Abstract
Deletions on chromosome 8p are common in human tumors, suggesting that one or more tumor suppressor genes reside in this region. Deleted in Liver Cancer 1 (DLC1) encodes a Rho-GTPase activating protein and is a candidate 8p tumor suppressor. We show that DLC1 knockdown cooperates with Myc to promote hepatocellular carcinoma in mice, and that reintroduction of wild-type DLC1 into hepatoma cells with low DLC1 levels suppresses tumor growth in situ. Cells with reduced DLC1 protein contain increased GTP-bound RhoA, and enforced expression a constitutively activated RhoA allele mimics DLC1 loss in promoting hepatocellular carcinogenesis. Conversely, down-regulation of RhoA selectively inhibits tumor growth of hepatoma cells with disabled DLC1. Our data validate DLC1 as a potent tumor suppressor gene and suggest that its loss creates a dependence on the RhoA pathway that may be targeted therapeutically.
Keywords: DLC1, HCC, RNAi, RhoA, mouse model
Tumor suppressor genes act in signaling networks that protect against tumor initiation and progression, and can be inactivated by deletions, point mutations, or promoter hypermethylation. Although tumor suppressors are rarely considered direct drug targets, they can negatively regulate pro-oncogenic signaling proteins that are amenable to small molecule inhibition. For instance, NF1 inhibits the Ras signaling pathway, which is deregulated in many cancers and has been pursued for its therapeutic potential (Downward 2003). Similarly, PTEN inhibits the PI3–kinase pathway, and inhibitors of PI3K pathway components such as PI3K, AKT, and mTORs have entered clinic trials (Luo et al. 2003).
Recurrent chromosomal deletions found in sporadic cancers often contain tumor suppressor genes. For example, PTEN loss on chromosome 10q23 frequently occurs in various cancers and promotes tumorigenesis by deregulating the PI3 kinase pathway (Maser et al. 2007). Similarly, heterozygous deletions on chromosome 8p22 in many hepatocellular carcinomas (HCC) (Jou et al. 2004) and other cancer types, including carcinomas of the breast, prostate, colon, and lung (Matsuyama et al. 2001; Durkin et al. 2007). Several genes, including DLC1, MTUS1, FGL1 and TUSC3, have been identified as candidate tumor suppressors in this region (Yan et al. 2004). Deleted in Liver Cancer 1 (DLC1) is a particularly attractive candidate owing to its genomic deletion, promoter methylation, and underexpressed mRNA in cancer (Yuan et al. 1998, 2003a; Ng et al. 2000; Wong et al. 2003; Guan et al. 2006; Seng et al. 2007; Ying et al. 2007; Zhang et al. 2007; Pike et al. 2008; for review, see Durkin et al. 2007).
Despite its potential importance, functional data implicating DLC1 loss in tumorigenesis are lacking. DLC1 encodes a RhoGAP protein that catalyzes the conversion of active GTP-bound RhoGTPase (Rho) to the inactive GDP-bound form and thus suppresses Rho activity (Yuan et al. 1998). DLC1 has potent GAP activity for RhoA and limited activity for CDC42 (Wong et al. 2003; Healy et al. 2008). When overexpressed, DLC1 inhibits the growth of tumor cells and xenografts (Yuan et al. 2003b, 2004; Zhou et al. 2004; Wong et al. 2005; Kim et al. 2007), but whether this requires its Rho-GAP activity or other functions remains unresolved (Qian et al. 2007; Liao et al. 2007). Most functional studies to date have relied on DLC1 overexpression and, as yet, none have documented that loss of DLC1 promotes transformation in vitro or tumorigenesis in vivo. Indeed, homozygous dlc1 knockout mice die around embryonic day 10.5 (E10.5), and there is no overt phenotype in dlc1 heterozygous mice (Durkin et al. 2005).
Our laboratory recently developed a “mosaic” mouse model whereby liver carcinomas can be rapidly produced with different genetic alterations by manipulation of cultured embryonic liver progenitor cells (hepatoblasts) followed by transplantation into the livers of recipient mice (Zender et al. 2005, 2006). We previously used this model to identify new oncogenes in HCC, which could be characterized in an appropriate biological and genetic context (Zender et al. 2006). Furthermore, using this system, we showed that shRNAs capable of suppressing gene function by RNAi could recapitulate the consequences of tumor suppressor gene loss on liver carcinogenesis (Zender et al. 2005; Xue et al. 2007). Here we combine this mosaic model and RNAi to validate DLC1 as a potent tumor suppressor gene and study its action in vivo.
