying this interaction. From the analysis of a trap assay for nuclear proteins, we have shown that the localization of ATRX to heterochromatin is encoded by its N-terminal region, which contains a conserved plant homeodomain-like finger and a coiled-coil domain. In addition to its association with heterochromatin, at metaphase ATRX clearly binds to the short arms of human acrocentric chromosomes, where the arrays of ribosomal DNA are located. The unexpected association of a putative transcriptional regulator with highly repetitive DNA provides a potential explanation for the variability in phenotype of patients with identical mutations in the ATRX gene.
ATRX is one of a burgeoning group of proteins that are thought to influence gene expression via an effect on chromatin structure and/or function, but as for many of these proteins, the evidence for its interaction with chromatin in vivo is circumstantial and its mechanism of action is unknown.
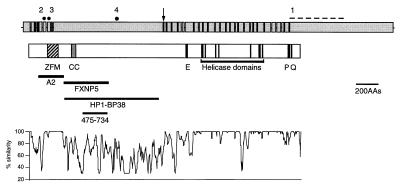
The X-encoded gene hATRX was identified originally because mutations give rise to a severe form of syndromal mental retardation characterized by the presence of α-thalassemia with urogenital abnormalities and facial dysmorphism (ATR-X syndrome; refs. 1 and 2). The predicted ATRX protein contains seven highly conserved, colinear domains (Fig. 1), which classify it as a member of the helicase/ATPase superfamily. Further analysis of the helicase domain and its flanking regions showed that it represents an entirely new subgroup of the SNF2 family that contains proteins with a wide range of cellular functions including DNA recombination and repair, mitotic recombination, and transcriptional regulation (3). It has been suggested that these proteins, including ATRX, may be incorporated variously into multicomponent complexes (e.g., the SWI/SNF complex) that utilize the energy from ATP hydrolysis to remodel chromatin and, thus, regulate protein/DNA interactions (4).
Figure 1.
Structure of the human ATRX gene and protein. The top of the figure shows exons 1–36 (excluding exon 7, which is alternatively spliced) of the human ATRX gene with the introns (not to scale). The position of mutations discussed in the text are marked by solid circles or a dashed line: 1, ped. 26; 2, ped. N29; 3, ped. 12; 4, ped.10. [The mutations are referred to by the pedigree (ped.) in which they were found. Affected members of the same family are distinguished by pedigree order.] The position at which insertion of the trap assay cassette occurred is indicated by an arrow. Below this, the main features of the ATRX protein structure are shown. ZFM, C2—C2 and PHD finger domains; CC, coiled coil; E, stretch of 21 glutamic acid residues; P, a region conserved in other SNF2 proteins; and Q, glutamine-rich region. The positions of the helicase domains are indicated. The locations of recombinant proteins (A2 and FXNP5) used to generate antibodies are shown. HP1-BP38 is the polypeptide that interacts with mHP1α in a two-hybrid screen (10). 475–734, Region that interacts with EZH2. At the bottom of the figure is a plot of percentage similarity between the predicted human and mouse ATRX proteins.
Further characterization of ATRX has shown that, in addition to the helicase domains, the N-terminal region of the protein, which includes a cysteine-rich domain related to previously described plant homeodomain (PHD) fingers (Cys4-His-Cys3), is highly conserved between mouse and man (Fig. 1). Approximately two-thirds of all natural mutations causing the ATRX syndrome lie in this region (5, 6). PHD-like fingers have been found in more than 40 proteins, many of which are thought to interact with chromatin to modify gene expression (7). In ATRX, this cysteine-rich region most closely resembles that found in the DNMT3 family of DNA methyltransferases (8). At present, the role of PHD-like domains is unknown, although some evidence suggests that they interact with the histone deacetylase HDAC1 (9).
Analysis of ATRX in a yeast two-hybrid screen has shown that it interacts with a murine homologue (mHP1α) of the Drosophila heterochromatic protein HP1 via an N-terminal region (delimited by HP1-BP38 in Fig. 1) that lies outside of the cysteine-rich domain (10). This region is poorly conserved between mouse and man but includes a coiled-coil motif that could mediate the interaction. Using a candidate protein approach, an interaction between a Polycomb group protein (EZH2) and residues 475–734 of ATRX (Fig. 1) also has been observed in a two-hybrid assay (11).
The consistent clinical features of ATRX syndrome (1) suggest that ATRX regulates expression of a discrete subset of genes, α-globin being one well defined example. Interestingly, hATRX mutations cause α-thalassemia by down-regulating α- but not β-globin expression. Although these coordinately expressed genes are regulated by a similar repertoire of transcription factors, they lie in very different chromatin environments in terms of their location (α, telomeric; β, interstitial), GC content, association with CpG islands, chromatin accessibility, and pattern of replication timing (12–14). If ATRX influences gene expression via an interaction with chromatin, this could explain its differential effect on these two gene clusters and point to other chromosomal regions that may be regulated similarly by ATRX.
