Abstract
Duchenne muscular dystrophy (DMD) is an inherited muscle-wasting disease caused by the absence of a muscle cytoskeletal protein, dystrophin. We have previously shown that utrophin, the autosomal homologue of dystrophin, is able to compensate for the absence of dystrophin in a mouse model of DMD; we have therefore undertaken a detailed study of the transcriptional regulation of utrophin to identify means of effecting its up-regulation in DMD muscle. We have previously isolated a promoter element lying within the CpG island at the 5′ end of the gene and have shown it to be synaptically regulated in vivo. In this paper, we show that there is an alternative promoter lying within the large second intron of the utrophin gene, 50 kb 3′ to exon 2. The promoter is highly regulated and drives transcription of a widely expressed unique first exon that splices into a common full-length mRNA at exon 3. The two utrophin promoters are independently regulated, and we predict that they respond to discrete sets of cellular signals. These findings significantly contribute to understanding the molecular physiology of utrophin expression and are important because the promoter reported here provides an alternative target for transcriptional activation of utrophin in DMD muscle. This promoter does not contain synaptic regulatory elements and might, therefore, be a more suitable target for pharmacological manipulation than the previously described promoter.
Duchenne muscular dystrophy (DMD) is a fatal muscle-wasting disease caused by mutations that abolish the expression of dystrophin (1). In normal skeletal muscle, dystrophin is localized to the cytoplasmic surface of the sarcolemma, where it is involved in a series of molecular interactions forming a mechanical link between the myofiber cytoskeleton and extracellular matrix (2). The absence of dystrophin in DMD results in loss of the dystrophin-associated protein complex from the sarcolemma (3) and muscle degeneration.
Utrophin is the autosomal homologue of dystrophin (4); the two proteins have similar functional domains and protein binding partners (5, 6). Unlike dystrophin, utrophin is ubiquitously expressed (7). In mature muscle fibers, utrophin is localized to the cytoplasmic face of the neuromuscular junctions (NMJ) and myotendinous junctions; it is found at the sarcolemma in developing and regenerating muscle (8, 9). The structural similarity between the two proteins led us to speculate that utrophin may be able to replace dystrophin in DMD (10). We subsequently showed that overexpression of utrophin in the skeletal muscle of the dystrophin-deficient mdx mouse completely prevented muscle pathology (11–13).
One strategy for therapeutic intervention in DMD involves augmenting transcription from the endogenous utrophin gene. This approach has the inherent attraction that problems surrounding gene delivery to muscle are circumvented. It is of crucial importance, therefore, to determine the processes regulating the expression of utrophin so that therapeutic up-regulation may be attempted in DMD patients.
We previously described a promoter within the CpG island at the 5′ end of the utrophin locus; this element is active in a broad range of cell types and tissues (14). The sequence contains a consensus N-box, a 6-bp motif important in the regulation of other genes expressed at the NMJ (15). Localization of utrophin at the NMJ in mature muscle is partially attributable to enhanced transcription of utrophin at subjunctional myonuclei, with consequent synaptic accumulation of mRNA (16, 17). The utrophin CpG-island promoter drives synaptic transcription of a reporter gene in vivo; this expression pattern is abolished by point mutations within the N-box (18).
Expression of full-length dystrophin is mediated by three independently regulated promoters driving tissue-specific transcription of unique first exons that splice into a common message (19–21). The similarity between utrophin and dystrophin extends to their genomic structure (22). We therefore hypothesized that utrophin might also be transcribed from more than one promoter, an important consideration for the following reasons. First, it may be undesirable to interfere with the mechanisms underlying synaptic regulation of genes, as this might affect the expression of other postsynaptic components and impair the structure and function of the NMJ; a promoter without synaptic regulatory elements might be a more suitable target for pharmacological manipulation. Second, cardiac dysfunction is a common feature of dystrophinopathies (23); manipulation of the CpG-island promoter alone may not be therapeutically sufficient if the cardiac utrophin message was transcribed from a different promoter. Finally, the inclusion of additional regulatory sequences might increase the yield of a screening program to identify small molecules capable of transcriptional activation of utrophin.
This paper describes the isolation and characterization of a second utrophin promoter that is highly regulated, expressed in a wide range of tissues, and has little similarity to the synaptically expressed promoter.
Materials and Methods
Oligonucleotides, PCR, Reverse Transcription–PCR (RT-PCR), and 5′ Rapid Amplification of cDNA Ends (RACE).