Results and Discussion
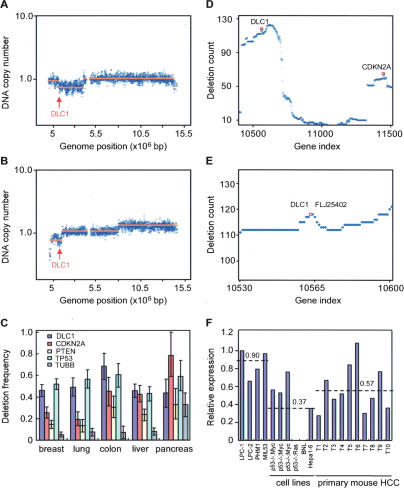
Studies using low-resolution genome scanning methods have identified chromosome 8p deletions as common lesions in liver carcinoma and other tumor types. To confirm and extend these observations, we examined a series of data sets of copy number alterations in HCC obtained using representational oligonucleotide microarray analysis (ROMA), a variation of array-based CGH that enables genome scanning at high resolution (Lucito et al. 2003). In a panel of 86 liver cancers, heterozygous deletions encompassing the DLC1 were observed in 59 tumors (Fig. 1A,B; data not shown). Consistent with previous reports, these deletions were large (>5 Mb), encompassing >20 annotated genes but invariably included the DLC1 locus. Indeed, heterozygous deletions of DLC1 occurred more frequently than those observed for the well-established tumor suppressors such as INK4a/ARF, PTEN, and TP53 (Fig. 1C). Furthermore, DLC1 deletions were nearly as common as those for TP53 in other major tumor types such as lung, colon, and breast (Fig. 1C). Again, most 8p deletions were large, although in breast cancer DLC1 resided at a local deletion epicenter reminiscent of that surrounding the INK4a/ARF locus on chromosome 9p21 (Fig. 1D,E). Although we did not examine the status of the remaining allele in this tumor cohort, studies suggest that it can be silenced by promoter methylation (Yuan et al. 2003a; for review, see Durkin et al. 2007). Together, these data suggest that DLC1 loss plays an important role in human cancer but, in the absence of functional validation, are not conclusive.
Figure 1.
DLC1 is a candidate tumor suppressor on chromosome 8p22. (A,B) DNA copy number profiles for chromosome 8 of two representative human HCC samples reveals chromosome 8p22 deletions containing DLC1. The blue data points represent the averaged fluorescent ratio (tumor vs. normal) and the orange lines correspond to the value determined by copy number segmentation. Arrows denote the DLC1 locus. (C) Deletion counts per case for DLC1, p16INK4a, PTEN, TP53, and β-Tubulin (TUBB) in five types of human carcinomas. The counts were obtained from ROMA profiles of 257 breast, 137 colon, 86 liver, 213 lung, and 46 pancreas cancers, respectively. β-Tubulin serves as a negative control. The error bars indicate the 90% confidence intervals for each quantity. (D) Deletion counts in ROMA profiles of 257 human breast cancers. A deletion count in each profile set was obtained by finding the maximal tier number for 24,719 genes across the genome in each profile in the set and summing the result over the set. The counts are plotted against the gene ordinal number, with the genes sorted by their genomic transcription start position. Shown is chromosome 8 and 9. The points in the vicinity of DLC1 and p16INK4A are highlighted by red circles. (E) High-magnification view of 8p22 region in D. DLC1 and FLJ25402 reside at a local deletion epicenter. FLJ25402 is an uncharacterized gene. (F) qPCR analysis of DLC1 expression in mouse embryonic liver progenitor cells (LPC), immortalized liver progenitor cells (MIL53 and PHM1), mouse HCC cells, and primary mouse HCC tumors. Numbers indicate the average value of each group.
Human liver tumor cells with 8p deletions invariably expressed low DLC1 mRNA (Supplemental Fig. S1). We also examined a series of murine liver carcinomas produced from embryonic hepatoblasts (Zender et al. 2006). None of the tumors we analyzed contained deletions on mouse chromosome 8qA4, the region containing the murine dlc1 gene (data not shown), although many displayed reduced dlc1 mRNA levels compared with parental hepatoblasts or immortalized liver progenitor cells (Fig. 1F). Thus, down-regulation of DLC1 expression can occur during murine hepatocellular carcinogenesis.
If DLC1 is a bona fide tumor suppressor in humans, we hypothesized that reductions in DLC1 levels should promote HCC in mice. To test this hypothesis and to avoid complications associated with the embryonic lethality of dlc1 knockout animals, we decided to knockdown DLC1 expression in hepatoblasts using RNAi and test the ability of these cells to form tumors following engraftment into the livers of recipient mice. We therefore generated microRNA-based shRNA capable of efficiently suppressing DLC1 expression at mRNA and protein levels (Fig. 2A,B).
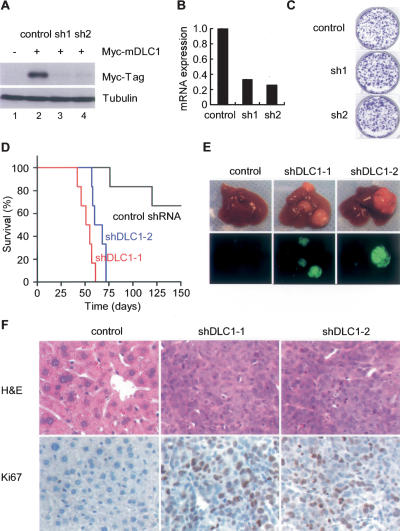
Figure 2.