To examine further the relationship between ATRX and chromatin in vivo, we have developed a panel of antibodies to determine its distribution in the nucleus during interphase and mitosis. Several lines of evidence show that a substantial proportion of ATRX associates with pericentromeric heterochromatin, and in human chromosomes at metaphase there is a striking and unexpected association between ATRX and the short arms of acrocentric chromosomes, where the GC-rich ribosomal DNA (rDNA) arrays are found.
Materials and Methods
Production of Antibodies, ELISA, and Immunocytochemistry.
BALB/c mice were immunized with the 26-kDa A2 fusion protein (residues 85–320), and mAbs were produced as described (15). Hybridomas were screened both by ELISA, by using the A2 protein, and by analyzing COS-1 cells transfected with construct pCDNA3/ZFM1, which expresses the ATRX cDNA fragment corresponding to the fusion protein. Positive clones were isolated after a further round of subcloning and screening. They were isotyped and found to be IgG1κ. Affinity-purified polyclonal antibodies (Affinity Biologicals, ON, Canada) were raised against the 88.5-kDa FXNP5 fusion protein (residues 320–730).
Analysis of Proteins from Nuclear Extracts by Western Blots.
Nuclei from Epstein–Barr virus (EBV) cell lines were prepared (16) and digested with DNase I (1 unit/μl, final concentration; Pharmacia) for 1 hr at room temperature. For optimal extraction (see Results), proteins were solubilized in RIPA buffer (50 mM Tris⋅HCl, pH 7.4/150 mM NaCl/0.2% SDS/1% Nonidet P-40/1% deoxycholic acid/1 mM PMSF). Protein extracts were run in a 4.5% SDS-polyacrylamide gel, immunoblotted, then incubated with dilutions of monoclonal (1:10) or polyclonal (1:500) anti-ATRX antibodies or TAF II p250 (1:400; Santa Cruz Biotechnology). Signals were detected with horseradish peroxidase-conjugated goat anti-mouse (1:1,000; Dako) or donkey anti-sheep (1:2,000; Sigma) antibodies by using enhanced chemiluminescence (ECL; Amersham).
Indirect Immunofluorescence.
Interphase analysis was performed by either culturing adherent cells on glass coverslips or by incubating suspension cells on coverslips treated with poly-l-lysine. Cells were fixed in 3% (vol/vol) paraformaldehyde/PBS (15 min) and permeablized in 0.1% (vol/vol) Triton X-100/PBS (10 min). Cells were washed in PBS and then incubated in 10% (vol/vol) FCS/PBS-T block (PBS and 0.1% vol/vol Tween 20) for 30 min. Primary antibodies anti-ATRX, 1:5; CREST serum, 1:1,000; rabbit anti-β-galactosidase (β-gal) (5′ → 3′), 1:100; and MAC353 and MAC385, which recognize M31 and M32, respectively (17, 18), were diluted in block buffer and incubated on cells at room temperature (45 min). After several washes in PBS-T, cells were incubated with block buffer (30 min). Secondary antibodies [Texas Red-conjugated anti-mouse IgG, FITC-conjugated anti-human IgG, anti-mouse IgG, or anti-rat IgG, 1:200 dilutions (Vector Laboratories); Texas Red-conjugated anti-rabbit IgG, 1:50 dilution (19)] were incubated on cells (45 min). After several washes in PBS-T, cells were fixed (10 min) in 10% (vol/vol) formaldehyde/PBS and then in 3:1 methanol/acetic acid (15 min). Coverslips were mounted in Vectashield (Vector Laboratories) containing 0.5–1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI; Sigma).
Metaphase analysis was performed by using a subconfluent flask of cells that subsequently were incubated with 0.1 μg/ml colcemid (30 min). Mitotic cells were washed in PBS and then incubated in 75 mM KCl at 7 × 104/ml (20 min). Cytospins were prepared by centrifuging cells (300 μl) onto Superfrost microscope slides (Merck) by using a Cytospin 3 (Shandon, Pittsburgh) for 10 min at 2,000 rpm. Slides were incubated immediately for 10 min in KCM buffer [10 mM Tris⋅HCl, pH 7.7/120 mM KCl/20 mM NaCl/0.1% (vol/vol) Triton X-100] and then washed twice in PBS. Unfixed chromosome spreads were examined by immunofluorescence, following the protocol for cells grown on coverslips.
Confocal Microscopy.
Images were obtained (20) on a Bio-Rad MRC-1000/1024 hybrid “confocal” microscope (running under comos 7.0a software). Z-series of optical sections at 0.5-μm nominal steps were obtained sequentially so that no “bleedthrough” was detected between channels. Raw images were “contrast-stretched” and presented or analyzed without further processing (i.e., without background subtraction).