PCR and RT-PCR were performed as described (24). Oligonucleotide sequences (5′ to 3′) were UM83, gatgttcctg tgaggccttc gag; UM82, cactcttgga aaatcgagcg t; U16, actatgatgt ctgccagagt tg; U107, gatccaatag cttccttcca tcttt; UBF, tggaaaaagt ggaggttgga; BR2, tccaacctcc actttttcca; BR4, gcctggagag ctacatgccc t; BF8, ctccacatct ttttcctcat catct; BF9, gattgtggtg atggttgtag aa; BR10, gattgtggtg atggttgtag aa; BR14, gatgatgagg aaaaagatgt ggag; BF15, aaacccaaaa taacacagga catc; BF16, agtgtaactt ctctctggtg; BF42, gctgcttttg ttgtccactt c; BR43, atagcttcct tccatctttg ag; 2ApF, gcgtgcagtg gaccattttt cagattta; 1BpF, cgctgcagca gccaccacat ttcgttg; and 3pR, gcgtgcagat cgagcgttta tccatttg. 5′ RACE was undertaken by using adapter-ligated mouse heart cDNA (Marathon-Ready, CLONTECH), following the manufacturer’s protocol, with nested mouse utrophin primers UM83 (exon 4) and UM82 (exon 3). Products were cloned in pGEM-T (Promega). Human exon 1B was isolated from skeletal muscle cDNA by PCR with mouse primers UBF and UM83. 5′ RACE was used to clone the 5′ end of human exon 1B, by using primers U107 and BR4. Semiquantitative RT-PCR was performed with primers BF42 and BR43 to amplify utrophin B, and with commercial primers (Stratagene) to amplify glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Exponential amplification was established by withdrawing samples from thermal cycling at one-cycle intervals; products were blotted and probed with labeled BR4 or a 600-bp GA3PDH probe, and band intensities were quantified by using a Storm phosphoimager (Molecular Dynamics).
Genomic Mapping and Clones.
Human yeast artificial chromosomes (YACs) are as previously described (22). Southern blots of restriction-digested YAC DNA were probed with end-labeled BR4. A 3.0-kb hybridizing XbaI fragment was cloned from 4X124H10 into pBlueScript (Stratagene), generating pBSX2.0. Mouse P1 artificial chromosomes (PACs) were identified from the RPCI21 library. A 398-bp exon 1B/promoter B DNA probe (UB400) encompassing human positions 1129 to 1527 was used for exon 1B mapping. Library filters were screened with probes to exons 1A–5 (14) and UB400. Eleven PACs were identified, and four of these arranged into a contig by restriction mapping. An 8.0-kb XbaI fragment from PAC 110C24, which hybridized with UB400, was cloned in pBlueScript, generating pBSX8.0.
Northern Blots and Probes.
A human multiple-tissue Northern blot and β-actin control cDNA probe were obtained from CLONTECH. A utrophin C-terminal cDNA probe, encompassing the last 4.0 kb of the utrophin message, was generated by PCR. The human exon 1B sequence between positions 1480 and 1596 was cloned into pGEM-T and an exon 1B antisense riboprobe (In Vitro Transcription Kit, Promega) was transcribed from the SP6 promoter after linearization of the plasmid with NcoI. Hybridization was carried out at 70°C in 50% formamide hybridization buffer (25), and the filter was washed at 75°C in 0.1× SSC/0.1% SDS for 2 hours.
RNase Protection.
Specific probes spanning the exon 1B/3 and exon 2A/3 boundaries were obtained by PCR amplification of mouse heart cDNA by using primers 2ApF, 1BpF, and 3pR. Products were cloned in the PstI site of pDP18 (Ambion, Austin TX) and sequenced. Plasmids were linearized with EcoRI (1B) or BamHI (2A); labeled antisense riboprobe was transcribed from the T7 promoter and gel purified. RNase protection was carried out by using an RPAIII kit (Ambion), following the manufacturer’s instructions (30 μg of total RNA unless stated, hybridization temperature 42°C, RNase A/T1 dilution 1:200). After electrophoretic separation, band intensities were quantified as above and corrected for the amount of label present in each protected fragment.
Promoter/Reporter Constructs.
Reporter constructs were generated by PCR amplification of the human sequence between positions 39 and 1503, by using pBSX2.0 as a template and primers BF9 and BR14. Products were cloned in pGEM-T. Clones were identified with product in both orientations and the insert, liberated by digestion with SacI/NcoI, cloned into the SacI/NcoI sites of a promoterless luciferase-reporter plasmid (pGL3 basic, Promega), generating constructs with insert in forward (pGL3/utroB/F) and reverse (pGL3/UtroB/R) orientation with respect to the coding sequence of luciferase. Deletions of the forward construct were generated by cleavage at SpeI, NdeI, EcoRI, and PvuII sites in the insert, followed by religation to sites in the 5′ or 3′ polylinker. All constructs were sequenced.
