Abstract
In the cyanobacterium Synechococcus elongatus PCC 7942, the KaiA, KaiB, and KaiC proteins are essential for the generation of circadian rhythms. Both in vivo and in vitro, phosphorylation of KaiC is regulated positively by KaiA and negatively by KaiB and shows circadian rhythmicity. The autonomous circadian cycling of KaiC phosphorylation is thought to be the basic pacemaker of the circadian clock and to control genome-wide gene expression in cyanobacteria. In this study, we found that temperature-compensated circadian oscillations of gene expression persisted even when KaiC was arrested in the phosphorylated state due to kaiA overexpression. Moreover, two phosphorylation mutants showed transcriptional oscillation with a long period. In kaiA-overexpressing and phosphorylation-deficient strains, KaiC oscillated and transient overexpression of phosphorylation-deficient kaiC reset the phase of the rhythm. These results suggest that transcription- and translation-based oscillations in KaiC abundance are also important for circadian rhythm generation in cyanobacteria. Furthermore, at low temperature, cyanobacteria can show circadian rhythms only when both the KaiC phosphorylation cycle and the transcription and translation cycle are intact. Our findings indicate that multiple coupled oscillatory systems based on the biochemical properties of KaiC are important to maintain robust and precise circadian rhythms in cyanobacteria.
Keywords: Circadian rhythm, cyanobacteria, transcription and translation oscillation, KaiC phosphorylation
Circadian rhythms, biological oscillations of physiological activities with a period of ∼24 h, are found in a wide spectrum of organisms and enhance their fitness in a day/night cycle. Transcription and translation feedback loops, in which the transcription of clock genes is inhibited by their own protein products, are thought to be essential for the generation of circadian rhythms (Young and Kay 2001; Dunlap et al. 2004). Cyanobacteria are the simplest organisms known to exhibit circadian rhythms (Golden et al. 1997). In the cyanobacterium Synechococcus elongatus PCC 7942 (Synechococcus, hereafter), a transcription- and translation-based autoregulatory loop of kaiBC gene expression has been proposed to drive circadian rhythms. Overexpression of kaiC represses kaiBC transcription, while overexpression of kaiA causes an increase in kaiBC transcription. Thus, KaiA and KaiC were, respectively, thought to act as positive and negative regulators of kaiBC expression (Ishiura et al. 1998). Moreover, promoter activity throughout the Synechococcus genome, assayed using bioluminescence reporters, shows circadian rhythmicity (Liu et al. 1995), and overexpression of kaiC eliminates the rhythmic component of all transcription (Nakahira et al. 2004). Thus, rhythmic kaiBC transcription and translation were thought to be essential to generate genome-wide circadian oscillation in Synechococcus transcription (Nakahira et al. 2004).
However, we found that circadian cycling of KaiC phosphorylation persisted when Synechococcus cells were maintained in continuous dark conditions in the absence of transcription and translation (Tomita et al. 2005). Moreover, self-sustained oscillation of KaiC phosphorylation was reconstituted in vitro by incubating KaiC with KaiA, KaiB, and ATP. The period of the in vitro oscillation was stable regardless of changes in temperature, and the period observed in vivo using KaiC mutants was consistent with that in vitro (Nakajima et al. 2005). Therefore, we proposed that the primary pacemaker for the cyanobacterial circadian system is not a transcription and translation feedback loop of kaiBC, but an autonomous oscillation of KaiC phosphorylation (Nakajima et al. 2005).
The mechanism of the in vitro circadian clock system has been studied using biochemical approaches. We determined that KaiA and KaiB rhythmically associate with KaiC (Kageyama et al. 2006). The phosphorylation of two adjoining residues of KaiC is sequentially programmed, and KaiC phosphorylation regulates Kai protein associations to maintain oscillation (Nishiwaki et al. 2007). The oscillation of KaiC phosphorylation is autonomously synchronized and very stable in vitro (Ito et al. 2007). Furthermore, KaiC has a temperature-compensated ATPase activity that correlates with the circadian period (Terauchi et al. 2007). Several theoretical and structural studies of the in vitro cyanobacterial circadian clock system have also been performed (Emberly and Wingreen 2006; Mehra et al. 2006; Takigawa-Imamura and Mochizuki 2006; Clodong et al. 2007; Miyoshi et al. 2007; Mori et al. 2007; Rust et al. 2007; van Zon et al. 2007; Yoda et al. 2007).
In addition to these analyses, it is necessary to analyze the role of the KaiC phosphorylation cycle in living cyanobacterial cells under permissive light conditions because the circadian clock controls many physiological activities under light conditions (Kondo et al. 1993; Liu et al. 1995). Here, we investigated the rhythms in a kaiA-overexpressing strain, in which the cycle of KaiC phosphorylation was impaired, and found that there were circadian rhythms of kaiBC gene expression and KaiB and KaiC protein accumulation. Our observations indicate that rhythmic KaiC phosphorylation is not the only driving force for rhythmicity, and that rhythmic KaiC accumulation may play a part in circadian rhythm generation in cyanobacteria. We also found that circadian rhythms were maintained at low temperature only when both the KaiC phosphorylation cycle and the transcription and translation cycle were intact. Thus, we propose that multiple coupled oscillatory systems are important to maintain precise and robust circadian rhythms in cyanobacteria.
Results
The transcription–translation loop oscillates even when KaiC is constitutively phosphorylated
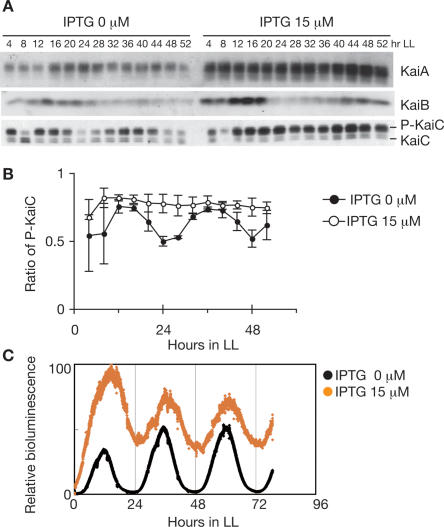
To examine the function of the KaiC phosphorylation cycle in circadian gene expression in Synechococcus, we studied kaiBC expression in a kaiA-inducible strain (ox-kaiA) because KaiA enhances the phosphorylation of KaiC both in vitro and in vivo (Iwasaki et al. 2002; Williams et al. 2002). As shown in Figure 1A, induction by IPTG elevated KaiA levels by 300%, and KaiB and KaiC levels were also increased, likely due to enhanced kaiBC expression.
Figure 1.
