Abstract
Circadian clocks regulate many different physiological processes and synchronize these to environmental light:dark cycles. In Drosophila, light is transmitted to the clock by a circadian blue light photoreceptor CRYPTOCHROME (CRY). In response to light, CRY promotes the degradation of the circadian clock protein TIMELESS (TIM) and then is itself degraded. To identify novel genes involved in circadian entrainment, we performed an unbiased genome-wide screen in Drosophila cells using a sensitive and quantitative assay that measures light-induced degradation of CRY. We systematically knocked down the expression of ∼21,000 genes and identified those that regulate CRY stability. These genes include ubiquitin ligases, signal transduction molecules, and redox molecules. Many of the genes identified in the screen are specific for CRY degradation and do not affect degradation of the TIM protein in response to light, suggesting that, for the most part, these two pathways are distinct. We further validated the effect of three candidate genes on CRY stability in vivo by assaying flies mutant for each of these genes. This work identifies a novel regulatory network involved in light-dependent CRY degradation and demonstrates the power of a genome-wide RNAi approach for understanding circadian biology.
Keywords: RNAi, circadian rhythms, clock proteins, degradation, photoreceptors
The molecular circadian clock consists of feedback loops in which specific clock proteins regulate transcription of their own mRNAs, thereby maintaining cycles of gene expression. The major loop in Drosophila is comprised of the PERIOD (PER) and TIMELESS (TIM) proteins, both of which are expressed cyclically and rhythmically regulate transcription by inhibiting the activity of transcriptional activators CLOCK (CLK) and CYCLE (CYC) (Ashmore and Sehgal 2003; Stanewsky 2003; Hardin 2006). The cycling of PER and TIM is essential for circadian rhythms and is thought to provide timekeeping cues for the organism. Consistent with the idea that levels of PER–TIM constitute time-of-day signals, light sets the time of the clock by altering levels of these molecules. Thus, entrainment of the molecular clock to light is mediated by light-induced degradation of the TIM protein. In response to light, TIM is targeted to the ubiquitin–proteasome pathway by an E3 ubiquitin ligase complex containing the F-box protein JETLAG (JET) (Naidoo et al. 1999; Koh et al. 2006). The ubiquitin–proteasome pathway may also affect light-independent PER–TIM degradation, which maintains cycling under free-running conditions (Grima et al. 2002; Ko et al. 2002). Degradation of TIM in response to light, as well as in free-running conditions, is affected by its phosphorylation state (Zeng et al. 1996; Naidoo et al. 1999; Grima et al. 2002; Yuan et al. 2005).
Photic signals are transmitted to TIM by a dedicated circadian photoreceptor, CRYPTOCHROME (CRY) (Cashmore 2003; Lin and Todo 2005). After signaling to TIM, CRY is itself degraded by the proteasome pathway (Lin et al. 2001), However the role of light-dependent CRY degradation in regulating circadian behavior and resetting is not clear. According to one report, clock neuron-specific overexpression of CRY increases circadian photo sensitivity (Emery et al. 2000), while another study found that overexpression of CRY reduces photo sensitivity (Ishikawa et al. 1999). A CRY mutant (CRYm) lacking the C-terminal domain required for the regulation of stability is able to transmit photic signals at low light intensity but is unstable and entrainment-defective at normal light intensity; thus, the requirement of light-induced CRY degradation for entrainment is not directly addressed (Busza et al. 2004). Regardless of the role for light-induced degradation of CRY, it is clear that activation of CRY by light, which leads to its degradation, is essential for entrainment of the clock. Interestingly, CRY activation involves intramolecular electron transfer and presumably subsequent conformational changes, so cellular redox status also regulates the transfer of photic information and CRY stability (Lin et al. 2001).
In addition to its role in entrainment, CRY functions as an integral component of peripheral clocks found outside the brain in Drosophila (Ivanchenko et al. 2001; Krishnan et al. 2001; Collins et al. 2006). Likewise in mammals, the two CRY orthologs are essential components of both central and peripheral clocks, where they, rather than TIM, act as partners of PER (Kume et al. 1999; Shearman et al. 2000). Importantly, both CRYs in mammals are expressed with a robust circadian rhythm, suggesting that they are central timekeeping molecules. Thus, the identification of molecules that regulate CRY is an important objective, since it should reveal mechanisms that sustain free-running clock function as well as those that entrain the clock.