Cell Culture and Transfections.
Human IN157, CL11T47, and HeLa cell lines were maintained as described (14). Two micrograms of pGL3/utroB/F or R, or its molar equivalent, mixed with 0.5 μg of β-galactosidase control plasmid (pSV-β-gal, Promega) was transfected in each well of 6-well plates by using Superfect (Qiagen, Chatsworth, CA), following the manufacturer’s protocol. Forty-eight hours later, cells were harvested and cell extracts were assayed for luciferase and β-galactosidase activity as described (14). Luciferase activity was standardized to β-galactosidase activity in each individual sample to control for transfection efficiency. Results are expressed as mean luciferase/β-galactosidase ratio for four individual transfections. Error bars indicate the standard error of the mean. For comparison of different constructs within the same cell line, results were standardized to those obtained with pGL3/utroB/F and are expressed as a percentage of this value. For comparison of constructs between cell lines, results were standardized to those obtained with a luciferase–simian virus 40 promoter/enhancer plasmid (pGL3 control, Promega).
Primer Extension.
Primer extension was carried out as described (14). End-labeled primer BR2 was annealed to 0, 30, or 50 μg of mouse heart total RNA at 58°C for 20 minutes, and extended at 42°C for 40 minutes. Products were separated on a 6% polyacrylamide gel, under denaturing conditions, alongside a sequencing ladder generated from pBSX8.0 by using primer BR2.
Results
An Alternative 5′ Exon in Utrophin mRNA.
Utrophin from a mouse heart cDNA library was amplified by 5′ RACE, and the resulting products were cloned and sequenced. Of 12 clones, 8 contained novel sequence 5′ of exon 3 that was not homologous to exon 2. Below, we present evidence that this sequence is a single alternative 5′ exon of utrophin containing a translational initiation codon. We refer to this sequence as “exon 1B” throughout the remainder of the paper to distinguish it from the previously described 5′ cDNA sequence comprising untranslated exon 1A and exon 2A, which contains the translational start (Fig. 4C).
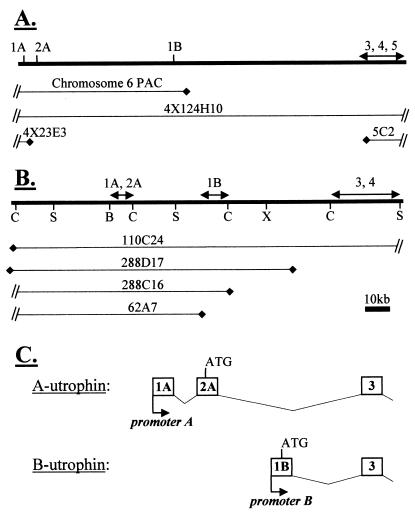
Figure 4.
Schematic representation of (A) human YAC and (B) mouse PAC contigs, showing position of exons within the genomic map. Mouse restriction sites: C, ClaI; S, SacII; B, BssHII; X, XhoI. (C) Nomenclature for utrophin promoters, exons, and transcripts.
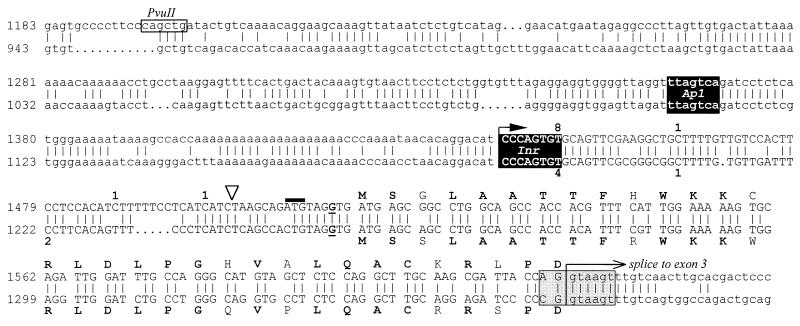
Fig. 1 shows a sequence comparison of human and mouse exons 1B and genomic flanking sequence. The position and phase of the splice junction at the 5′ end of exon 3 is identical for both exon 1B- and exon 2A-containing transcripts. Exon 1B contains a putative ATG translation-initiation codon and ORF, in-frame with that of exon 3, predicting a 31-amino acid N terminus to the utrophin protein. The context of the ATG codon is predicted to be favorable for translation in that there is a purine at position −3 (bold in Fig. 1; ref. 26). Human and mouse exons 1B show 82% nucleotide identity. The predicted translations are 77% identical and 81% similar. The position and context of the ATG codon are conserved. The human sequence contains a second putative ATG codon immediately 5′ (position 1511, solid bar in Fig. 1), followed by a TAG stop codon. As this ATG does not adhere to the Kozak consensus, is not associated with an ORF, and is not present in the mouse sequence, we predict that this is not a functional translation start. A similar feature is present in human exon 2A, where the 5′ untranslated region (UTR) contains a short ORF prior to the true translation start.