Transcription–translation circadian oscillation in ox-kaiA. Cells were grown in a continuous culture system in continuous light (LL). After two 12-h light:12-h dark cycles, cells were returned to LL. Bioluminescence was measured continuously, and cells were collected at the indicated times. (A) Temporal profiles of Kai protein accumulation and KaiC phosphorylation in ox-kaiA cells harboring the PkaiBC∷luxAB reporter with or without 15 μM IPTG. Whole-cell extracts (2.5 μg for KaiA and KaiB, 1.5 μg for KaiC) were subjected to SDS-PAGE and Western blotting. In the bottom panel, the top band corresponds to phosphorylated KaiC (P-KaiC) and the bottom band corresponds to unphosphorylated KaiC (KaiC). (B) Quantification of KaiC phosphorylation in ox-kaiA cells with (open circles) or without (closed circles) 15 μM IPTG. The ratio of phosphorylated KaiC to total KaiC is plotted. The results are shown as means ± SEM (n = 4). (C) Bioluminescence rhythms of ox-kaiA cells under turbidostatic cultivation conditions with (orange circles) or without (black circles) 15 μM IPTG.
In the absence of IPTG, KaiC phosphorylation oscillated (Fig. 1B) with an amplitude smaller than that observed in the wild-type strain (Figs. 4B, 5A, below), likely due to leaky expression of kaiA. As in vitro KaiC phosphorylation oscillation disappears at a higher KaiA concentration (M. Nakajima and T. Kondo, unpubl.), overproduction of KaiA was expected to eliminate the KaiC phosphorylation cycle in vivo. As expected, when ox-kaiA was cultured with 15 μM IPTG, KaiC was constitutively hyperphosphorylated and no rhythmic KaiC phosphorylation was observed (Fig. 1A,B).
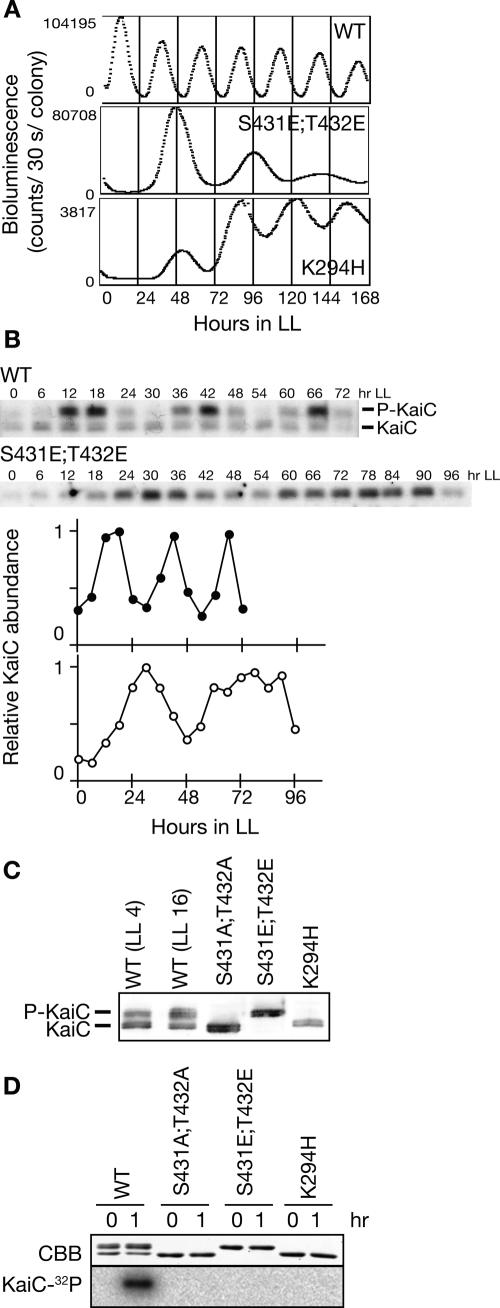
Figure 4.
Transcription–translation oscillation and phosphorylation. (A) Bioluminescence profiles of PkaiBC promoter activity in the wild-type strain (WT) and mutants in the phosphorylation sites [S431E;T432E] and in the Walker-A motif (K294H) are shown. Cells carrying the PkaiBC reporter cassette were grown on solid medium under LL. After a 12-h dark treatment, cells were returned to LL, and bioluminescence was measured. (B) Circadian profile of KaiC accumulation in wild-type and KaiC [S431E;T432E] mutant cells. Cells were grown in a continuous culture system in LL. After two 12-h light:12-h dark cycles, cells were returned to LL and collected at the indicated times. Whole-cell extracts (1.5 μg) were subjected to SDS-PAGE and Western blotting. The top band corresponds to phosphorylated KaiC (P-KaiC) and the bottom band corresponds to unphosphorylated KaiC (KaiC). Densitometric data for wild-type (closed circles) and KaiC [S431E;T432E] (open circles) are shown. Values at peak times were normalized to 1.0. (C) Phosphorylation state of KaiC protein mutated at the autophosphorylation sites ([S431A;T432A] and [S431E;T432E]) or the Walker-A motif (K294H). Proteins were extracted from wild-type (at LL4 and LL16) and mutant (at LL16) strains, and subjected to immunoblotting analysis to determine the phosphorylation state of KaiC. The top bands represent phosphorylated KaiC while the bottom bands represent unphosphorylated KaiC. (D) Autokinase activity of wild-type and KaiC mutant proteins. Wild-type and mutant KaiC were incubated with [γ-32P]ATP for 2 h in the presence of KaiA. The products were separated by SDS-PAGE followed by Coomassie Brilliant Blue (CBB)-staining (top panel) and autoradiography visualized using a BAS-2000 (bottom panel).
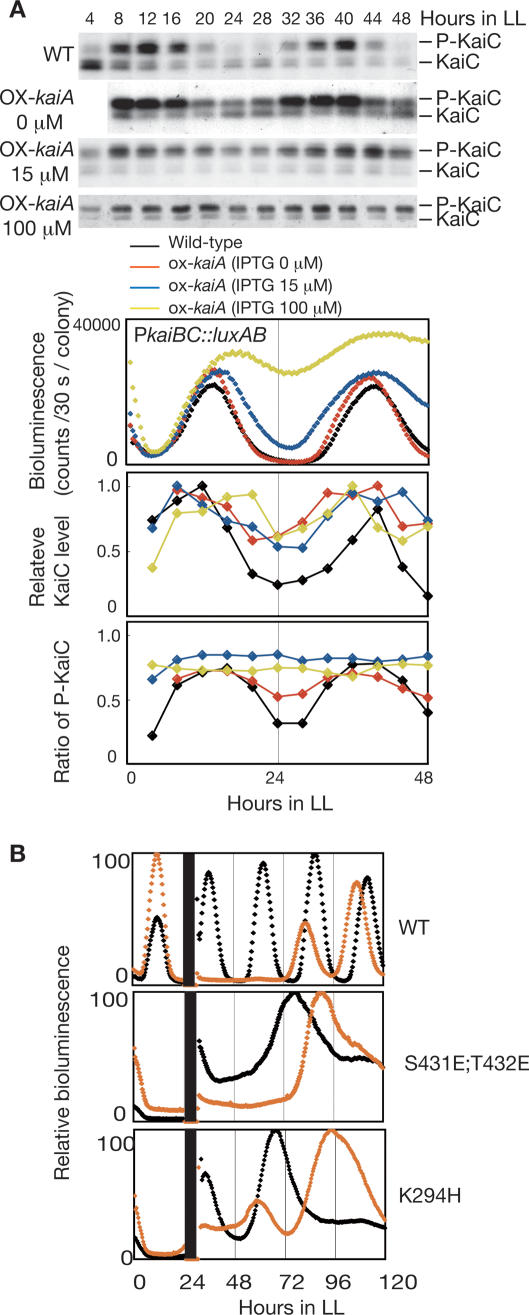
Figure 5.