DLC1 loss triggered by in vivo RNAi promotes tumorigenesis in a mouse liver cancer model. (A) Immunoblots of 293T cells cotransfected with a 6xMyc-tagged murine dlc1 cDNA (lanes 2–4) and a control shRNA (lane 2) or DLC1 shRNAs (lanes 3,4). Tubulin serves as a loading control. (B) DLC1 qRT–PCR of p53-null liver progenitor cells coinfected with Myc and a control shRNA or two DLC1 shRNAs. (C) Cells as in B were plated at equal number and stained with crystal violet after 8 d. Shown are representative results from three experiments. (D) DLC1 loss cooperates with Myc and p53 loss to accelerate liver tumor formation. Kaplan-Meier survival curve of mice transplanted with p53-null liver progenitor cells coinfected with Myc and DLC1 shRNAs (n = 6 for each group). (E) Representative images of explanted livers. GFP imaging identifies shRNA transduced cells. (F) Histopathology of representative tumors. Proliferating cells were labeled by Ki67 staining.
Our genomic analyses, together with previous reports, identify amplifications of the Myc oncogene and inactivation of the p53 tumor suppressor as common events in HCC (Staib et al. 2003; data not shown). We therefore decided to test the impact of DLC1 loss in cells coexpressing Myc and lacking p53, which together are only weakly oncogenic in this model (Zender et al. 2006). p53-deficient liver progenitor cells were cotransduced with one retrovirus expressing Myc and another coexpressing a DLC1 shRNA with a GFP reporter, thus enabling imaging of cells with reduced DLC1 expression (Supplemental Fig. S2A,B). Interestingly, DLC1 knockdown had little impact on the colony forming ability of liver progenitor cells in vitro (Fig. 2C; Supplemental Fig. S3A).
Genetically modified liver progenitors were seeded into the livers of syngeneic recipients to assess their ability to form tumors in situ. In contrast to the modest impact of DLC1 loss in vitro, DLC1 shRNAs significantly accelerated tumor onset in vivo (P value < 0.0001 for shDLC1-1 and P < 0.0005 for shDLC1-2) (Fig. 2D,E). In fact, at 57 d post-transplantation, GFP-positive tumor nodules were observed in the livers of most animals receiving cells harboring DLC1 shRNAs, whereas the control animals showed no macroscopically detectable tumor burden (Fig. 2E). Furthermore, the pathology of tumors derived from DLC1 knockdown resembled aggressive human HCC and displayed a high proliferative index as assessed by Ki67 immunohistochemistry (Fig. 2F). Tumors also expressed the HCC markers α-fetoprotein (AFP) and albumin (Supplemental Fig. S3B). These data demonstrate that loss of DLC1 can efficiently promote the development of HCC.
We also ectopically expressed the murine dlc1 gene in mouse hepatoma cells and tested their ability to form tumors orthotopically. To this end, we cloned a Myc-tagged murine dlc1 cDNA and confirmed its ability to produce a protein of the correct molecular weight (Fig. 3A). A mouse hepatoma cell line harboring a luciferase reporter and expressing oncogenic Ras and undetectable DLC1 (see Fig. 1F, lane 8) was infected with the DLC1-expressing retrovirus or an empty vector. Consistent with the literature (Ng et al. 2000), reintroduction of DLC1 produced a modest effect on proliferation in colony formation assays (Supplemental Fig. S4A,B).
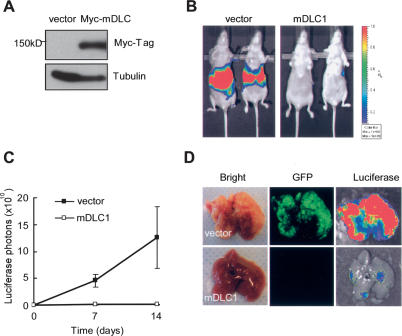
Figure 3.
Reintroduction of DLC1 reverts tumorigenesis. (A) Immunoblots of Ras-driven hepatoma cells infected with control retrovirus (v) or retrovirus expressing DLC1 cDNA. (B) Bioluminescence imaging of in situ liver tumor. Hepatoma cells as in A are transplanted into livers of NCR nu/nu mice and imaged at day 14. (C) Quantification of luciferase signal as in B. Error bars denote SD (n = 3). (D) Bioluminescence and GFP imaging of explanted livers at day 14.
At 2 wk post-liver transplantation, animals receiving control cells developed aggressive liver tumors revealed by whole-body bioluminescence imaging (Fig. 3B, left). Mice transplanted with cells expressing DLC1 had a greatly reduced tumor burden (Fig. 3B [right], C). Accordingly, explanted livers from control animals harbored disseminated tumor nodules that coexpressed both luciferase and GFP (Fig. 3D, top panel), whereas livers from mice receiving cells overexpressing DLC1 showed little evidence of tumor formation even using these sensitive imaging reporters (Fig. 3D, bottom).