Figure 1.
Sequence alignment of human (Upper) and mouse (Lower) exon 1B (in uppercase) and promoter B (in lowercase). Numbering corresponds to the inserts of pBSX2.0 and pBSX8.0, respectively. The human PvuII site (see Fig. 6) is indicated. The open triangle indicates the position at which the luciferase coding sequence was inserted to make pGL3/UtroB/F. The deduced translation of exon 1B is shown; amino acids marked in boldface type are identical between the human and mouse sequences. The conserved splice donor consensus is shown in gray. A putative Ap1 site and an initiator-like element (Inr) are 100% conserved and indicated in black. A solid arrow marks the single transcription start indicated by primer extension; numbers adjacent to the sequence indicate the number of individual 5′ RACE clones that terminated at the positions shown.
The Transcript Associated with Exon 1B.
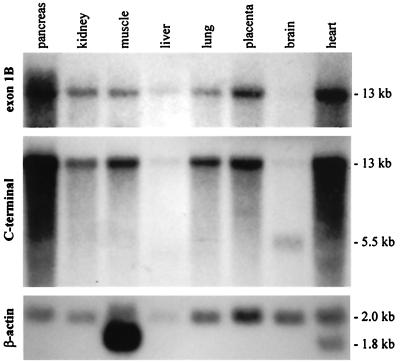
A human multiple-tissue Northern blot was probed with an exon 1B antisense riboprobe. A single hybridizing 13-kb band was observed, identical to that produced by probing the same blot with a cDNA encompassing 4 kb of the utrophin C terminus (Fig. 2), indicating that exon 1B is exclusively associated with a full-length utrophin mRNA. Exon 1B is ubiquitously expressed, and appears most abundant in the heart and pancreas, relative to β-actin.
Figure 2.
Human multiple-tissue Northern blot probed sequentially with exon 1B, utrophin C-terminal, and β-actin probes. A single 13-kb transcript hybridizes with both exon 1B and utrophin C-terminal probes. The 5.5-kb brain transcript is G-utrophin (31). The β-actin probe indicates that the gel was not loaded evenly, accounting for some of the apparent differences in transcript abundance noted in the upper two panels.
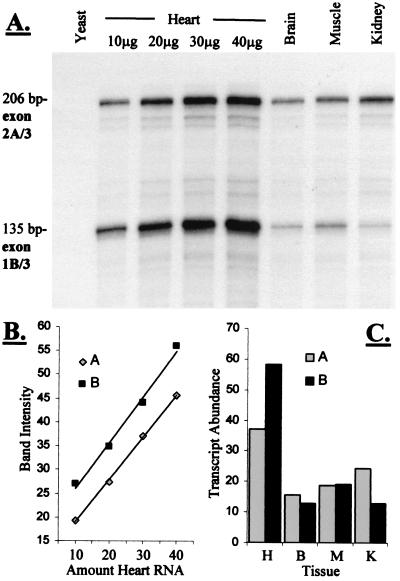
The expression profiles of exons 1B and 2A were examined by using RNase protection (Fig. 3). Specific riboprobes corresponding to the exon 1B/3 and 2A/3 boundaries were simultaneously hybridized with total RNA, allowing direct quantitation of transcript abundance. B-utrophin is the most abundant form in the heart, whereas exon 2A-containing transcripts predominate in the kidney. Approximately equal amounts of exons 1B and 2A were observed both in the brain and in skeletal muscle.
Figure 3.
RNase protection to quantify expression of exons 1B and 2A. (A) Specific protected fragments of 206 bp (exon 2A/3) and 135 bp (exon 1B/3) were generated by simultaneous hybridization of two probes with mouse, but not yeast, RNA. (B) The signal generated is linearly related to the amount of RNA for both probes under these conditions, allowing quantitative analysis. (C) Amount of each transcript present (in arbitrary units) in 30 μg of total RNA from four different tissues. H, heart; B, brain; M, muscle; K, kidney.
Mapping and Cloning of Genomic Sequence Associated with Exon 1B.