For analysis of colocalization between ATRX and CREST foci, images were incorporated into confocal assistant 4.02, build 101 (T. C. Brelje, University of Minnesota, Minneapolis), “contrast-stretched,” pseudocolored red and green, and merged into an AVI file. Chromatic aberration in the imaging system was measured by using 0.1-μm TetraSpeck microspheres (fluorescent in green and red; Molecular Probes) and corrected, when necessary, by merging the appropriate optical sections. Colocalization of green and red foci was assessed as follows: (i) determine the total number of green (CREST) and red (ATRX) foci by scrolling through the z series; (ii) establish whether green and red signals colocalize (yellow/orange) or lie adjacent in the XY plane or in sequential Z sections; and (iii) express the number of colocalized and adjacent signals as a proportion of the total number of centromeric signals. Sufficient cells were analyzed to ensure that progressive means given by the last 50% of cells analyzed always lay within 10% of the final mean.
Isolation of the F9/18D6 Gene-Trap Cell Line.
A gene-trap screen for nuclear proteins was performed (19) with modification. Male F9 embryonic carcinoma cells, maintained in α-MEM and 10% FCS, were transfected with pGT1, 2, and 3, and neomycin-resistant clones were selected with 400 μg/ml G418. Resistant clones were stained with 5-bromo-4-chloro-3-indolyl β-d-galactoside to identify those in which β-gal activity was confined to the nucleus. One of these (F9/18D6) exhibited intense foci of nuclear β-gal staining. The integration site of the gene trap in F9/18D6 was ascertained by fluorescence in situ hybridization to metaphase chromosomes by using pGT1 as a probe. 5′ Rapid amplification of cDNA end products (19) from the integrant were directly sequenced.
Results
Development of Anti-ATRX Antibodies and Biochemical Fractionation of ATRX Protein.
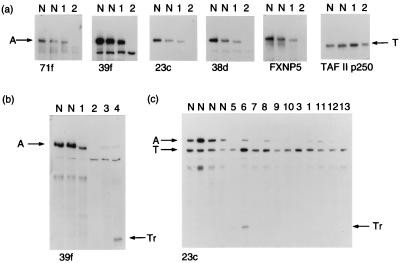
Using recombinant proteins FXNP5 and A2 (Fig. 1), we developed sheep polyclonal and mouse mAbs against N-terminal segments of the predicted human ATRX protein. To validate the antibodies, we analyzed Western blots of optimized cellular extracts (see below) from EBV-transformed B lymphocytes obtained from normal individuals and patients with previously defined mutations in the ATRX gene (Fig. 2). In normal individuals (lane N, Fig. 2a), all of the antibodies detected the same ≈280-kDa fragment, consistent with the predicted size of the ATRX protein (3, 6). A slightly smaller band (lane 1, Fig. 2 a and b) was detected in extracts from a patient (ped. 26) with a 1,973-bp deletion from the 3′ end of the ATRX gene (2), predicted to produce a truncated protein of 272 kDa. No signal (lane 2, Fig. 2 a and b) was detected in extracts from a patient (ped. N29, unpublished case) with a 2-bp deletion, which leads to a frameshift within the region of the protein (A2) from which the mAbs were generated (Fig. 1). However, by using a polyclonal antibody (FXNP5) that recognizes the protein beyond this region, a faint band was visible, presumably detecting a previously described splice form that removes exon 6 (with the 2-bp deletion) (3). A reduced signal (lane 3; Fig. 2b) was seen in another patient with a splice-site mutation, removing 21 aa from the region from which antibodies were made (3). Using the anti-ATRX antibody 39f with extracts from two patients (ped. 10 and ped. 58, in ref. 21) with identical premature in-phase termination codons lying beyond the region from which antibodies were made (Fig. 1), truncated proteins of the predicted size were seen (lane 4, Fig. 2b, and lane 6, Fig. 2c). Finally, comparisons with a similar-sized nuclear protein (TAF II p250) showed significantly reduced levels of ATRX protein in all ATRX patients tested (Fig. 2c).
Figure 2.
Western blot analyses by using a panel of anti-ATRX antibodies. (a) Nuclear extracts from EBV-transformed lymphocytes probed with polyclonal (FXNP5) and monoclonal (71f, 39f, 23c, and 38d) antibodies. An antibody against TAF II p250 was used as a control. Lanes: N, normal individual; 1, ped. 26; 2, ped. N29. (b) Nuclear extracts from EBV-transformed lymphocytes probed with the anti-ATRX antibody 39f. Annotations are as above; 3, ped. 12; 4, ped. 10 (II-3). (c) Nuclear extracts probed with anti-ATRX antibody 23c and anti-TAF II p250. Annotations are as before; 5, ped. 10 (II-2); 6, ped. 58; 7, ped. 59 (III-10); 8, ped. 59 (III-1); 9, ped. N1 (III-5); 10, ped. N1 (III-4); 11, ped. 38; 12, ped. 17; 13, ped. 27. Arrows indicate: A, full-length ATRX protein; T, TAF II p250; Tr, truncated ATRX protein.