Since DLC1 has GAP activity for RhoA (Wong et al. 2003), we tested whether DLC1 knockdown increased RhoA-GTP levels and mimicked activated RhoA in promoting stress fiber formation, a hallmark of RhoA activity (Wong et al. 2005). p53-deficient liver progenitor cells harboring Myc and DLC1 shRNAs displayed increased levels of GTP-bound RhoA as assessed by a pull-down assay using a Rhotekin-RBD domain (Kim et al. 2007), which selectively binds RhoA-GTP (Fig. 4A,B). Concordantly, hepatoblasts with reduced DLC1 levels showed a prominent actin stress fiber network as assessed by fluorescent phalloidin labeling actin filaments (Fig. 4C). Thus, loss of DLC1 recapitulates the effects of activated RhoA on liver epithelial cells in vitro.
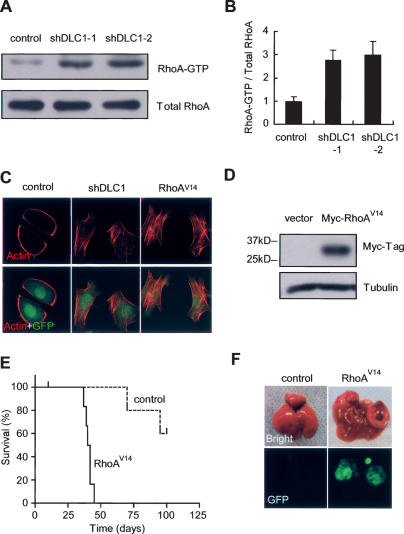
Figure 4.
DLC1 knockdown deregulates RhoA activity, which is sufficient to accelerate tumorigenesis. (A) RhoA-GTP pull-down assay of p53-null liver progenitor cells coinfected with Myc and a control shRNA or DLC1 shRNAs. (B) Quantification of A. Error bars denote SD (n = 3). (C) DLC1 knockdown increases actin stress fiber formation. p53−/−;Myc Liver progenitor cells infected with shDLC1 or RhoAV14 were serum starved and stained with fluorescent phalloidin. (D) Immunoblots of p53−/−-null hepatoblasts infected with Myc and a constitutively active RhoAV14 allele with 6xMyc tag at the N terminus. (E) Activated RhoA cooperates with Myc and loss of p53 to accelerate liver tumor formation. Kaplan-Meier survival curve of syngeneic mice transplanted with p53-null liver progenitor cells coinfected with Myc and RhoAV14 (n = 6 for each group). (F) Representative images of explanted livers at day 40 following cell transplantation. GFP imaging identifies retrovirally transduced cells.
To test whether activated RhoA, like loss of DLC1, can promote liver carcinoma formation, we infected p53−/− liver progenitor cells with retrovirus expressing Myc and a constitutively active RhoA (RhoAV14) (Fig. 4D) and transplanted the cells into the livers of syngeneic mice. Indeed, enforced expression of RhoAV14 dramatically accelerated tumor formation (Fig. 4E [P < 0.0001], F; see also Supplemental Fig. S5), producing tumors with the pathological features of HCC (data not shown). Thus, DLC1 loss and RhoA activation have similar effects on hepatocellular carcinogenesis.
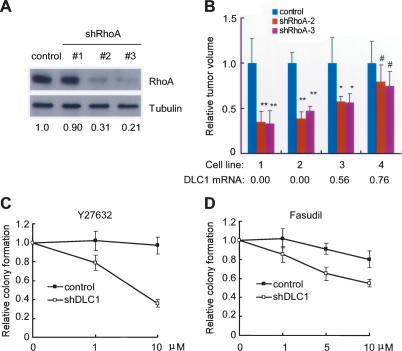
Although RhoA has been identified as a DLC1 effector, overexpression studies suggest that other DLC1 functions can contribute to its anti-proliferative activities (Liao et al. 2007; Qian et al. 2007). To determine whether RhoA is required for maintaining tumorigenesis stimulated by DLC1 loss, we tested whether suppression of RhoA in DLC1-suppressed hepatoma lines would impact their expansion as subcutaneous tumors in immunocompromised mice. shRNAs capable of down-regulating RhoA to varying degrees (Fig. 5A) decreased the in vivo growth of two independent murine hepatoma lines with undetectable DLC1 (Fig. 5B, cell lines 1,2; Supplemental Fig. S6A,B). Of note, none of the shRNAs completely suppressed RhoA expression, and their ability to limit tumor expansion was proportional to their knockdown efficiency (Supplemental Fig. S6A). The impact of these shRNAs was less pronounced in hepatoma cell lines with higher DLC1 levels (Fig. 5B, cell lines 3,4; Supplemental Fig. S6C,D). Although complete inhibition of RhoA activity might be generally cytostatic (see Piekny et al. 2005), these data suggest that RhoA is required for maintaining the growth of tumors with attenuated DLC1 activity.