By using probe BR4, exon 1B was mapped within our previously described human YAC contig (26), encompassing the 5′ end of the utrophin locus (Fig. 4A). A hybridizing band was seen with YAC 4X124H10 but not 4X23E3 or 5C2 (data not shown), indicating that exon 1B lies within the 120-kb intron 2 of the utrophin gene. A subsequent database search identified a clone from the Sanger Centre human chromosome 6 sequencing project, containing exons 1A, 2A, and 1B. This indicated that exon 1B lies 52.2 kb 3′ of exon 2A (Fig. 4A). Probing the RPCI21 library with utrophin exons 1A, 1B, and 2–5, inclusive, identified a series of genomic PACs spanning the 5′ end of the mouse utrophin gene. Four of these PACs were assembled into a contig of the region. Hybridization with UB400 confirmed that exon 1B lies within intron 2 in the mouse (Fig. 4B), approximately 50 kb 3′ of exon 2.
Two kilobases of genomic sequence encompassing exon 1B was obtained from human and mouse plasmid clones, pBSX2.0 and pBSX8.0, respectively. The longest unique cDNA sequence matched a single contiguous region of genomic sequence, compatible with an absence of further 5′ exons in the 1B-containing transcript, and suggesting that the genomic flanking sequence contained the promoter element responsible for exon 1B expression. Our nomenclature for utrophin 5′ exons, transcripts, and promoters appears in Fig. 4C.
Promoter B.
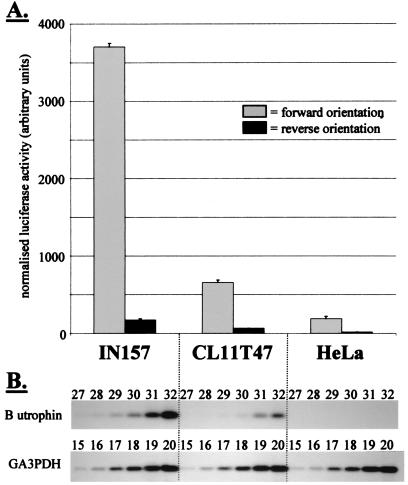
One-and-a-half kilobases of human genomic sequence 5′ of exon 1B, including the 5′ UTR of exon 1B, was cloned in both orientations into a promoterless luciferase reporter vector. Three human cell lines (IN157 rhabdomyosarcoma, CL11T47 kidney epithelial, and HeLa cervical epithelial) were transiently transfected with these constructs. Reporter activity was detected at significantly higher levels in cells transfected with the forward than with the reverse orientation construct, indicating promoter activity (Fig. 5A). Interestingly, the level of activity varied between cell lines by an order of magnitude. Semiquantitative RT-PCR demonstrated that the variation of luciferase expression mimicked the transcription profile of endogenous utrophin exon 1B (Fig. 5B). These data show that the 1.5 kb of genomic sequence 5′ of exon 1B used in these reporter clones contains both the necessary signals to initiate transcription of exon 1B and regulatory elements that determine the level of expression in these cell lines.
Figure 5.
In vitro activity of utrophin promoter B. (A) Normalized luciferase activity after transfection of three different human cell types with either pGL3/utroB/F (forward orientation) or pGL3/utroB/R (reverse orientation). (B) Semiquantitative RT-PCR to estimate the relative abundance of B-utrophin in the three cell lines. Successive PCR cycles are shown; the numbers above each band correspond to the cycle number at which PCR was stopped. The GA3PDH control shows identical amplification in all cDNA samples, indicating that the differences seen in B-utrophin amplification have arisen from differences in the level of expression of the endogenous B-utrophin transcript in these cell lines. The levels are commensurate with the level of reporter gene activity seen in A.
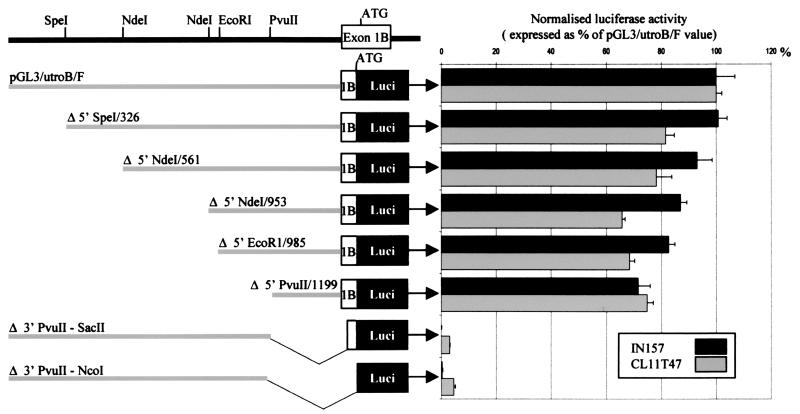
To further delineate important elements within this region, a series of 5′ and 3′ deletions of promoter B were made, and the in vitro activity of each one was assayed (Fig. 6). A 300-bp element, contained within clone pGL3/utroB/F/Δ5′ PvuII/1199, retains 70% activity of the full 1.5-kb construct in expressing cell lines and shows 74% sequence identity between human and mouse (Fig. 1). Homology falls to 50% when sequence further 5′ of the human PvuII site is compared with the corresponding mouse sequence (data not shown).