Immunocytochemical analysis (not shown) and indirect immunolocalization (see below) demonstrated that ATRX is a nuclear protein. This was confirmed by analyzing nuclear and cytoplasmic fractions from a variety of cells; all ATRX protein was found in the nuclear fraction. ATRX was initially extracted from unsynchronized HeLa cells by using a high-salt buffer (400 mM NaCl) in the absence of detergents (22). However, analysis of the pellet from these extracts indicated that less than 50% of ATRX protein had been solubilized. To analyze ATRX partitioning more rigorously, we performed sequential, stepwise extractions of the nuclear pellet after detergent treatment with nucleases, with 250 mM ammonium sulfate, and, finally, with 2 M NaCl. These extractions solubilize most nuclear proteins, and the insoluble high-salt pellet contains proteins associated with components of the nuclear matrix (23). This demonstrated that most of the ATRX protein was extracted from the nuclear pellet after treatment with nucleases and <10% was found in the fraction containing the nuclear lamins. All subsequent extractions thus were performed in RIPA buffer after nuclease digestion. These observations suggest that a substantial proportion (>50%) of ATRX is tightly associated with DNA and/or chromatin in the nucleus, consistent with its localization to pericentromeric heterochromatin (see below).
ATRX Is a Nuclear Protein Associated with Pericentromeric Heterochromatin and Localized at the Short Arms of Acrocentric Chromosomes.
The antibodies developed here detect both the human and mouse ATRX proteins by indirect immunofluorescence, and, therefore, we examined its localization in unsynchronized human and mouse cells during interphase and in mitosis by using the mAbs 39f and 23c.
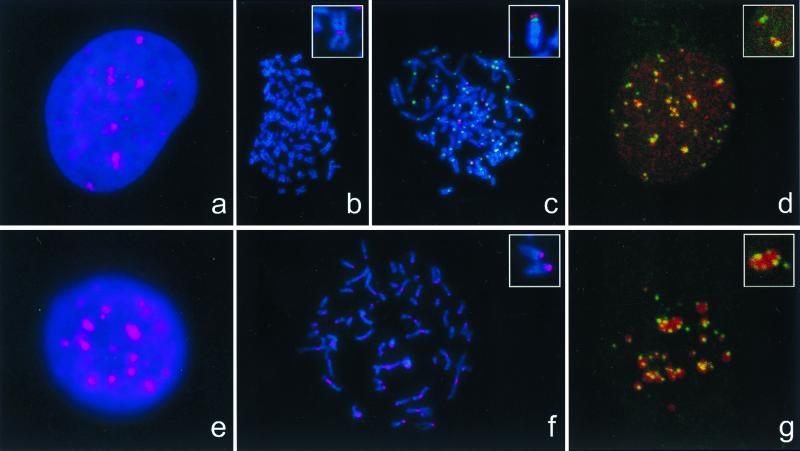
In mouse chromosomes (L929, MEL, P388D) ATRX protein is concentrated at, or close to, the centromeres (Fig. 3f and Inset), where it is associated with signals detected by CREST serum, which recognizes the centromeric protein CENP-C (24) (data not shown). In mouse cells at interphase, the anti-ATRX antibody produced a nonuniform pattern of staining in the nucleus, with between 20 and 35 discrete areas of dense staining against a more diffuse distribution of ATRX protein (Fig. 3e). The densely staining areas correspond to DAPI bright regions of the nucleus, which are known to represent pericentromeric heterochromatin and are associated with staining by the CREST serum (Fig. 3g). In each nucleus ATRX densely staining regions colocalized with DAPI bright regions. The association between ATRX and pericentromeric heterochromatin was quantitated by counting the coincidence of signals from anti-ATRX and CREST serum on serial sections obtained by confocal microscopy (Fig. 3g and Inset). In this assay, 93–95% of centromeric signals were closely associated with ATRX. Occasionally in interphase nuclei, two or three distinct ATRX foci did not colocalize with DAPI bright heterochromatin.
Figure 3.
Localization of ATRX protein in interphase and metaphase. HeLa (a–d) or L929 (e–g) cells were grown on coverslips (a, d, e, and g) or cytospun onto glass slides (b, c, and f), and ATRX (pseudocolored red) and CREST (pseudocolored green) were indirectly immunolabeled. Nucleic acids were counterstained with DAPI (blue). Images of ATRX, centromere staining, and nucleic acids were obtained in a charge-coupled device camera (a–c, e, and f) or on confocal images, without DAPI (d and g), and merged. d and g are projections of z series of optical sections taken across whole nuclei. Insets in d and g show the detail observed in a single optical section in which CREST-stained, centromeric heterochromatin is found closely associated with densely staining regions of ATRX. Identical results were obtained by using either 39f or 23c anti-ATRX antibodies.