Figure 5.
HCC mediated by DLC1 loss requires RhoA and is sensitized to Rho inhibitors. (A) Hepatoma cell line BNL is infected by retroviruses expressing control shRNA (control) or three RhoA shRNAs and immunoblotted for RhoA protein. Tubulin serves as a loading control. Numbers denote relative protein abundance. (B) RhoA shRNAs selectively suppress tumors harboring low levels of DLC1. Relative tumor volume is the last measurement in Figure S6A-D. (*) P < 0.05; (**) P < 0.0005; (#) P > 0.05. DLC1 mRNA level is measured by qRT–PCR and normalized to control liver progenitor cells. (C,D) Loss of DLC1 sensitizes liver cells to ROCK kinase inhibitors Y27632 and Fasudil. Error bars denote SD (n = 3).
RhoA signals through ROCK kinase and other mediators to activate a downstream kinase cascade regulating cell mobility and cytoskeleton remodeling (Boettner and Van Aelst 2002; Sahai and Marshall 2002; Benitah et al. 2004; Jaffe and Hall 2005). As DLC1 loss leads to increased RhoA activation and RhoA is important for DLC1-mediated tumorigenesis, we hypothesized that DLC1 loss might sensitize tumor cells to inhibitors of RhoA effectors. As an initial test of this idea, we compared colony formation of p53−/−;Myc liver progenitor cells infected with control or DLC1 shRNAs in the presence of Y27632 or Fasudil, two distinct small molecule compounds capable of blocking ROCK kinase activity. These agents selectively suppress colony formation in cells harboring a DLC1 shRNA (Fig. 5C,D), although another putative ROCK inhibitor (H1152) was generally toxic (data not shown). Still, these pharmacologic results are consistent with the genetic studies indicating a dependency on Rho signaling in DLC1-deficient cells.
In this study, we combined in vivo RNAi and a mosaic mouse model of HCC to study the impact of DLC1 loss on liver carcinogenesis in mice, which to date has not been possible owing to the embryonic lethality of DLC1 knockout animals. We show that DLC1 loss, when combined with other oncogenic lesions, promotes HCC in vivo and that RhoA activation is both necessary and sufficient for its effects. In our survey of copy number alterations in human tumors, 8p22 deletions encompassing DLC1 occurred in >60% of heptocellular carcinomas as well as a large portion of human lung, breast, and colon carcinomas (see also Durkin et al. 2007). Similarly, RhoA is up-regulated in HCC and many other tumor types (Sahai and Marshall 2002; Fukui et al. 2006). Although other tumor suppressor genes may also reside in the 8p region, our results demonstrate that DLC1 is functionally important and highlight the potential importance of the RhoA signaling network in epithelial cancers.
Molecularly targeted therapies have been devised for inhibiting several oncogenic pathways, including those affected by BCR-ABL, activated Ras and PI3kinase (Downward 2003; Luo et al. 2003). Although tumor suppressors are generally not amenable to direct therapeutic targeting, their mutation may confer a cellular dependency on downstream oncogenic proteins that can be inhibited with small molecule drugs. In this regard, the impact of DLC1 loss may parallel that produced by loss of PTEN, which deregulates the PI3K pathway and can sensitize cells to pharmacological inhibitors of downstream effectors such as mTOR (Maser et al. 2007). Our data indicate that RhoA is required for maintaining at least some tumors driven by DLC1 loss, and that cells with disabled DLC1 are particularly sensitive to inhibitors that target at least one RhoA effector. Clearly, more studies will be required to confirm and extend these observations; nevertheless, the high frequency of DLC1 loss in human cancer implies that pharmacologic intervention of the signaling pathways modulated by DLC1 may have broad therapeutic utility.
Material and methods
Plasmid construction
Two miR30 design shRNAs (codex accession HP_260153 and HP_255554) targeting mouse DLC1 were subcloned from the pSM2 RNAi codex library vector into the MSCV-SV40-GFP vector. The full-length mouse DLC1 was amplified from a RIKEN cDNA (M5C1068G17) and cloned into the MSCV-PGK-PIG vector harboring 6xMyc tag at the N terminus. Constitutive active RhoA (RhoAV14) was cloned into the MSCV-IRES-GFP. Myc was cloned into pWZL-Neo.