In the current study, we sought to identify novel genes involved in the photic input pathway in Drosophila, using light-induced CRY degradation as an end point assay. We performed a genome-wide RNAi screen in an S2 cell culture system and identified 190 candidate genes involved in the regulation of CRY in response to light. Most of these genes are components of the proteasome, ubiquitin ligases, signal transduction molecules, or redox molecules. We validated the effects of the selected candidates on CRY degradation through Western blot analysis. To further understand their role in the circadian light entrainment pathway, we examined their effect on TIM degradation in response to light. Using mutant flies, we obtained in vivo validation for the function of an E2–E3 ubiquitin ligase, Bruce; a dual specific phosphatase, slingshot (ssh); and a novel HECT domain-containing E3 ligase, CG17735, in the regulation of CRY. Since the screen identified several ubiquitin ligases that specifically affect CRY degradation without affecting TIM degradation, we propose that once CRY is activated by light, it targets TIM for degradation, following which the pathways for CRY and TIM degradation are distinct.
Results
A high-throughput assay for light-dependent CRY degradation in Drosophila Schneider 2 (S2) cells
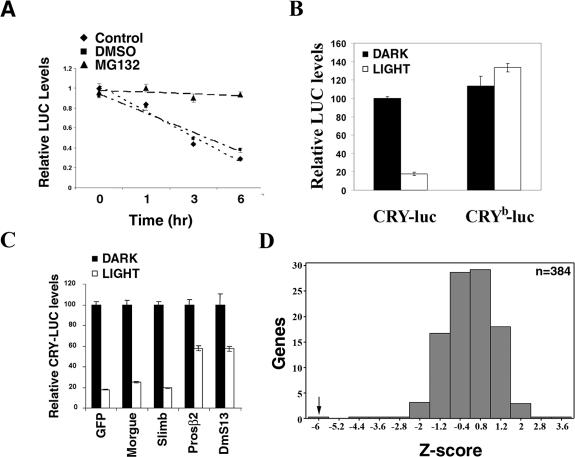
We previously reported that CRY is degraded by light in Drosophila S2 cells (Lin et al. 2001). This suggested an assay for a high-throughput cell-based genomic screen. To develop a robust, high-throughput assay for determining CRY stability in S2 cells, we generated N-terminal or C-terminal luciferase fusion proteins of CRY and monitored luciferase activity driven by these constructs in the presence and absence of light. As seen for CRY itself, luciferase activity from each of the fusion proteins decreased in a light-dependent manner (Fig. 1A; data not shown). Incubation with a proteasome inhibitor, MG132, blocked this light-dependent CRY-luc degradation (Fig. 1A). We also quantified protein levels by Western blot analysis and observed a corresponding decrease in the levels of the CRY-luc fusion proteins (data not shown). However, the luciferase assay was more sensitive and amenable to high-throughput screening. As a control, we generated a luciferase fusion protein of an inactive form of CRY, CRYb, which has a point mutation (D410N) in the flavin-binding pocket that renders it insensitive to light (Stanewsky et al. 1998; Ceriani et al. 1999; Busza et al. 2004). Consistent with other assays indicating lack of light responsiveness of the CRYb protein, the CRYb-luc fusion protein was insensitive to light (Fig. 1B).
Figure 1.
Development of a primary screen to identify novel components involved in light-dependent CRY degradation. (A) Light-dependent degradation of a CRY-luc fusion protein: S2R+ cells were transfected with expression constructs for a Cry-luc fusion protein and Actin-Renilla luciferase. Cells were exposed to light in the presence of DMSO (control) or proteasome inhibitor MG132 (100 μM) for the indicated times, and luciferase activity was monitored. Luciferase activity was normalized to that of cells kept in darkness, set as 1. Average values from three samples are plotted, and the error bars depict standard error of the mean (SEM). The results are representative of two independent experiments. Light-dependent degradation of the CRY-luciferase fusion protein was blocked by proteasome inhibitor MG132. (B) A single point mutation in the flavin-binding pocket of CRY, CRYb, renders it insensitive to light. S2R+ cells were transfected with expression constructs for CRY-luc or CRYb-luc fusion protein in 384-well plates and assayed for luciferase activity after light exposure for 6 h. Luciferase activity from three samples is plotted relative to the CRY activity in transfected cells kept in the dark (set to 100%); error bars represent SEM. In response to light exposure, activity of the CRY-luc fusion protein was reduced by 83% (P-value = 2 × 10−9), while activity of the CRYb-luc fusion protein was unchanged. Similar results were obtained in three experiments with different cell densities. (C) RNAi of 26S proteasome components blocks light-dependent CRY degradation. S2R+ cells were cotransfected with the CRY-luc expression construct and dsRNAs, as indicated, in 384-well plates. Act-Renilla luc was included as a transfection control. Ninety-six hours post-transfection, cells were exposed to light and luciferase activity was measured as indicated above. Relative luciferase activity from three samples is plotted; error bars represent SEM. The value obtained with dsRNA