KaiC accumulation and phase resetting of gene expression rhythms. (A) Comparison of the changes in promoter activity, protein accumulation, and phosphorylation. Temporal profiles of bioluminescence, KaiC accumulation, and phosphorylation were examined in kaiA-overexpressing cells induced with 0, 15, or 100 μM IPTG, and wild-type cells. Whole-cell extracts (1.5 μg) were subjected to SDS-PAGE and Western blotting. The top band corresponds to phosphorylated KaiC (P-KaiC) and the bottom band corresponds to unphosphorylated KaiC (KaiC). Bioluminescence was measured under LL conditions. KaiC accumulation (middle) and KaiC phosphorylation (bottom) profiles were analyzed as in Figure 1A. Values at peak times were normalized to 1.0. The average accumulation of KaiC cannot be compared with each IPTG concentration because assays were performed by independent experiments with different exposure times. (B) Phase shifting after kaiC overexpression by IPTG induction. Bioluminescence rhythms of wild-type cells carrying a wild-type kaiC overexpression construct (top panel), KaiC [S431E;T432E] mutant cells carrying a kaiC [S431E;T432E] overexpression construct (middle panel), and KaiC [K294H] mutant cells carrying a kaiC [K294H] overexpression construct (bottom panel) are shown. Cells were treated with water (black line) or 1 mM IPTG (orange line) for 6 h at LL24.
Since the KaiC phosphorylation cycle is thought to be the pacemaker of the circadian clock in Synechococcus (Nakajima et al. 2005), we did not expect to find rhythmicity in the cells under these conditions. Surprisingly, however, kaiBC promoter activity in liquid culture containing 15 μM IPTG, monitored as bioluminescence, showed an evident rhythm with a circadian period, although the trough of the rhythm was considerably elevated (Fig. 1C). As shown in Figure 1A, the levels of KaiB and KaiC appeared to oscillate by rhythmic gene expression in a similar fashion as in the absence of IPTG or as in the wild-type strain (Figs. 4B, 5A, below; Xu et al. 2000).
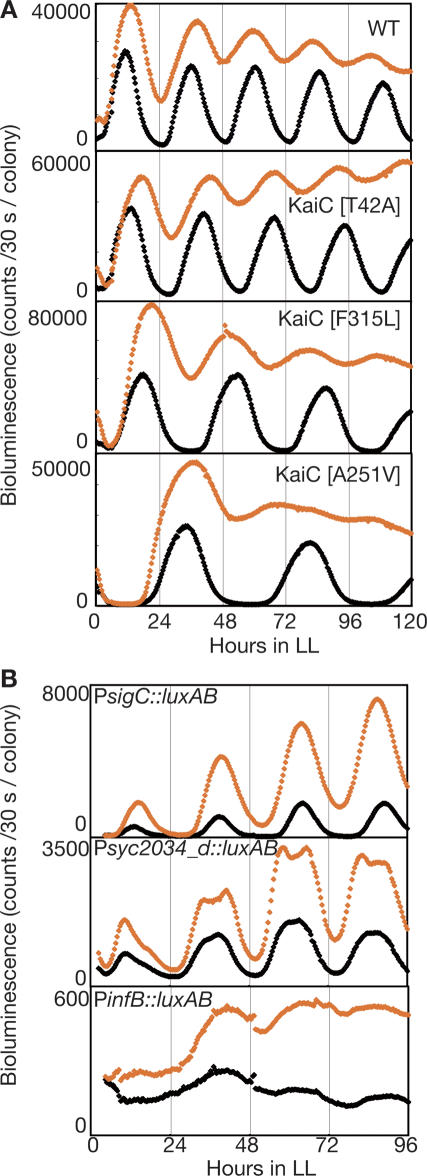
We examined the bioluminescence rhythms of several kaiC mutants in which the free-running periods of gene expression and KaiC phosphorylation were altered similarly (Nakajima et al. 2005; Terauchi et al. 2007; K. Imai and T. Kondo, unpubl.). When kaiA was overexpressed, the rhythms of kaiBC expression in all three mutants had periods that were ∼15% shorter than those observed without kaiA overexpression, as was observed in the wild-type background (Fig. 2A). These results imply that the period of transcriptional rhythm under constitutive KaiC phosphorylation is still regulated by KaiC activity.
Figure 2.
KaiC regulates global circadian transcription rhythms in ox-kaiA. (A) The bioluminescence rhythms of wild-type (WT) and three kaiC point mutants in kaiA-overexpressing cells. Bioluminescence was monitored on solid media with (orange diamonds) or without (black diamonds) 15 μM IPTG under LL. (B) The bioluminescence rhythms of three clones carrying promoter∷luxAB reporters isolated by promoter trapping from ox-kaiA cells. Bioluminescence was monitored on solid media with (orange diamonds) or without 15 μM IPTG (black diamonds) under LL. We confirmed that the upstream regions of sigC, syc2034_d (CYORF ID. Cyanobacteria Gene Annotation Database; http://cyano.genome.jp), or infB were fused to luxAB by sequencing the clones.
We examined the expression of three genes that represent major groups of circadian profiles in cyanobacteria (a high-amplitude group represented by sigC, a low-amplitude group represented by syc2034, and an unstable and low-expression group represented by infB) by bioluminescence reporters under moderate induction of kaiA. As shown in Figure 2B, the expression of all three genes was elevated by moderate induction of kaiA (15 μM IPTG), and the trough of the rhythm was elevated even for high-amplitude genes, such as sigC and kaiBC. However, evident circadian rhythms persisted in the PsigC and Psyc2034 reporter strains, and a weak but significant rhythm was found even in PinfB. These results suggest that KaiC controls global transcription–translation rhythms independent of the KaiC phosphorylation cycle.