Figure 6.
Deletion analysis of promoter B. The 1.5-kb insert of pGL3/utroB/F was deleted at its 5′ and 3′ ends by using the internal restriction sites indicated. Reporter activity was assayed after transient transfection of IN157 and CL11T47 cells.
Promoter B Transcription Start Site.
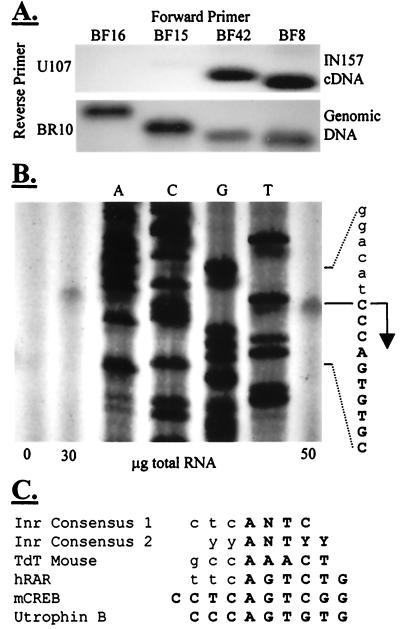
The 5′ ends of eight human and four mouse 5′ RACE clones clustered around a putative cap site in the genomic sequence (Fig. 1). None of the 5′ RACE clones generated by amplification across the exon 3/exon 1B boundary extended further upstream. RT-PCR was carried out using forward primers around this region with a reverse primer in exon 4 (Fig. 7A). A product of expected size was amplified from IN157 cDNA by primers BF42 and BF8, but not BF16 or BF15, indicating that the transcription start is within the 18 bp that separate the two primers BF15 and BF42. These 18 bases contain the putative cap site and the cluster of RACE clone 5′ ends.
Figure 7.
Transcription start site mapping. (A) RT-PCR with cDNA from IN157 cells. Only BF42 and BF8 generate a specific product, hybridizing with BR4. The sequences and annealing conditions of all primers are correct, however, as products of expected sizes are amplified from genomic DNA. The transcription start, therefore, is between the sequences corresponding to BF15 and BF42. (B) Primer extension analysis of mouse heart RNA indicates a single transcription start on the C residue at mouse position 1183. (C) The region around the transcription start is homologous to the initiators of other promoters. Consensus 1, initiator consensus derived from sequence comparison of Inr+ genes (32); consensus 2, experimentally derived consensus for functional initiator (33); TdT, terminal deoxynucleotidyl transferase; hRAR, human retinoic acid receptor α; mCREB, mouse cAMP response element binding protein. Transcribed sequence is indicated in bold uppercase.
To map the start site accurately, primer extension with an exon 1B reverse primer and mouse heart RNA was employed. This yielded a single product indicative of a single transcription start site (Fig. 7B). Transcription initiates at mouse position 1183 within a 25-bp motif, which is 100% conserved between human and mouse. Part of this motif, spanning the cap site, is a 6/7 base match for the initiator consensus, and correspondingly shows homology to the initiators of other genes (Fig. 7C). The transcription start site is at an analogous position to that of the mouse cAMP response element binding protein (mCREB) initiator, 5′ to the consensus initiator cap site. Sequence analysis shows absence of consensus TATA or CAAT motifs, and we consider this promoter to be of the TATA− Inr+ type.
Discussion
We have demonstrated that there is a second promoter within intron 2 of the utrophin gene, driving expression of a unique first exon that splices into a common 13-kb mRNA. These data are important, both in terms of understanding the molecular physiology of utrophin expression, and in view of their potential application to therapeutic intervention in DMD.
There are many reported genes with more than one promoter, and various functional consequences have been postulated (reviewed in ref. 27). A single gene may achieve a complex temporal and spatial expression pattern through the interaction of different promoters with discrete subsets of transcription factors. Dystrophin is an example: three dissimilar promoters are active at different levels in specific cell types within the heart, skeletal muscle, and the brain (21, 28, 29). RNase protection, however, indicates that utrophin promoters A and B are coexpressed, although independently regulated, in a number of tissues. It is conceivable that examination of transcript distribution in whole tissue samples has masked cell type-specific patterns of expression, a possibility supported by in vitro data demonstrating 10-fold variation in promoter B activity between different cell lines. Alternatively, the two promoters may be spatially regulated at a subcellular level. Within adult skeletal muscle fibers, promoter A is synaptically driven (16), yet aggregates of utrophin mRNA are detectable at up to 25% extrasynaptic nuclei (17). Expression of promoter B in the extrasynaptic compartment might be invoked as one possible explanation.