against gfp in the dark was set as 100% and used for normalization. dsRNA against cry, which dramatically reduced luciferase activity (data not shown), was used as a positive control. RNAi of the 26S proteasome core components Prosβ2 and DmS13 (also called as Rpn11) significantly blocked light-dependent CRY-luc degradation. Similar results were obtained in two independent experiments. (D) A pilot screen demonstrates the validity of the CRY-luc assay. The light-dependent CRY-luc degradation assay was performed in S2R+ cells with 380 randomly picked dsRNAs from the genomic collection. dsRNA against the proteasome component Prosβ2 was used as a positive control; thread dsRNA was included as a control to verify the efficiency of RNAi. Normalized luciferase levels were compared between the dark- and light-treated plates. Z-scores, standard deviations from the plate mean (see the Materials and Methods for details), are plotted. While dsRNA against Prosβ2 significantly blocked light-dependent CRY-luc degradation (Z-score = −6.2), most dsRNAs had no effect. Similar results were obtained in two independent experiments.
We next optimized conditions for RNA-mediated interference using various S2 cell derivatives. Since other high-throughput screens in Drosophila have used S2R+ cells with dsRNA libraries plated in 384-well plates, we needed to develop a robust CRY degradation assay under these conditions. We optimized assay conditions in S2R+ cells using dsRNA against proteasome components Prosβ2 and DmS13. DmS13 (also called Rpn11) is a component of the 19S regulatory lid complex, and Prosβ2 is an essential component of the 20S proteolytic core complex. dsRNA against each of these proteasome components significantly blocked light-dependent CRY degradation. On the other hand, dsRNAs against Slimb, an E3 ligase for PER (Ko et al. 2002), or morgue, a putative SCF component that may be involved in the circadian entrainment pathway (Murad et al. 2007), had no effect (Fig. 1C). The robustness of the assay was tested in a miniscreen in which we examined the effect of dsRNAs against a random collection of 380 genes on light-dependent CRY degradation. dsRNA against the proteasome component Prosβ2 was used as a positive control. In order to quantify the effects of the dsRNAs we determined Z-scores as a measure of the effect on CRY degradation (see the Materials and Methods for details regarding Z-scores). The Z-score for the Prosβ2 dsRNA indicated that the efficiency of CRY degradation was reduced such that it was 6.2 standard deviations away from the mean (Fig. 1D), reflecting the sensitivity and robustness of this assay. As expected, most of the dsRNAs did not score positive, but dsRNAs against four genes showed small effects, including one against a mono-oxygenase in the ubiquitination pathway (CG7277) that gave a Z-score of less than −2 (Fig. 1D). Similar results were observed in a duplicate experiment (data not shown). dsRNA against thread, which is essential for S2 cell survival, promoted light-dependent CRY degradation (Z-score +3.2) (Fig. 1D), suggesting that dsRNAs that promote apoptosis and cell death may indirectly promote CRY-luc reporter degradation. Since it is very difficult to distinguish the effect of dsRNAs that affect cell health from those that actively promote CRY degradation, we confined our analysis to dsRNAs that block light-dependent CRY degradation (i.e., Z-score less than −2). Our ability to identify ubiquitin–proteasome components from a random selection of dsRNAs validated our assay, indicating that it was capable of identifying novel genes involved in regulating CRY stability.
Genome-wide RNAi screen for components involved in the light-dependent CRY degradation pathway
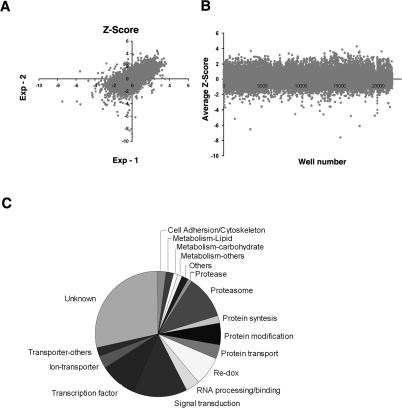
Having developed a robust assay, a genome-wide RNAi library was screened in duplicate using S2R+ cells to identify dsRNAs that block light-dependent CRY degradation. Analysis of the Z-scores from duplicate assays showed that the results were reproducible, with good correlation between the two experiments (Fig. 2A). The comparison of dark and light samples identified dsRNAs that inhibited CRY degradation and we selected those with a Z-score of less than or equal to −2. In addition, those dsRNAs that elevated CRY-luc levels under light conditions, with a Z-score of 2 or more, were also scored as positives. Of the 21,000 dsRNAs in the library, after filtering out dsRNAs harboring homology with multiple repeat elements with predicted off-target effects (Kulkarni et al. 2006), the primary screen identified 303 annotated genes that inhibited CRY degradation (Fig. 2B; Supplemental Table S1).