Circadian characteristics of the rhythm in the absence of the KaiC phosphorylation cycle
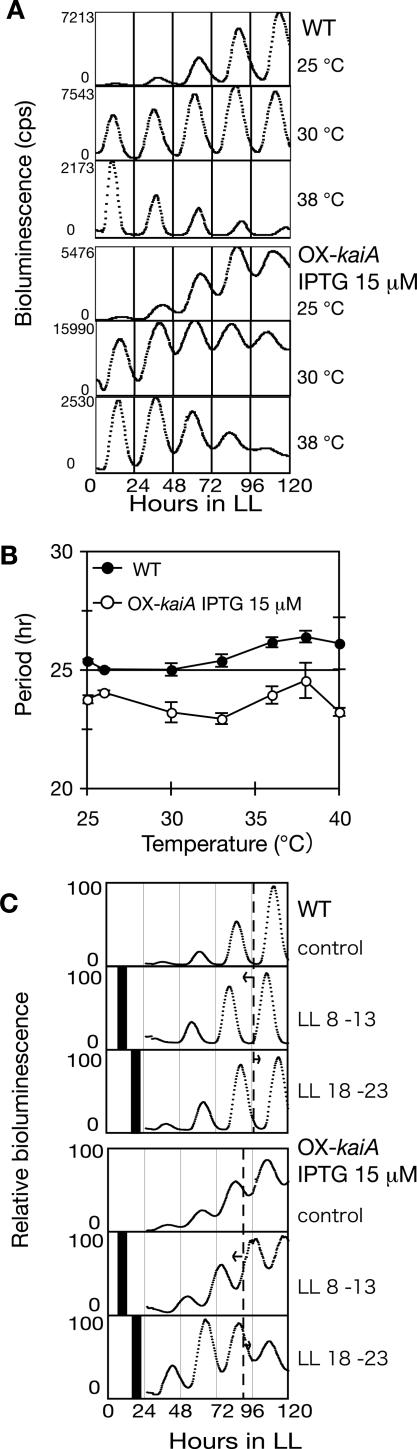
Circadian rhythms are defined as self-sustaining biological rhythms that have ∼24-h periods, which are stable against changes in ambient temperature and can be reset by external cues. The oscillation in transcription and translation observed in this study persisted with a period of ∼24 h. To confirm circadian characteristics of the rhythm in the ox-kaiA strain treated with 15 μM IPTG, we examined the rhythms at different temperatures. The periods in the ox-kaiA strain were slightly shorter than those in the wild-type strain at all temperatures examined, but temperature changes did not seem to alter the period further (Fig. 3A,B).
Figure 3.
Oscillation without rhythmic KaiC phosphorylation has the characteristics of a circadian rhythm. (A) Temperature compensation of period length. Bioluminescence rhythms in the wild-type strain (top panel) and ox-kaiA strain induced with IPTG (bottom panel) at different temperatures. (B) The period of the bioluminescence rhythm was plotted against the culture temperature. Results are shown as means ± SEM (n = 3–6) for wild-type without IPTG (closed circles) and ox-kaiA induced with 15 μM IPTG (open circles). (C) Phase shifting of transcription–translation oscillation by a dark pulse. Cyanobacterial cells were inoculated onto agar plates and kept in LL for 1 d. After synchronization with a single 12-h dark period, the plates were returned to LL. Eight hours or 18 h after the return to LL, samples were placed in the dark for 5 h. After dark treatment, bioluminescence rhythms of wild-type (top panel) and ox-kaiA induced with IPTG (bottom panel) were monitored in LL. Maximum bioluminescence was standardized to 100. Black bars indicate the 5-h dark pulses.
We next examined the effect of a 5-h dark pulse on the phase of bioluminescence rhythms in ox-kaiA cells to determine whether the rhythm could be reset. As shown in Figure 3C, the phase of the rhythm was shifted similarly to the rhythm in wild-type cells. Thus, rhythmicity of transcription–translation in cyanobacteria satisfied the circadian characteristics even when the KaiC phosphorylation cycle was arrested; however, the robustness of the rhythm was weakened.
Transcription–translation can oscillate independent of KaiC phosphorylation
It was formally possible that undetectable levels of the KaiC phosphorylation cycle were sufficient to drive circadian oscillation of transcription–translation. To account for this possibility, we analyzed gene expression in KaiC phosphorylation mutants. KaiC is phosphorylated at Ser-431 and Thr-432 (Nishiwaki et al. 2004). In a constitutively unphosphorylated mutant, in which Ala is substituted for Ser-431 and Thr-432 (S431A;T432A), kaiBC promoter activity fluctuated arrhythmically (Nishiwaki et al. 2004). However, we found that kaiBC promoter activity in a KaiC [S431E;T432E] mutant, in which the phosphorylation sites are substituted by Glu to mimic phosphorylation, showed a dampened but clear rhythm with a period of 48 h (Fig. 4A). The levels of KaiC and KaiB in this mutant fluctuated with a period corresponding to the bioluminescence rhythm (Fig. 4B; data not shown). KaiC phosphorylation did not occur in this mutant, as shown by the detection of KaiC as a single band in vivo (Fig. 4B,C) and lack of γ-32P incorporation into the mutant KaiC in vitro (Fig. 4D).
As shown in Figure 1, transcription–translation oscillated when KaiC was constitutively hyperphosphorylated in induced ox-kaiA cells. These observations implied that phosphorylated KaiC is essential for rhythm generation. Thus, we examined a kaiC [K294H] mutant that lacks autokinase activity in vitro (Fig. 4D; Hayashi et al. 2004). As expected, the KaiC protein in the kaiC [K294H] mutant was detected as a single band corresponding to unphosphorylated KaiC (Fig. 4C). However, as shown in Figure 4A, even in kaiC [K294H] mutant cells, kaiBC promoter activity showed weak but evident rhythms with a long period. Similar rhythms were also reported previously (Nishiwaki et al. 2000; Hayashi et al. 2004). These observations indicate that transcription–translation oscillates even in the absence of the KaiC phosphorylation cycle and that the oscillation can persist regardless of the phosphorylation state and kinase activity of KaiC.
Note that ox-kaiA shows circadian period and KaiC [S431E;T432E] shows long period phenotype (∼48 h); nevertheless, KaiC is hyperphosphorylated in both strains. Furthermore, in a constitutively unphosphorylated mutant KaiC [S431A;T432A], kaiBC promoter activity fluctuated arrhythmically (Nishiwaki et al. 2004), while KaiC [K294H] shows rhythmicity of long period. These data indicate that the period of the transcription–translation oscillation is not regulated by the phosphorylation state of KaiC but by unknown activities of KaiC that were affected by KaiC mutations.