A second proposed function of alternative promoters is the generation of transcripts with interchangeable 5′ exons, giving rise to mRNAs with alternative 5′ UTRs or proteins with unique N-terminal domains. Unlike exon 1B, utrophin exon 1A contains a long, GC-rich 5′ UTR. In some cases, GC-rich 5′ UTRs are not translated efficiently (26), and there are examples of genes in which alternative use of GC-rich and non-GC-rich 5′ UTRs has been implicated in posttranscriptional regulation of protein synthesis (30). In addition, the predicted 31 amino acids encoded by exon 1B are different from the 26 amino acids of exon 2A; the functions of the resulting N-termini may be different.
The identification of a second promoter provides a new target for the up-regulation of utrophin to ameliorate the DMD phenotype. Promoter B is highly regulated, probably by different factors than promoter A. Elucidation of the mechanisms responsible for the large difference in promoter B activity between IN157 and HeLa cells might lead to the identification of a factor that can be delivered to muscle to activate utrophin expression. Importantly, as the N-box motif is absent from promoter B, this is unlikely to carry any risk of NMJ disruption potentially inherent in the pharmacological manipulation of synaptically regulated promoter A.
Acknowledgments
Mouse PAC clones were kindly supplied by Prof. P. deJong (http://bacpac.med.buffalo.edu). The expert technical assistance of Mrs. A. Potter is gratefully acknowledged. E.A.B. is an Action Research Training Fellow. This work was funded by the Muscular Dystrophy Group (U.K.), the Muscular Dystrophy Association (U.S.A.), Association Francaise Contre les Myopathies, and the Medical Research Council (U.K.).
Abbreviations
- DMD
Duchenne muscular dystrophy
- NMJ
neuromuscular junction
- RT-PCR
reverse transcription–PCR
- RACE
rapid amplification of cDNA ends
- YAC
yeast artificial chromosome
- PAC
P1 artificial chromosome
- UTR
untranslated region
Footnotes
References
- 1.Koenig M, Hoffman E P, Bertelson C J, Monaco A P, Feener C, Kunkel L M. Cell. 1987;50:509–517. doi: 10.1016/0092-8674(87)90504-6. [DOI] [PubMed] [Google Scholar]
- 2.Tinsley J M, Blake D J, Zuellig R A, Davies K E. Proc Natl Acad Sci USA. 1994;91:8307–8313. doi: 10.1073/pnas.91.18.8307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ervasti J M, Ohlendieck K, Kahl S D, Gaver M G, Campbell K P. Nature (London) 1990;345:315–319. doi: 10.1038/345315a0. [DOI] [PubMed] [Google Scholar]
- 4.Love D R, Hill D F, Dickson G, Spurr N K, Byth B C, Marsden R F, Walsh F S, Edwards Y H, Davies K E. Nature (London) 1989;339:55–58. doi: 10.1038/339055a0. [DOI] [PubMed] [Google Scholar]
- 5.Tinsley J M, Blake D J, Roche A, Fairbrother U, Riss J, Byth B C, Knight A E, Kendrick-Jones J, Suthers G K, Love D R, et al. Nature (London) 1992;360:591–593. doi: 10.1038/360591a0. [DOI] [PubMed] [Google Scholar]
- 6.Blake D J, Tinsley J M, Davies K E. Brain Pathol. 1996;6:37–47. doi: 10.1111/j.1750-3639.1996.tb00781.x. [DOI] [PubMed] [Google Scholar]
- 7.Love D R, Byth B C, Tinsley J M, Blake D J, Davies K E. Neuromuscul Disord. 1993;3:5–21. doi: 10.1016/0960-8966(93)90037-k. [DOI] [PubMed] [Google Scholar]
- 8.Khurana T S, Watkins S C, Chafey P, Chelly J, Tome F M S, Fardeau M, Kaplan J-C, Kunkel L M. Neuromuscul Disord. 1991;1:185–194. doi: 10.1016/0960-8966(91)90023-l. [DOI] [PubMed] [Google Scholar]
- 9.Clerk A, Morris G E, Dubowitz V, Davies K E, Sewry C A. Histochem J. 1993;25:554–561. [PubMed] [Google Scholar]
- 10.Tinsley J M, Davies K E. Neuromuscul Disord. 1993;3:537–539. doi: 10.1016/0960-8966(93)90111-v. [DOI] [PubMed] [Google Scholar]