A variety of human cells (HeLa, G-CCM, HT29, HepG2, fibroblasts, EBV-transformed B lymphocytes) were analyzed by indirect immunofluorescence. Consistent with Western blot analysis (see above), ATRX protein was readily detected in the nuclei of EBV-transformed B lymphocytes from normal individuals but not in a cell line from an ATR-X patient (ped. N29, data not shown) expressing ATRX protein lacking the epitopes detected by these antibodies. During interphase, the pattern of ATRX staining in human cells (Fig. 3a) was similar to that seen in the murine cells. However, its relationship to pericentromeric heterochromatin was less clear. In all of the human cells studied, DAPI bright, heterochromatic regions were less prominent than in mouse cells. Nevertheless, quantitation by confocal microscopy showed that in unsynchronized HeLa cells, between 39–95% of centromeric signals detected by using CREST serum were closely associated with ATRX (Fig. 3d). The lack of distinct ATRX foci in some CREST-positive cells may reflect differences in the distribution of ATRX during the cell cycle. ATRX is localized at centromeres during mitosis, for example, at metaphase (Fig. 3b and Inset) and telophase (Fig. 4), although this localization appeared less consistent and more sensitive to the method of preparation than for the mouse chromosomes. However, a striking observation in metaphase preparations was that ATRX antibodies consistently localized to the short arms of acrocentric chromosomes (Fig. 3c and Inset). Dual-staining studies with the CREST antibody demonstrated that ATRX appears to localize adjacent to the centromere on the stalks of the short arms (Fig. 3c Inset and data not shown), where the arrays of rDNA are found. Weak signals also were seen at several chromosome arms, but their significance is not clear.
Figure 4.
The distribution of ATRX in mitosis. A Hela cell in telophase stained with DAPI (a), CREST serum (b), anti-ATRX antibody (c), and CREST and anti-ATRX antibody (d).
The N-Terminal Region of the ATRX Protein, Containing a Conserved PHD Domain, Is Sufficient for Localization to Heterochromatin.
Further evidence for the association between ATRX protein and pericentromeric heterochromatin came from an entirely independent line of investigation. Using a gene-trap screen designed to detect nuclear- and, in particular, chromatin-associated proteins (19), we (H.S. and W.B.) identified one cell line (F9/18D6, derived from a male F9 embryonic carcinoma) that showed intense staining foci in the nucleus by using the 5-bromo-4-chloro-3-indolyl β-d-galactoside readout for this assay (data not shown). Western analysis of nuclear extracts from F9/18D6 demonstrated a truncated ATRX protein of ≈260 kDa (data not shown).
To identify the “trapped” nuclear protein, approximately 170 bp of sequence from the exon upstream of the integration site in F9/18D6 was obtained by 5′ rapid amplification of cDNA ends. A blast search revealed that it closely matched mouse and human ATRX mRNAs. Confirmation that the gene trapped in F9/18D6 cells was ATRX came from mapping of the integration site by fluorescence in situ hybridization, with both the gene-trap vector and a mouse X chromosome paint (Cambio, Cambridge, U.K.) as probes. Using the gene-trap vector as a probe, a signal was seen within X chromosome material on an abnormal (translocation) chromosome of the F9 karyotype (data not shown). Thus, the male F9/18D6 cells retain an intact copy of ATRX on the normal X chromosome and have a targeted copy of ATRX on the translocated chromosome. In F9/18D6, the gene-trap vector has integrated within intron 11 (after nucleotide 3893) of the ATRX gene. Therefore, the fusion protein contains the PHD-like domain and the putative HP1-interacting domain of ATRX, but lacks the glutamic acid and glutamine-rich regions and the helicase domains (Fig. 1).
To test whether the subnuclear distribution of the β-gal fusion protein reflects that of endogenous ATRX, coimmunofluorescence experiments with anti-β-gal and anti-ATRX antibodies were performed in F9/18D6 cells. Both fusion protein and endogenous ATRX colocalized with the DAPI bright foci in the nucleus (Fig. 5c), although there were also a few spots of ATRX signal that were not coincident with either concentrations of β-gal or DAPI signal. The significance of this is unclear, though there may be subtle differences between the distribution of the fusion protein and ATRX if domains in the C-terminal end of the protein direct interactions with other nuclear structures. On F9/18D6 metaphase chromosomes, both the fusion protein and endogenous ATRX were completely coincident with the pericentromeric heterochromatin (Fig. 6 Lower).
Figure 5.