Overexpression of KaiC can reset the phase of rhythm independent of the phosphorylation state
Without the KaiC phosphorylation cycle, what parameter regulates the circadian rhythm of transcription–translation? The oscillation in KaiC abundance accompanied by gene expression rhythms (Figs. 1, 4B) could be a candidate. We therefore compared the changes in kaiBC expression with KaiC accumulation in ox-kaiA induced by various doses of IPTG. As shown in Figure 5A, the KaiC phosphorylation rhythm was lost at IPTG concentrations of 15 μM or higher. In contrast, weak rhythms of kaiBC expression and KaiC accumulation were observed even in the presence of 100 μM IPTG. This observation implies that KaiC accumulation is responsible for the generation of gene expression rhythms.
Previous studies have shown that overproduction of KaiC represses most promoters and abolishes gene expression rhythmicity in Synechococcus (Ishiura et al. 1998; Nakahira et al. 2004), and transient overexpression of KaiC shifted the phase of bioluminescence rhythms (Fig. 5B; Ishiura et al. 1998), indicating that negative feedback regulation of gene expression by KaiC is important for circadian transcription in vivo under physiologically permissive conditions. Thus, we examined whether the expression of phosphorylation-independent mutant KaiC (KaiC [S431E;T432E] or KaiC [K294H]) could regulate gene expression rhythms in respective mutants. Interestingly, overexpression of these KaiC mutants caused large phase shifts in bioluminescence rhythms (Fig. 5B), although the direction and extent of the phase shifts were opposite. These observations suggest that regulation by KaiC abundance is important for gene expression rhythms when the KaiC phosphorylation cycle is absent.
Two oscillations cooperatively function to maintain cellular circadian rhythm
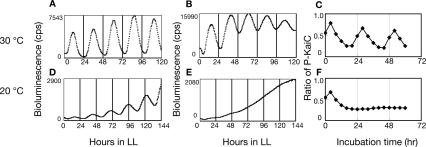
Finally, we examined the relationship between the transcription and translation cycle and the KaiC phosphorylation cycle by changing the ambient temperature. We measured rhythms under three different conditions: (1) in vitro, in which only the KaiC phosphorylation cycle functioned (Fig. 6C,F); (2) in ox-kaiA cells induced with 15 μM IPTG, in which only the transcription and translation cycle functioned (Fig. 6B,E); and (3) in wild-type cells, in which both cycles were intact (Fig. 6A,D). At the standard temperature, 30°C, circadian rhythms were observed for KaiC phosphorylation in vitro and for transcription and translation activity in both ox-kaiA and wild-type strains (Fig. 6A–C). However, at 20°C, no rhythms were observed either in vitro or in ox-kaiA cells induced with 15 μM IPTG (Fig. 6E,F). In contrast, circadian rhythms of kaiBC expression were observed in the wild-type strain even at 20°C (Fig. 6D). These observations indicate that the transcription and translation cycle and the KaiC phosphorylation cycle act cooperatively to maintain circadian rhythm at low temperature.
Figure 6.
Relationship between transcription–translation oscillation and phosphorylation oscillation. Bioluminescence profiles of wild-type cells (A,D), or ox-kaiA cells induced with 15 μM IPTG (B,E) under LL. Bioluminescence profiles were analyzed as in Figure 4A at 30°C (A,B) or 20°C (D,E). (C,F) KaiC phosphorylation profiles in vitro. Recombinant KaiC was incubated with KaiA and KaiB. Aliquots of reaction mixture were taken and subjected to SDS-PAGE and CBB staining. The ratio of phosphorylated KaiC to total KaiC is plotted. Assays were performed at 30°C (C) or 20°C (F).
Discussion
In this study, we demonstrated that KaiA-overexpressing cyanobacteria, which have constitutively phosphorylated KaiC, show circadian gene expression. The period of the rhythm was temperature-compensated and the phase of the rhythm could be reset by a light/dark signal. Using KaiC ([S431E;T432E] and [K294H]) mutants, we confirmed that oscillation of gene expression was generated regardless of the phosphorylation state or kinase activity of KaiC. Furthermore, even in these mutants, KaiC accumulated rhythmically and a pulse of KaiC overexpression reset the phase of the rhythm. Based on these observations, we propose that transcription- and translation-based oscillation in KaiC abundance is an essential contributor to circadian rhythm generation in cyanobacteria.
We have shown previously that circadian cycling of KaiC phosphorylation persists even in the absence of transcription and translation (Tomita et al. 2005). Moreover, we reconstituted the circadian oscillation of KaiC phosphorylation in vitro by mixing the three Kai proteins and ATP (Nakajima et al. 2005). As the period of the KaiC phosphorylation cycle in vitro was temperature-compensated and the periods of KaiC mutants observed in vivo and in vitro were consistent, we assumed that the post-translational oscillator functioned as the basic pacemaker of cyanobacteria (Nakajima et al. 2005). However, in this study, we show that circadian gene expression can be generated in the absence of the KaiC phosphorylation cycle. These results indicate that there are at least two pathways that regulate circadian rhythms in cyanobacteria.
In Neurospora, FRQ-less oscillators (FLO) and FRQ-based oscillators have been proposed (Bell-Pedersen et al. 2005). Although the existence of a Kai-less oscillator cannot be excluded, it should be noted that the transcription and translation rhythm observed here is still dependent on KaiC, because the periods of the transcription and translation rhythm in the absence or presence of the phosphorylation cycle were affected similarly by mutation of KaiC (Fig. 2A). We showed recently that an extremely weak but temperature-compensated KaiC ATPase activity correlated with the period in the cyanobacterial circadian system (Terauchi et al. 2007). This ATP hydrolysis is assumed to be coupled to phosphorylation of KaiC to generate the self-sustained KaiC phosphorylation cycle (Terauchi et al. 2007). As this activity is the most elemental reaction in cyanobacteria reported to satisfy the essential features of a circadian clock, it might be possible to regulate the transcription and translation cycle without driving the KaiC phosphorylation cycle. For example, KaiC is a member of the RecA/DnaB family (Leipe et al. 2000) that hydrolyzes ATP to mechanically manipulate DNA structure (Ye et al. 2004); therefore, KaiC might have ATP-driven DNA-related activity, and its quantitative alteration might regulate rhythmic chromosome compaction (Smith and Williams 2006). Alternatively, KaiC might act as an ATP-driven protease or molecular chaperone in vivo to control the accumulation of Kai proteins. Therefore, we assume that the intrinsic activity of KaiC serves as a basic pacemaker of the circadian clock in cyanobacteria by regulating two oscillations: the KaiC phosphorylation cycle and the transcription and translation cycle.