- 11.Tinsley J M, Potter A C, Phelps S R, Fisher R, Trickett J I, Davies K E. Nature (London) 1996;384:349–353. doi: 10.1038/384349a0. [DOI] [PubMed] [Google Scholar]
- 12.Deconinck N, Tinsley J, De Backer F, Fisher R, Kahn D, Phelps S, Davies K E, Gillis J M. Nat Med. 1997;3:1216–1221. doi: 10.1038/nm1197-1216. [DOI] [PubMed] [Google Scholar]
- 13.Tinsley J, Deconinck N, Fisher R, Kahn D, Phelps S, Gillis J M, Davies K E. Nat Med. 1998;4:1441–1444. doi: 10.1038/4033. [DOI] [PubMed] [Google Scholar]
- 14.Dennis C L, Tinsley J M, Deconinck A E, Davies K E. Nucleic Acids Res. 1996;24:1646–1652. doi: 10.1093/nar/24.9.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koike S, Schaeffer L, Changeux J-P. Proc Natl Acad Sci USA. 1995;92:10624–10628. doi: 10.1073/pnas.92.23.10624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gramolini A O, Dennis C L, Tinsley J M, Robertson G S, Cartaud J, Davies K E, Jasmin B J. J Biol Chem. 1997;272:8117–8120. doi: 10.1074/jbc.272.13.8117. [DOI] [PubMed] [Google Scholar]
- 17.Vater R, Young C, Anderson L V B, Lindsay S, Blake D J, Davies K E, Zuellig R, Slater C. Mol Cell Neurosci. 1998;10:229–242. [PubMed] [Google Scholar]
- 18.Gramolini A O, Burton E A, Tinsley J M, Ferns M J, Cartaud A, Cartaud J, Davies K E, Lunde J A, Jasmin B J. J Biol Chem. 1998;273:736–743. doi: 10.1074/jbc.273.2.736. [DOI] [PubMed] [Google Scholar]
- 19.Klamut H, Gangopadhyay S, Worton R, Ray P. Mol Cell Biol. 1990;10:193–205. doi: 10.1128/mcb.10.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boyce F M, Beggs A H, Feener C, Kunkel L M. Proc Natl Acad Sci USA. 1991;88:1276–1280. doi: 10.1073/pnas.88.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorecki D C, Monaco A P, Derry J M J, Walker A P, Barnard E A, Barnard P J. Hum Mol Genet. 1992;1:505–510. doi: 10.1093/hmg/1.7.505. [DOI] [PubMed] [Google Scholar]
- 22.Pearce M, Blake D J, Tinsley J M, Byth B C, Campbell L, Monaco A P, Davies K E. Hum Mol Genet. 1993;2:1765–1772. doi: 10.1093/hmg/2.11.1765. [DOI] [PubMed] [Google Scholar]
- 23.Hoogerwaard E M, de Voogt W G, Wilde A A, van der Wouw P A, Bakker E, van Ommen G J, de Visser M. J Neurol. 1997;244:657–663. doi: 10.1007/s004150050163. [DOI] [PubMed] [Google Scholar]
- 24.Blake D J, Nawrotski R, Peters M F, Froehner S C, Davies K E. J Biol Chem. 1996;271:7802–7810. doi: 10.1074/jbc.271.13.7802. [DOI] [PubMed] [Google Scholar]
- 25.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley; 1999. [Google Scholar]
- 26.Kozak M. J Cell Biol. 1991;115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ayoubi T A Y, Van de Ven W J M. FASEB J. 1996;10:453–460. [PubMed] [Google Scholar]
- 28.Barnea E, Zuk D, Simantov R, Nudel U, Yaffe D. Neuron. 1990;5:881–888. doi: 10.1016/0896-6273(90)90348-j. [DOI] [PubMed] [Google Scholar]
- 29.Holder E, Maeda M, Bies R D. Hum Genet. 1996;97:232–239. doi: 10.1007/BF02265272. [DOI] [PubMed] [Google Scholar]
- 30.Nielsen F C, Gammeltoft S, Christiansen J. J Biol Chem. 1990;265:13431–13434. [PubMed] [Google Scholar]
- 31.Blake D J, Schofield J N, Zuellig R A, Gorecki D C, Phelps S R, Barnard E A, Edwards Y H, Davies K E. Proc Natl Acad Sci USA. 1995;92:3697–3701. doi: 10.1073/pnas.92.9.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Azizkhan J C, Jensen D E, Pierce A J, Wade M. Crit Rev Eukaryotic Gene Expression. 1993;3:229–254. [PubMed] [Google Scholar]
- 33.Javahery R, Khachi A, Lo K, Zenzie-Gregory B, Smale S T. Mol Cell Biol. 1994;14:116–127. doi: 10.1128/mcb.14.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]