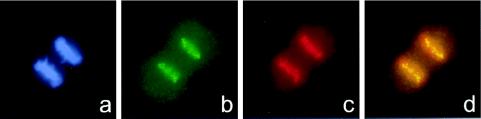
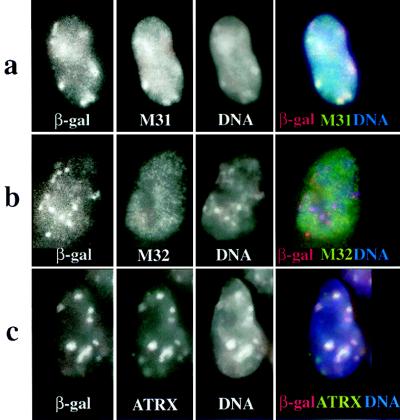
Localization of β-gal/18D6 fusion, M31, M32, and ATRX proteins in mouse (F9/18D6) nuclei. Immunostaining of F9/18D6 nuclei with antibody against β-gal (images on the far left and red in merged images), together with antibodies against M31 (a), M32 (b), and ATRX (c) (second from left and green in merged images), is shown. DNA was counterstained with DAPI (second from right and blue in merged images). Colocalized signals appear as white.
Figure 6.
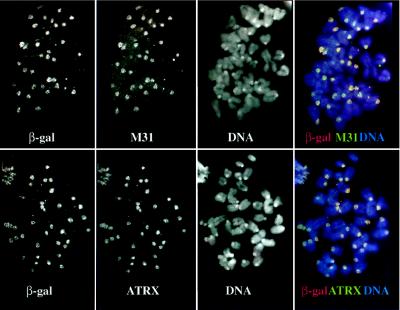
Localization of β-gal/18D6 fusion, M31, and ATRX proteins on mouse (F9/18D6) metaphase chromosomes. Immunostaining of F9/18D6 metaphase chromosomes with antibody against β-gal (images on the far left and red in merged images), together with antibodies against M31 (Upper, second from left) and ATRX (Lower, second from left and green in merged images), is shown. DNA was counterstained with DAPI (second from right and blue in merged images). Note the intense DAPI staining at the pericentromeric heterochromatin of the mouse chromosomes. Colocalized signals appear as white.
To examine this further, F9/18D6 cells were stained both with anti-β-gal antibody and with antibodies raised against either M31 or M32, murine homologues of the Drosophila heterochromatin protein 1 (HP1). M31 is known to localize at sites of constitutive heterochromatin within the nuclei of mouse cells (17), and Fig. 5a shows that in F9/18D6 cells, the foci of β-gal fusion protein colocalize with the concentrations of M31. By contrast, M32 is localized to euchromatin in interphase nuclei (18), and there was no correspondence between the β-gal and M32 immunofluorescence signals in F9/18D6 nuclei (Fig. 5b). In mitotic cells (data not shown) and in cytospin preparations of unfixed F9/18D6 metaphase chromosomes, β-gal immunostaining was concentrated at the pericentromeric heterochromatin together with M31 (Fig. 6 Upper). These data suggest that the first 10 exons of ATRX contain sufficient information for protein localization to constitutive heterochromatin.
Discussion
Using indirect immunofluorescence and by the analysis of a nuclear protein-trap assay, we have shown that a substantial proportion of ATRX protein is associated with pericentromeric heterochromatin both in mitosis and during interphase. This was unexpected because previous observations suggested that ATRX is involved in remodeling chromatin within euchromatic regions, such as that containing the α-globin locus.
Similar patterns of pericentromeric localization now have been reported for a number of proteins that play multiple roles in modifying chromosomal structure and function. In Saccharomyces cerevisiae, the centromere promoter factor (called CP1 or CPF1) may modify chromatin structure at many promoters, where it is necessary for optimal gene expression, but it is also found at centromeres where it is thought to facilitate the binding of kinetechore proteins (25). CP1 mutants show abnormalities in centromere function and gene expression (26). In Drosophila, GAGA factor, a product of the Trithorax-like locus, binds hundreds of euchromatic loci, where, in association with the nucleosomal remodeling factor (NURF), it facilitates transcriptional activation (27). GAGA factor is also associated with pericentromeric heterochromatin in diploid cells, where it plays a role in chromosome condensation and segregation (28). In mammals, it has been shown recently that Ikaros, a transcriptional regulator required for normal lymphocyte development, may act as repressor when localized with Mi-2 and HDAC to heterochromatin (29, 30) and as an activator with SWI/SNF in euchromatic regions (29). Similarly, ATRX may have multiple (positive and negative) effects on chromatin-mediated processes, although at present there is no evidence that patients with inactivating mutations of the ATRX gene have any disturbance in chromosome division or segregation.