How do these two pathways function to control the circadian rhythm of global gene expression in cyanobacteria? The signal from the KaiC phosphorylation cycle is transferred to the SasA–RpaA two-component system to generate genome-wide transcriptional rhythms (Takai et al. 2006). Although the particular signaling pathway remains unknown, information about the quantity of KaiC should also be transmitted to genome-wide transcriptional rhythms. For example, cyanobacteria have kaiC-dependent but sasA-independent circadian chromosome compaction; changes in KaiC abundance may regulate this process, resulting in changes in global gene expression (Smith and Williams 2006). LabA, a modulator of negative feedback regulation of KaiC (Taniguchi et al. 2007), may also function to transmit changes in KaiC abundance to transcriptional rhythms.
The KaiC phosphorylation cycle and the transcription and translation cycle appear to influence each other. The period and amplitude of KaiC phosphorylation cycle in vitro change when the concentration or molar ratio of the Kai proteins changes (Kageyama et al. 2006). The period and amplitude of the transcription and translation cycle became shorter and lower in the absence of the KaiC phosphorylation cycle (Figs. 1, 3). High-amplitude rhythm was observed only when both the KaiC phosphorylation cycle and the transcription and translation cycle were intact (Figs. 1, 5). At low temperature, circadian rhythms were observed only when both transcription–translation oscillation and phosphorylation oscillation were intact (Fig. 6). This synergetic effect on cellular rhythmicity indicates that these two processes mutually maintain robust circadian rhythms in cyanobacteria. We previously proposed that post-translational biochemical oscillators, such as the KaiC phosphorylation cycle, act as a timekeeping process to maintain circadian rhythms even in the dark or under metabolically limiting conditions that perturb transcriptional or translational processes (Tomita et al. 2005). On the other hand, the transcription and translation cycle may compensate for the weakness of the KaiC phosphorylation oscillator. In cyanobacterial cells, the concentration and stoichiometry of the Kai proteins fluctuate dynamically due to repeated synthesis and degradation (Imai et al. 2004) and subcellular localization (Kitayama et al. 2003). It is also possible that the proportion of kaiA mRNA to kaiBC mRNA changes under some conditions, as they are expressed from different promoters (Ishiura et al. 1998) and kaiA expression is specifically regulated by Pex, a light/dark resetting protein (Arita et al. 2007; Kutsuna et al. 2007). Although changes in concentration and stoichiometry of the Kai proteins disrupt the circadian rhythm of KaiC phosphorylation in vitro (Kageyama et al. 2006), cyanobacteria show stable and precise circadian rhythms in vivo (Kondo et al. 1993; Mihalcescu et al. 2004). Thus, the transcription and translation cycle may enhance a stability of timekeeping processes when the phosphorylation cycle is subjected to intracellular noises. At low temperature, some processes of both cycles may be blocked in different phases, and they may not oscillate separately; however, they act as “time memory” processes for each other.
Another recent study showed that the KaiC phosphorylation cycle was very stable in vitro and proposed that this resilience contributes to the robustness of the circadian rhythm in cyanobacteria (Ito et al. 2007). We demonstrate here that the actual cellular circadian clock, which operates in a fluctuating and complex environment, requires dual integrated oscillatory processes to maintain robust and precise circadian rhythm. Further study of the mechanisms that control each oscillatory process and elucidation of the communication between them are essential for understanding the entire circadian system.
Materials and methods
Bacterial strains
The Synechococcus elongatus PCC 7942 strains used in this study were wild-type (Nishimura et al. 2002); ox-kaiA, which carries an inducible kaiA gene, in wild-type background (Kutsuna et al. 2007); KaiC [K294H] (Nishiwaki et al. 2000); and KaiC [S431E;T432E] (this study). The KaiC ORF in pCkaiABC (Ishiura et al. 1998) was mutagenized by PCR using the overlap extension method to change residues S431 and T432 to glutamate as described previously (Nishiwaki et al. 2000). The primers used for mutagenesis were 5′-TCCCATATCGAAGAAAT TACGGATACG-3′ and 5′-CGTATCCGTAATTTCTTCGATA TGGGA-3′. The mutated pCkaiABC was introduced into the kaiABC-deficient strain, NUC43 (Nishimura et al. 2002).
For overexpression of kaiC, wild-type, KaiC [S431E;T432E], or KaiC [K294H] strains were transformed with overexpression plasmids for wild-type or mutated kaiC, which were constructed as reported previously (Nishiwaki et al. 2004).
KaiC [T42S] (Nakajima et al. 2005), KaiC [F315L] (K. Imai and T. Kondo, unpubl.), and KaiC [A251V] (Terauchi et al. 2007) mutants were generated by PCR-based mutagenesis as described (Nakajima et al. 2005). Briefly, kaiC was amplified by PCR, using pCkaiABC as a template. The mutated pCkaiABC was introduced to NUC43. We also generated kaiA-overexpressing cells from KaiC [T42S], KaiC [F315L], and KaiC [A251V] mutant backgrounds.
All strains carry the PkaiBC∷luxAB reporter construct at neutral site I. KaiC overexpression was performed as described previously (Ishiura et al. 1998)
Culture conditions and measurement of bioluminescence
Cells were cultured at 30°C in a continuous dilution culture system in BG-11 liquid medium to maintain an OD730 of 0.2 under continuous light (LL) as described previously (Iwasaki et al. 2002). Cultures were synchronized in 12-h light and 12-h dark cycles (12L12D) and returned to free-running LL conditions. Cells were then collected at 4-h or 6-h intervals. To measure bioluminescence rhythms, aliquots were drawn continuously using a peristaltic pump and were passed automatically under the detector of a photomultiplier tube. The bioluminescence of colonies grown on solid BG-11 plates was monitored with a photomultiplier tube-based system (Kondo et al. 1993). Briefly, Synechococcus cells were inoculated onto BG-11 agar plates with or without IPTG and incubated under LL conditions to form colonies. The cells were then subjected to 12 h of darkness to synchronize the circadian clock, and bioluminescence was monitored automatically under LL.
Western blotting analysis
Synechococcus whole-cell extracts were prepared as described previously (Kitayama et al. 2003). Protein concentration was determined by the BCA method using bovine serum albumin as a standard. Whole-cell extracts were subjected to SDS-PAGE and transferred onto nitrocellulose membranes. The blots were then incubated with anti-KaiA, anti-KaiB, and anti-KaiC antibodies (Iwasaki et al. 1999), and protein was detected by enhanced chemiluminescence.
Promoter trap screening
ox-kaiA without the luciferase reporter gene was generated by transforming reporter-less wild-type Synechococcus with pTS2Kptrc∷kaiA. The strain was then transformed with the promoter trap library (Nakahira et al. 2004). Bioluminescent colonies were screened using a cooled CCD camera system (Kondo et al. 1994). Among 2.5 × 104 Cm-resistant colonies, 342 bright bioluminescent colonies were isolated.