Figure 2.
Results of the primary screen to identify genes required for light-dependent CRY degradation. (A) Correlation of the replicate Z-scores for the primary screen plates. Artificially elevated Z-scores >10 were eliminated from A and B. (B) Scatter plot of the average Z-score from replicate experiments. Each plate contained a positive control, Prosβ2 dsRNA (with Z-score near −5), which was eliminated from the plot. (C) Functional classification of the 173 hits that were reconfirmed by the CRY-luc degradation assay. The genes were assigned to different functional classes based on the GO annotation and plotted.
To confirm our results, we conducted further analysis with the positives identified in the primary screen. Of the 303 candidates, we were able to synthesize dsRNA against 287 genes and confirmed 173 (60%) as positive by rescreening them using the CRY-luc light response assay. Functional annotation and clustering analysis identified a high enrichment of proteasome complex genes (EASE score 10−14) (Hosack et al. 2003). Notably, we identified 17 genes encoding proteasome components, including Prosβ2 and DmS13. Among other proteasome components, we identified seven of the eight AAA-ATPases from the 19S base complex. These AAA-ATPases are essential for substrate anchoring, unfolding, and the gating of the 20S proteolytic core.
Apart from components of the core–proteasome complex, we isolated five genes encoding components of the ubiquitin-conjugating complex. This type of complex includes E3 ligases that perform an essential/rate limiting role in targeting specific proteins for ubiquitination and proteasome degradation. We identified three novel HECT or RING domain-containing E3 ligases (CG17735, CG11321, and CG5604), as well as a composite E2–E3 ligase, Bruce, which contains both ubiquitin-conjugating enzyme activity and a putative substrate-binding domain.
Previous studies have indicated a role for electron transfer and redox in light-dependent CRY degradation (Lin et al. 2001). Our screen identified eight genes involved in regulating cellular redox status, including Glutathione S-transferase (GstE7), Thioredoxin (Txl), lactate/malate dehydrogenase (ImpL3), and biopterin-binding thyrosine hydroxylase (TH) gene pale (ple). We also identified 13 genes involved in signal transduction, of which three are kinases and two are phosphatases. Given earlier studies suggesting a role of tyrosine kinases in light entrainment (Naidoo et al. 1999), it is interesting that the two phosphatases (slingshot and CG7556) are predicted to recognize phospho-tyrosine residues.
Effects of candidate genes on light-dependent CRY and TIM degradation
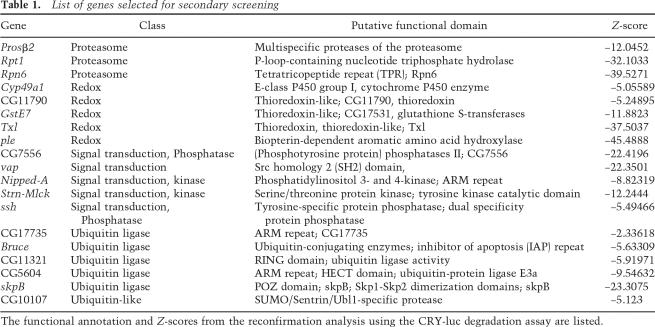
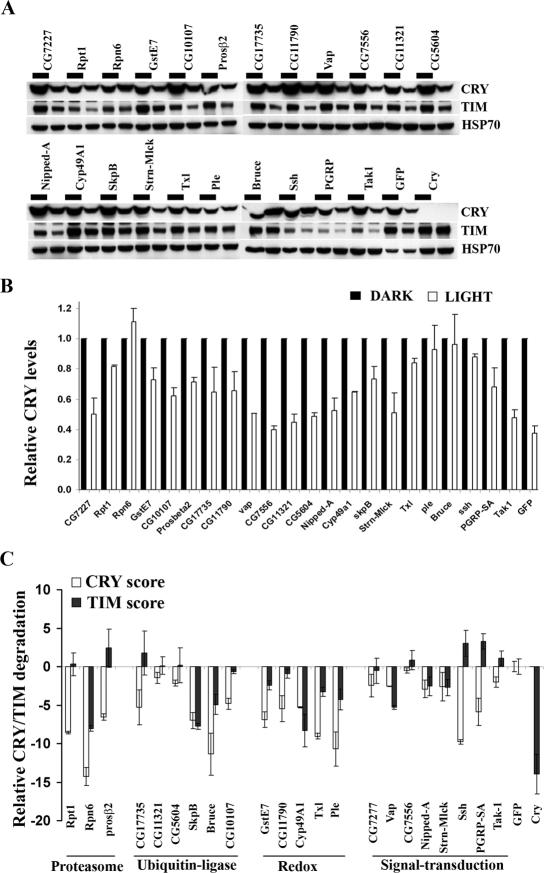
To confirm the effect of the candidate dsRNAs on CRY degradation and to place these molecules in the circadian entrainment pathway, we extended our analysis for a select set of 22 genes. These genes included ubiquitin ligases, redox molecules, and signal transduction molecules that significantly blocked light-dependent CRY degradation (Table 1; Supplemental Table S1). Three genes involved in the 26S proteasome complex were also included. After light activation, CRY binds TIM and then both proteins are targeted for proteasome-mediated degradation (Ceriani et al. 1999; Lin et al. 2001). To investigate if the candidates identified