The conditions required to solubilize ATRX from cellular extracts suggest that it is tightly associated with DNA and/or chromatin: it has been suggested previously that ATRX may bind DNA (6). However, the association of ATRX and mHP1α in a two-hybrid assay (10) and colocalization of ATRX with M31 antibodies (against HP1β) described here offer an alternative, but not exclusive, explanation for its association with DNA. Emerging evidence suggests that HP1, a known chromatin-binding protein, is a structural adapter whose role is to assemble macromolecular complexes in chromatin. HP1-like proteins interact with many nuclear proteins, including the lamin B receptor (an integral protein of the nuclear envelope) components of the SWI/SNF complex, the nuclear receptor cofactor TIF1-α, the product of Su(var)3–7 (a heterochromatic protein of unknown function), Orc1 and Orc2 (two members of the origin recognition complex), SP100 (a component of PML bodies, nuclear structures that are disrupted in promyelocytic leukemia), and transcriptional regulators such as Ikaros (30, 31). Therefore, an interaction between ATRX and HP1 may be sufficient to colocalize ATRX within heterochromatin via a protein/protein interaction. The trap assay suggests that such an interaction could occur via the N-terminal region of ATRX, and this is substantiated by the previously described two-hybrid assay (10).
The localization of ATRX to the short arms of acrocentric chromosomes suggests that, in addition to binding heterochromatic satellites, this protein also binds at, or close to, rDNA repeats. Preliminary data also suggest that a proportion of ATRX/heterochromatin may concentrate around, but not within, the nucleolus in some cells during interphase (T.L.M., unpublished data). At present, it is not clear whether ATRX plays a role in regulating chromatin at pericentromeric heterochromatin or ribosomal arrays or binds these repetitive elements nonspecifically as a “passenger” protein. It is of interest that, like the α-globin cluster, where ATRX clearly has an effect on gene expression, the rDNA repeats are GC-rich and contain a high density of unmethylated CpG dinucleotides (32).
At present, we do not know whether ATRX plays a direct, functional role at heterochromatin and rDNA repeats. However, by binding ATRX, the repeats could provide buffers that regulate the amount of the protein available to its biologically important sites. This may be analogous to previous observations in Drosophila in which deletion or amplification of rDNA or satellite DNA repeats enhance or suppress position effect variegation by varying the amount of the modifier proteins that are bound (33, 34). Polymorphic variation in the numbers of such repeats would provide a plausible explanation for the clinical variability seen in patients with identical mutations of the ATRX gene producing a structurally normal but reduced amount of ATRX (3, 35).
Acknowledgments
We are grateful to Dr. Prim Singh (Roslin Institute) for the M31 and M32 antibodies. This work was supported by the Medical Research Council, the Wellcome Trust (R.J.G. and A.P.), Action Research (D.M.O.), and the Royal Society (A.P.). H.S. is supported by a long-term European Molecular Biology Organization Fellowship. D.J.P. is a Scholar of the Medical Research Council of Canada.
Abbreviations
- EBV
Epstein–Barr virus
- DAPI
4′,6-diamidino-2-phenylindole
- β-gal
β-galactosidase
- rDNA
ribosomal DNA
- PHD
plant homeodomain
References
- 1.Gibbons R J, Brueton L, Buckle V J, Burn J, Clayton-Smith J, Davison B C C, Gardner R J M, Homfray T, Kearney L, Kingston H M, et al. Am J Med Genet. 1995;55:288–299. doi: 10.1002/ajmg.1320550309. [DOI] [PubMed] [Google Scholar]
- 2.Gibbons R J, Picketts D J, Villard L, Higgs D R. Cell. 1995;80:837–845. doi: 10.1016/0092-8674(95)90287-2. [DOI] [PubMed] [Google Scholar]
- 3.Picketts D J, Higgs D R, Bachoo S, Blake D J, Quarrell O W J, Gibbons R J. Hum Mol Genet. 1996;5:1899–1907. doi: 10.1093/hmg/5.12.1899. [DOI] [PubMed] [Google Scholar]
- 4.Kingston R E, Bunker C A, Imbalzano A N. Genes Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- 5.Gibbons R J, Bachoo S, Picketts D J, Aftimos S, Asenbauer B, Bergoffen J, Berry S A, Dahl N, Fryer A, Keppler K, et al. Nat Genet. 1997;17:146–148. doi: 10.1038/ng1097-146. [DOI] [PubMed] [Google Scholar]