Purification of recombinant Kai proteins
Recombinant KaiA, KaiB, and KaiC proteins were purified as described by Nishiwaki et al. (2007). Briefly, recombinant GST fusion Kai proteins were produced in Escherichia coli BL21, followed by purification with glutathione-Sepharose beads (GE Healthcare). After removal of the GST tag with the PreScission Protease (GE Healthcare), proteins were further purified by Resource Q anion exchange chromatography (GE Healthcare) and gel filtration chromatography.
Reconstitution of KaiC phosphorylation in vitro
Reconstitution of the KaiC phosphorylation cycle in vitro was performed as described (Nakajima et al. 2005). Briefly, KaiC was incubated at 30°C or 20°C with KaiA and KaiB in buffer (20 mM Tris-HCl at pH 8.0, 150 mM NaCl, 5 mM MgCl2) containing 1 mM ATP. The final concentrations of KaiA, KaiB, and KaiC were 1.2, 3.5, and 3.5 μM, respectively. Every 4 h, aliquots (3 μL) of the reaction mixture were taken and subjected to SDS-PAGE and Coomassie Brilliant Blue (CBB) staining.
KaiC autokinase assay
Autophosphorylation assays were performed at 30°C. We incubated KaiC (210 pmol) and KaiA (72 pmol) in 30 μL of reaction buffer consisting of 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1 mM [γ-32P]ATP (12.3 GBq/mmol), and 5 mM MgCl2. The reaction was terminated by adding SDS sample buffer, and the protein was separated by SDS-PAGE and detected by autoradiography using a BAS-2000 bioimaging analyzer system (Fuji).
Acknowledgments
We thank Reiko Kiyohara and Takafumi Okada for technical assistance, Keiko Imai for providing the kaiC point mutants, and other members of the laboratory for helpful comments. This research was supported in part by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan (15GS0308 to T.K., 19770029 to Y.K., and 19042011 to K.T.). Analysis of DNA sequences was performed in conjunction with the Life Research Support Center at Akita Prefectural University.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1661808.
References
- Arita K., Hashimoto H., Igari K., Akaboshi M., Kutusna S., Sato M., Shimizu T. Structural and biochemical characterization of a cyanobacterium circadian clock-modifier protein. J. Biol. Chem. 2007;282:1128–1135. doi: 10.1074/jbc.M608148200. [DOI] [PubMed] [Google Scholar]
- Bell-Pedersen D., Cassone V.M., Earnest D.J., Golden S.S., Hardin P.E., Thomas T.L., Zoran M.J. Circadian rhythms from multiple oscillators: Lessons from diverse organisms. Nat. Rev. Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clodong S., Duhring U., Kronk L., Wilde A., Axmann I., Herzel H., Kollmann M. Functioning and robustness of a bacterial circadian clock. Mol. Syst. Biol. 2007;3:90. doi: 10.1038/msb4100128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap J.C., Loros J.J., DeCoursey P.J. Chronobiology: Biological timekeeping. Sinauer; Sunderland, MA: 2004. [Google Scholar]
- Emberly E., Wingreen N.S. Hourglass model for a protein-based circadian oscillator. Phys. Rev. Lett. 2006;96:038303. doi: 10.1103/PhysRevLett.96.038303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden S.S., Ishiura M., Johnson C.H., Kondo T. Cyanobacterial circadian rhythm. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997;48:327–354. doi: 10.1146/annurev.arplant.48.1.327. [DOI] [PubMed] [Google Scholar]
- Hayashi F., Itoh N., Uzumaki T., Iwase R., Tsuchiya Y., Yamakawa H., Morishita M., Onai K., Itoh S., Ishiura M. Roles of two ATPase-motif-containing domains in cyanobacterial circadian clock protein KaiC. J. Biol. Chem. 2004;279:52331–52337. doi: 10.1074/jbc.M406604200. [DOI] [PubMed] [Google Scholar]
- Imai K., Nishiwaki T., Kondo T., Iwasaki H. Circadian rhythms in the synthesis and degradation of a master clock protein KaiC in cyanobacteria. J. Biol. Chem. 2004;279:36534–36539. doi: 10.1074/jbc.M405861200. [DOI] [PubMed] [Google Scholar]
- Ishiura M., Kutsuna S., Aoki S., Iwasaki H., Andersson C.R., Tanabe A., Golden S.S., Johnson C.H., Kondo T. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science. 1998;281:1519–1523. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- Ito H., Kageyama H., Mutsuda M., Nakajima M., Oyama T., Kondo T. Autonomous synchronization of the circadian KaiC phosphorylation rhythm. Nat. Struct. Mol. Biol. 2007;14:1084–1088. doi: 10.1038/nsmb1312. [DOI] [PubMed] [Google Scholar]
- Iwasaki H., Taniguchi Y., Ishiura M., Kondo T. Physical interactions among circadian clock proteins KaiA, KaiB and KaiC in cyanobacteria. EMBO J. 1999;18:1137–1145. doi: 10.1093/emboj/18.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H., Nishiwaki T., Kitayama Y., Nakajima M., Kondo T. KaiA-stimulated KaiC phosphorylation in circadian timing loops in cyanobacteria. Proc. Natl. Acad. Sci. 2002;99:15788–15793. doi: 10.1073/pnas.222467299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama H., Nishiwaki T., Nakajima M., Iwasaki H., Oyama T., Kondo T. Cyanobacterial circadian clock: Kai protein complex dynamics in KaiC phosphorylation cycle in vitro. Mol. Cell. 2006;23:161–171. doi: 10.1016/j.molcel.2006.05.039. [DOI] [PubMed] [Google Scholar]
- Kitayama Y., Iwasaki H., Nishiwaki T., Kondo T. KaiB functions as and attenuation of KaiC phosphorylation in the cyanobacterial circadian clock system. EMBO J. 2003;22:2127–2134. doi: 10.1093/emboj/cdg212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T., Strayer C.A., Kulkarni R.D., Taylor W., Ishiura M., Golden S.S., Johnson C.H. Circadian rhythms in prokaryotes: Luciferase as a reporter of circadian gene expression in cyanobacteria. Proc. Natl. Acad. Sci. 1993;90:5672–5676. doi: 10.1073/pnas.90.12.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T., Tsinoremas N.F., Golden S.S., Johnson C.H., Kustuna S., Ishiura M. Circadian clock mutant of cyanobacteria. Science. 1994;266:1233–1236. doi: 10.1126/science.7973706. [DOI] [PubMed] [Google Scholar]
- Kutsuna S., Kondo T., Ikegami H., Uzumaki T., Katayama M., Ishiura M. The circadian clock-related gene pex regulates a negative cis element in the kaiA promoter region. J. Bacteriol. 2007;189:7690–7696. doi: 10.1128/JB.00835-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipe D.D., Aravid L., Grishin N.V., Koonin E.V. The bacterial replicative helicase DnaB evolved from a RecA duplication. Genome Res. 2000;10:5–16. [PubMed] [Google Scholar]