Generation and characterization of liver carcinomas
Embyronic heptoblasts were isolated from E13.5 embryos, enriched by an E-cadherin immunoselection, and cultured as described (Zender et al. 2005). Cells with transduced with retroviral vectors (Xue et al. 2007), expanded briefly in culture, and transplanted into the spleen of retrorsine pretreated mice where the progenitor cells engraft the liver (Zender et al. 2005). Animals were treated with CCl4 to stimulate tumor growth (Zender et al. 2006). For subcutaneous tumor growth, 2 × 106 cells were injected into the flanks of nude mice and monitored as described (Zender et al. 2006). Fluorescence and bioluminescence imaging of tumor-bearing mice was as described (Xue et al. 2007). For colony formation assays, 5000 cells were plated; after 6–8 d, colony formation was quantified by the Multigauge software (FujiFilm). Histopathological evaluation of murine liver carcinomas was performed by an experienced pathologist (S.S.). Ki67 staining was performed using standard protocols on paraffin-embedded tumor sections.
Gene activity and expression
RhoA-GTP pull-down assay was performed using EZ-detect GTPase activation kit (Pierce). For immunoblotting, fresh tumor tissue or cell pellets were lysed in Laemmli buffer using a tissue homogenizer. Equal amounts of protein (16 μg) were separated on 10% SDS–polyacrylamide gels and transferred to PVDF membranes. Blots were probed with antibodies against RhoA (1:1000; Santa Cruz Biotechnologies, 26C4), Myc-Tag (1:1000; Abcam, 9E10), or Tubulin (1:5000; B-5-1-2, Sigma). For immunofluorescence, cells were serum-starved for 24 h, fixed, blocked with 5% goat serum, and incubated with 1:1000 Alexa 594-conjugated phalloidin (Invitrogen). Images were captured under a 40× lens of a confocal microscope (Zeiss). To assess mRNA expression, hepatoma cells or tumors were freshly homogenized in Trizol (Gibco) and RNA was purified with Qiagen RNeasy columns and converted to cDNA using TaqMan reverse transcription reagents (Applied Biosystems). qPCR reactions were done in triplicate with SYBR Green PCR Master Mix (Applied Biosystems). The expression level of each gene was normalized to β-actin. Primer sequences can be found in the Supplemental Material.
Gene copy number analysis of human tumors
The ROMA data sets used in this study were obtained by analyzing panels of primary tumors (Hicks et al. 2006; S. Powers, S.W. Lowe, R. Lucito, and M. Wigler, in prep.). Copy-number profiles underwent normalization, segmentation, and masking of frequent copy number polymorphisms (Hicks et al. 2006). Deletion frequencies for DLC1 and other relevant genes were compared with all other genes found in the NCBI Entrez Gene database as described in the Supplemental Material and as well were described in Krasnitz et al. (A. Krasnitz and M. Wigler, in prep.).
Acknowledgments
We thank L. Bianco, B. Ma, M. Yang, and J. Simon for excellent technical assistance. We also thank J. Hicks, N. Popescu (NCI), M. McCurrach, P. Paddison, G. Hannon, P. Schirmacher, and other members of the Lowe and Hannon laboratories for advice and discussions. W.X. is in the MCB graduate program at Stony Brook University. L.Z. is a Seligson clinical fellow. S.L. is a Howard Hughes Medical Institute investigator. This work was generously supported by grant CA13106 from the National Institutes of Health, the German Research Foundation and the Don Monti Foundation.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1672608.
References
- Benitah S.A., Valeron P.F., Van A.L., Marshall C.J., Lacal J.C. Rho GTPases in human cancer: An unresolved link to upstream and downstream transcriptional regulation. Biochim. Biophys. Acta. 2004;1705:121–132. doi: 10.1016/j.bbcan.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Boettner B., Van Aelst L. The role of Rho GTPases in disease development. Gene. 2002;286:155–174. doi: 10.1016/s0378-1119(02)00426-2. [DOI] [PubMed] [Google Scholar]
- Downward J. Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- Durkin M.E., Avner M.R., Huh C.G., Yuan B.Z., Thorgeirsson S.S., Popescu N.C. DLC-1, a Rho GTPase-activating protein with tumor suppressor function, is essential for embryonic development. FEBS Lett. 2005;579:1191–1196. doi: 10.1016/j.febslet.2004.12.090. [DOI] [PubMed] [Google Scholar]