- 6.Villard L, Lossi A-M, Cardoso C, Proud V, Chiaroni P, Colleaux L, Schwartz C, Fontés M. Genomics. 1997;43:149–155. doi: 10.1006/geno.1997.4793. [DOI] [PubMed] [Google Scholar]
- 7.Aasland R, Gibson T J, Stewart A F. Trends Biochem Sci. 1995;20:56–59. doi: 10.1016/s0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- 8.Xie S, Wang Z, Okano M, Nogami M, Li Y, He W-W, Okumura K, Lli E. Gene. 1999;236:87–95. doi: 10.1016/s0378-1119(99)00252-8. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, LeRoy G, Seelig H-P, Lane W S, Reinberg D. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 10.Le Douarin B, Nielsen A L, Garnier J-M, Ichinose H, Jeanmougin F, Losson R, Chambon P. EMBO J. 1996;15:6701–6715. [PMC free article] [PubMed] [Google Scholar]
- 11.Cardoso C, Timsit S, Villard L, Khrestchatisky M, Fontés M, Colleaux L. Hum Mol Genet. 1998;7:679–684. doi: 10.1093/hmg/7.4.679. [DOI] [PubMed] [Google Scholar]
- 12.Higgs D R, Sharpe J A, Wood W G. Semin Hematol. 1998;35:93–104. [PubMed] [Google Scholar]
- 13.Vyas P, Vickers M A, Simmons D L, Ayyub H, Craddock C F, Higgs D R. Cell. 1992;69:781–793. doi: 10.1016/0092-8674(92)90290-s. [DOI] [PubMed] [Google Scholar]
- 14.Smith Z E, Higgs D R. Hum Mol Genet. 1999;8:1373–1386. doi: 10.1093/hmg/8.8.1373. [DOI] [PubMed] [Google Scholar]
- 15.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 16.Higgs D R, Wood W G, Jarman A P, Sharpe J, Lida J, Pretorius I-M, Ayyub H. Genes Dev. 1990;4:1588–1601. doi: 10.1101/gad.4.9.1588. [DOI] [PubMed] [Google Scholar]
- 17.Wreggett K A, Hill F, James P S, Hutchings A, Butcher G W, Singh P B. Cytogenet Cell Genet. 1994;66:99–103. doi: 10.1159/000133676. [DOI] [PubMed] [Google Scholar]
- 18.Horsely D, Hutchings A, Butcher G W, Singh P B. Cytogenet Cell Genet. 1996;73:308–311. doi: 10.1159/000134363. [DOI] [PubMed] [Google Scholar]
- 19.Tate P, Lee M, Tweedie S, Skarnes W C, Bickmore W A. J Cell Sci. 1998;111:2575–2585. doi: 10.1242/jcs.111.17.2575. [DOI] [PubMed] [Google Scholar]
- 20.Pombo A, Cuello P, Schul W, Yoon J-B, Roeder R G, Cook P R, Murphy S. EMBO J. 1998;17:1768–1778. doi: 10.1093/emboj/17.6.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gibbons R J, Higgs D R. In: Disorders of Hemoglobin. Forget B G, Higgs D R, Nagel R L, Steinberg M H, editors. Cambridge, U.K.: Cambridge Univ. Press; 1999. , in press. [Google Scholar]
- 22.Wu C. Nature (London) 1984;309:229–234. doi: 10.1038/309229a0. [DOI] [PubMed] [Google Scholar]
- 23.Capco D G, Wan K M, Penman S. Cell. 1982;29:847–856. doi: 10.1016/0092-8674(82)90446-9. [DOI] [PubMed] [Google Scholar]
- 24.Earnshaw W C, Ratrie H, Stetten G. Chromosoma. 1989;98:1–12. doi: 10.1007/BF00293329. [DOI] [PubMed] [Google Scholar]
- 25.Mellor J, Jiang W, Funk M, Rathjen J, Barnes C A, Hegemann J H, Philippsen P. EMBO J. 1990;9:4017–4026. doi: 10.1002/j.1460-2075.1990.tb07623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masison D C, O’Connell K F, Baker R E. Nucleic Acids Res. 1993;21:4133–4141. doi: 10.1093/nar/21.17.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilkins R C, Lis J T. Nucleic Acids Res. 1997;15:3963–3968. doi: 10.1093/nar/25.20.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Platero J S, Csink A K, Quintanilla A, Henikoff S. J Cell Biol. 1998;140:1297–1306. doi: 10.1083/jcb.140.6.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, et al. Immunity. 1999;10:345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- 30.Brown K E, Guest S S, Smale S T, Hahm K, Merkenschlager M, Fisher A G. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- 31.Lamond A I, Earnshaw W C. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- 32.Brock G J R, Bird A. Hum Mol Genet. 1997;6:451–456. doi: 10.1093/hmg/6.3.451. [DOI] [PubMed] [Google Scholar]
- 33.Hilliker A J, Appels R. Chromosoma. 1982;86:469–490. doi: 10.1007/BF00330122. [DOI] [PubMed] [Google Scholar]
- 34.Spofford J B, DeSalle R. Genet Res. 1991;57:245–255. doi: 10.1017/s0016672300029396. [DOI] [PubMed] [Google Scholar]
- 35.Villard L, Toutain A, Lossi A-M, Gecz J, Houdayer C, Moraine C, Fontés M. Am J Hum Genet. 1996;58:499–505. [PMC free article] [PubMed] [Google Scholar]