- Liu Y., Tsinoremas N.F., Johnson C.H., Lebedeva N.V., Golden S.S., Ishiura M., Kondo T. Circadian orchestration of gene expression in cyanobacteria. Genes & Dev. 1995;9:1469–1478. doi: 10.1101/gad.9.12.1469. [DOI] [PubMed] [Google Scholar]
- Mehra A., Hong C.I., Shi M., Loros J.J., Dunlap J.C., Ruoff P. Circadian rhythmicity by autocatalysis. PLoS Comput. Biol. 2006;2:e96. doi: 10.1371/journal.pcbi.0020096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihalcescu I., Hsing W., Leibler S. Resilient circadian oscillator revealed in individual cyanobacteria. Nature. 2004;430:81–85. doi: 10.1038/nature02533. [DOI] [PubMed] [Google Scholar]
- Miyoshi F., Nakayama Y., Kaizu K., Iwasaki H., Tomita M. A mathematical model for the Kai-protein-based chemical oscillator and clock gene expression rhythms in cyanobacteria. J. Biol. Rhythms. 2007;22:69–80. doi: 10.1177/0748730406295749. [DOI] [PubMed] [Google Scholar]
- Mori T., Williams D.R., Byrne M.O., Qin X., Egli M., Mchaourab H.S., Stewart P.L., Johnson C.H. Elucidating the ticking of an in vitro circadian clockwork. PLoS Biol. 2007;5:e93. doi: 10.1371/journal.pbio.0050093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahira Y., Katayama M., Miyashita H., Kutsuna S., Iwasaki H., Oyama T., Kondo T. Global gene repression gy KaiC as a master process of prokaryotic circadian system. Proc. Natl. Acad. Sci. 2004;101:881–885. doi: 10.1073/pnas.0307411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima M., Imai K., Ito H., Nishiwaki T., Murayama Y., Iwasaki H., Oyama T., Kondo T. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- Nishimura H., Nakahira Y., Imai K., Tsuruhara A., Kondo H., Hayashi H., Hirai M., Saito H., Kondo T. Mutations in KaiA, a clock protein, extend the period of circadian rhythm in the cyanobacterium Synechococcus elongatus PCC 7942. Microbiology. 2002;148:2903–2909. doi: 10.1099/00221287-148-9-2903. [DOI] [PubMed] [Google Scholar]
- Nishiwaki T., Iwasaki H., Ishiura M.T., Kondo T. Role of KaiC nucleotide binding and autophosphorylation of the clock protein KaiC as a circadian timing process of cyanobacteria. Proc. Natl. Acad. Sci. 2000;97:495–499. doi: 10.1073/pnas.97.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiwaki T., Satomi Y., Nakajima M., Lee C., Kiyohara R., Kageyama H., Kitayama Y., Temamoto M., Yamaguchi A., Hijikata A., et al. Role of KaiC phosphorylation in the circadian clock system of Synechococcus elongatus PCC 7942. Proc. Natl. Acad. Sci. 2004;101:13927–13932. doi: 10.1073/pnas.0403906101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiwaki T., Satomi Y., Kitayama Y., Terauchi K., Kiyohara R., Takao T., Kondo T. A sequential program of dual phosphorylation of KaiC as a basis for circadian rhythm in cyanobacteria. EMBO J. 2007;26:4029–4037. doi: 10.1038/sj.emboj.7601832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust M.J., Markson J.S., Lane W.S., Fisher D.S., O’Shea E.K. Ordered phosphorylation governs oscillation of a three-protein circadian clock. Science. 2007;318:809–812. doi: 10.1126/science.1148596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R.M., Williams S.B. Circadian rhythm in gene transcription imparted by chromosome compaction in the cyanobacterium Synechococcus elongatus. Proc. Natl. Acad. Sci. 2006;103:8564–8569. doi: 10.1073/pnas.0508696103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai N., Nakajima M., Oyama T., Kito R., Sugita C., Sugita M., Kondo T., Iwasaki T. A KaiC-associating SasA–RpaA two-component regulatory system as a major circadian timing mediator in cyanobacteria. Proc. Natl. Acad. Sci. 2006;103:12109–12114. doi: 10.1073/pnas.0602955103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takigawa-Imamura H., Mochizuki A. Predicting regulation of the phosphorylation cycle of KaiC clock protein using mathematical analysis. J. Biol. Rhythms. 2006;21:405–416. doi: 10.1177/0748730406291329. [DOI] [PubMed] [Google Scholar]
- Taniguchi Y., Katayama M., Ito R., Takai N., Kondo T., Oyama T. labA: A novel gene required for negative feedback regulation of the cyanobacterial circadian clock protein KaiC. Genes & Dev. 2007;21:60–70. doi: 10.1101/gad.1488107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terauchi K., Kitayama Y., Nishiwaki T., Miwa K., Murayama Y., Oyama T., Kondo T. The ATPase activity of KaiC determines the basic timing for circadian clock of cyanobacteria. Proc. Natl. Acad. Sci. 2007;104:16377–16381. doi: 10.1073/pnas.0706292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita J., Nakajima M., Kondo T., Iwasaki H. No transcription–translation feedback in circadian rhythm of KaiC phosphorylation. Science. 2005;307:251–254. doi: 10.1126/science.1102540. [DOI] [PubMed] [Google Scholar]
- van Zon J.S., Lubensky D.K., ten Wolde Altena P.R. An allosteric model of circadian KaiC phosphorylation. Proc. Natl. Acad. Sci. 2007;104:7420–7425. doi: 10.1073/pnas.0608665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S.B., Vakonakis I., Golden S.S., LiWang A.C. Structure and function from the circadian clock protein KaiA of Synechococcus elongatus reveals a clock input mechanism. Proc. Natl. Acad. Sci. 2002;99:15357–15362. doi: 10.1073/pnas.232517099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Mori T., Johnson C.H. Circadian clock-protein expression in cyanobacteria: Rhythms and phase setting. EMBO J. 2000;19:3349–3357. doi: 10.1093/emboj/19.13.3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Osborne A.R., Groll M., Rapoport T.A. RecA-like motor ATPase lessons from structures. Biochim. Biophys. Acta. 2004;1659:1–18. doi: 10.1016/j.bbabio.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Yoda M., Eguchi K., Terada T.P., Sasai M. Monomer-shuffling and allosteric transition in KaiC circadian oscillation. PLoS ONE. 2007;2:e408. doi: 10.1371/journal.pone.0000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M.W., Kay S.A. Time zones: A comparative genetics of circadian clocks. Nat. Rev. Genet. 2001;2:702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]