- Durkin M.E., Yuan B.Z., Zhou X., Zimonjic D.B., Lowy D.R., Thorgeirsson S.S., Popescu N.C. DLC-1: A Rho GTPase-activating protein and tumour suppressor. J. Cell. Mol. Med. 2007;11:1185–1207. doi: 10.1111/j.1582-4934.2007.00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui K., Tamura S., Wada A., Kamada Y., Sawai Y., Imanaka K., Kudara T., Shimomura I., Hayashi N. Expression and prognostic role of RhoA GTPases in hepatocellular carcinoma. J. Cancer Res. Clin. Oncol. 2006;132:627–633. doi: 10.1007/s00432-006-0107-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan M., Zhou X., Soulitzis N., Spandidos D.A., Popescu N.C. Aberrant methylation and deacetylation of deleted in liver cancer-1 gene in prostate cancer: Potential clinical applications. Clin. Cancer Res. 2006;12:1412–1419. doi: 10.1158/1078-0432.CCR-05-1906. [DOI] [PubMed] [Google Scholar]
- Healy K.D., Hodgson L., Kim T.Y., Shutes A., Maddileti S., Juliano R.L., Hahn K.M., Harden T.K., Bang Y.J., Der C.J. DLC-1 suppresses non-small cell lung cancer growth and invasion by RhoGAP-dependent and independent mechanisms. Mol. Carcinog. 2008;47:326–337. doi: 10.1002/mc.20389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks J., Krasnitz A., Lakshmi B., Navin N.E., Riggs M., Leibu E., Esposito D., Alexander J., Troge J., Grubor V., et al. Novel patterns of genome rearrangement and their association with survival in breast cancer. Genome Res. 2006;16:1465–1479. doi: 10.1101/gr.5460106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe A.B., Hall A. Rho GTPases: Biochemistry and biology. Annu. Rev. Cell Dev. Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- Jou Y.S., Lee C.S., Chang Y.H., Hsiao C.F., Chen C.F., Chao C.C., Wu L.S., Yeh S.H., Chen D.S., Chen P.J. Clustering of minimal deleted regions reveals distinct genetic pathways of human hepatocellular carcinoma. Cancer Res. 2004;64:3030–3036. doi: 10.1158/0008-5472.can-03-2320. [DOI] [PubMed] [Google Scholar]
- Kim T.Y., Lee J.W., Kim H.P., Jong H.S., Kim T.Y., Jung M., Bang Y.J. DLC-1, a GTPase-activating protein for Rho, is associated with cell proliferation, morphology, and migration in human hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 2007;355:72–77. doi: 10.1016/j.bbrc.2007.01.121. [DOI] [PubMed] [Google Scholar]
- Liao Y.C., Si L., Vere White R.W., Lo S.H. The phosphotyrosine-independent interaction of DLC-1 and the SH2 domain of cten regulates focal adhesion localization and growth suppression activity of DLC-1. J. Cell Biol. 2007;176:43–49. doi: 10.1083/jcb.200608015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucito R., Healy J., Alexander J., Reiner A., Esposito D., Chi M., Rodgers L., Brady A., Sebat J., Troge J., et al. Representational oligonucleotide microarray analysis: A high-resolution method to detect genome copy number variation. Genome Res. 2003;13:2291–2305. doi: 10.1101/gr.1349003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Manning B.D., Cantley L.C. Targeting the PI3K–Akt pathway in human cancer: Rationale and promise. Cancer Cell. 2003;4:257–262. doi: 10.1016/s1535-6108(03)00248-4. [DOI] [PubMed] [Google Scholar]
- Maser R.S., Choudhury B., Campbell P.J., Feng B., Wong K.K., Protopopov A., O’Neil J., Gutierrez A., Ivanova E., Perna I., et al. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;447:966–971. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama H., Pan Y., Oba K., Yoshihiro S., Matsuda K., Hagarth L., Kudren D., Naito K., Bergerheim U.S.R., Ekman P. Deletions on Chromosome 8p22 may predict disease progression as well as pathological staging in prostate cancer. Clin. Cancer Res. 2001;7:3139–3143. [PubMed] [Google Scholar]
- Ng I.O., Liang Z.D., Cao L., Lee T.K. DLC-1 is deleted in primary hepatocellular carcinoma and exerts inhibitory effects on the proliferation of hepatoma cell lines with deleted DLC-1. Cancer Res. 2000;60:6581–6584. [PubMed] [Google Scholar]
- Piekny A., Werner M., Glotzer M. Cytokinesis: Welcome to the Rho zone. Trends Cell Biol. 2005;15:651–658. doi: 10.1016/j.tcb.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Pike B.L., Greiner T.C., Wang X., Weisenburger D.D., Hsu Y.H., Renaud G., Wolfsberg T.G., Kim M., Weisenberger D.J., Siegmund K.D., et al. DNA methylation profiles in diffuse large B-cell lymphoma and their relationship to gene expression status. Leukemia. 2008 doi: 10.1038/leu.2008.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X., Li G., Asmussen H.K., Asnaghi L., Vass W.C., Braverman R., Yamada K.M., Popescu N.C., Papageorge A.G., Lowy D.R. Oncogenic inhibition by a deleted in liver cancer gene requires cooperation between tensin binding and Rho-specific GTPase-activating protein activities. Proc. Natl. Acad. Sci. 2007;104:9012–9017. doi: 10.1073/pnas.0703033104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai E., Marshall C.J. RHO-GTPases and cancer. Nat. Rev. Cancer. 2002;2:133–142. doi: 10.1038/nrc725. [DOI] [PubMed] [Google Scholar]
- Seng T.J., Low J.S., Li H., Cui Y., Goh H.K., Wong M.L., Srivastava G., Sidransky D., Califano J., Steenbergen R.D., et al. The major 8p22 tumor suppressor DLC1 is frequently silenced by methylation in both endemic and sporadic nasopharyngeal, esophageal, and cervical carcinomas, and inhibits tumor cell colony formation. Oncogene. 2007;26:934–944. doi: 10.1038/sj.onc.1209839. [DOI] [PubMed] [Google Scholar]
- Staib F., Hussain S.P., Hofseth L.J., Wang X.W., Harris C.C. TP53 and liver carcinogenesis. Hum. Mutat. 2003;21:201–216. doi: 10.1002/humu.10176. [DOI] [PubMed] [Google Scholar]
- Wong C.M., Lee J.M., Ching Y.P., Jin D.Y., Ng I.O. Genetic and epigenetic alterations of DLC-1 gene in hepatocellular carcinoma. Cancer Res. 2003;63:7646–7651. [PubMed] [Google Scholar]
- Wong C.M., Yam J.W., Ching Y.P., Yau T.O., Leung T.H., Jin D.Y., Ng I.O. Rho GTPase-activating protein deleted in liver cancer suppresses cell proliferation and invasion in hepatocellular carcinoma. Cancer Res. 2005;65:8861–8868. doi: 10.1158/0008-5472.CAN-05-1318. [DOI] [PubMed] [Google Scholar]
- Xue W., Zender L., Miething C., Dickins R.A., Hernando E., Krizhanovsky V., Cordon-Cardo C., Lowe S.W. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Yu Y., Wang N., Chang Y., Ying H., Liu W., He J., Li S., Jiang W., Li Y., et al. LFIRE-1/HFREP-1, a liver-specific gene, is frequently downregulated and has growth suppressor activity in hepatocellular carcinoma. Oncogene. 2004;23:1939–1949. doi: 10.1038/sj.onc.1207306. [DOI] [PubMed] [Google Scholar]
- Ying J., Li H., Murray P., Gao Z., Chen Y.W., Wang Y., Lee K.Y., Chan A.T., Ambinder R.F., Srivastava G., et al. Tumor-specific methylation of the 8p22 tumor suppressor gene DLC1 is an epigenetic biomarker for Hodgkin, nasal NK/T-cell and other types of lymphomas. Epigenetics. 2007;2:15–21. doi: 10.4161/epi.2.1.3883. [DOI] [PubMed] [Google Scholar]
- Yuan B.Z., Miller M.J., Keck C.L., Zimonjic D.B., Thorgeirsson S.S., Popescu N.C. Cloning, characterization, and chromosomal localization of a gene frequently deleted in human liver cancer (DLC-1) homologous to rat RhoGAP. Cancer Res. 1998;58:2196–2199. [PubMed] [Google Scholar]
- Yuan B.Z., Durkin M.E., Popescu N.C. Promoter hypermethylation of DLC-1, a candidate tumor suppressor gene, in several common human cancers. Cancer Genet. Cytogenet. 2003a;140:113–117. doi: 10.1016/s0165-4608(02)00674-x. [DOI] [PubMed] [Google Scholar]
- Yuan B.Z., Zhou X., Durkin M.E., Zimonjic D.B., Gumundsdottir K., Eyfjord J.E., Thorgeirsson S.S., Popescu N.C. DLC-1 gene inhibits human breast cancer cell growth and in vivo tumorigenicity. Oncogene. 2003b;22:445–450. doi: 10.1038/sj.onc.1206064. [DOI] [PubMed] [Google Scholar]
- Yuan B.Z., Jefferson A.M., Baldwin K.T., Thorgeirsson S.S., Popescu N.C., Reynolds S.H. DLC-1 operates as a tumor suppressor gene in human non-small cell lung carcinomas. Oncogene. 2004;23:1405–1411. doi: 10.1038/sj.onc.1207291. [DOI] [PubMed] [Google Scholar]
- Zender L., Xue W., Cordon-Cardo C., Hannon G.J., Lucito R., Powers S., Flemming P., Spector M.S., Lowe S.W. Generation and analysis of genetically defined liver carcinomas derived from bipotential liver progenitors. Cold Spring Harb. Symp. Quant. Biol. 2005;70:251–261. doi: 10.1101/sqb.2005.70.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zender L., Spector M.S., Xue W., Flemming P., Cordon-Cardo C., Silke J., Fan S.T., Luk J.M., Wigler M., Hannon G.J., et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Ying J., Zhang K., Li H., Ng K.M., Zhao Y., He Q., Yang X., Xin D., Liao S.K., et al. Aberrant methylation of the 8p22 tumor suppressor gene DLC1 in renal cell carcinoma. Cancer Lett. 2007;249:220–226. doi: 10.1016/j.canlet.2006.08.019. [DOI] [PubMed] [Google Scholar]
- Zhou X., Thorgeirsson S.S., Popescu N.C. Restoration of DLC-1 gene expression induces apoptosis and inhibits both cell growth and tumorigenicity in human hepatocellular carcinoma cells. Oncogene. 2004;23:1308–1313. doi: 10.1038/sj.onc.1207246. [DOI] [PubMed] [Google Scholar]