by our screen also block light-dependent TIM degradation, we cotransfected S2R+ cell with V5 tagged cry along with tim. An expression construct for JET, a recently identified E3 ligase for TIM, was also included in the assay since this is required for the TIM response to light in S2 cells. Following light exposure, TIM and CRY levels were examined by Western blot analysis (Fig. 3). For ease of comparison, relative Z-scores were calculated and plotted for CRY and TIM degradation (Fig. 3C). dsRNAs against all three core components of the proteasome included in the list of 22 genes (Rpt1, Rpn6, and Prosβ2) significantly inhibited light-dependent CRY degradation (Fig. 3). RNAi of Rpn6, the component of the proteasome lid complex, was most effective in blocking light-dependent CRY degradation, and it also affected TIM degradation. However, dsRNAs against Rpt1 and Prosβ2 were ineffective against TIM degradation. The role of the 26S proteasome complex in regulating light-dependent TIM degradation has been well documented (Naidoo et al. 1999; Koh et al. 2006). It is possible that when E3 ligase (JET) levels are in excess, TIM degradation is accomplished by limiting amounts of the proteasome complex. In general, our results show that proteasome-mediated degradation is essential for CRY and TIM degradation.
Table 1.
List of genes selected for secondary screening
The functional annotation and Z-scores from the reconfirmation analysis using the CRY-luc degradation assay are listed.
Figure 3.
Effect of select candidates on light-dependent CRY and TIM degradation in S2R+ cells. (A) Western blot analysis showing effects of primary screen positives on light-dependent CRY and TIM degradation. S2R+ cells were transfected with expression constructs for cry, tim, and jet and the indicated dsRNA and then either exposed to light for 2 h or maintained in constant darkness (black bar). CRY (top) and TIM (middle) were visualized via Western analysis using V5 and TIM antibodies, respectively. (Bottom) Endogenous HSP-70 levels were used as a loading control. (B) Quantification of the effects of different dsRNAs on CRY degradation. CRY levels were quantified, and differences in average intensities from two experiments were normalized to the dark values and plotted. Error bars represent SEM. Similar results were obtained by using a myc-tagged CRY (data not shown). (C) Quantification of the effects of different dsRNAs on CRY degradation. Relative light-dependent TIM degradation was measured by similar quantification of the TIM band. Based on these values, a relative TIM or CRY degradation score (see the Materials and Methods) was obtained and plotted.
The selected 22 genes included five that regulate electron transfer or redox status of proteins. Previous studies using small molecule regulators of cellular redox suggested that electron transfer plays an important role in the light-dependent activation of CRY (Lin et al. 2001; Froy et al. 2002). However the molecules involved in regulating this process have not been identified. Our rescreen included three thioredoxin domain-containing proteins (GstE7, Txl, and CG11790) that are small disulphide-containing proteins that regulate redox status of many proteins (Maher 2006). RNAi-mediated knockdown of all three genes (GstE7, Txl, and CG11790) blocked light-dependent CRY degradation (Fig. 3), thus strongly supporting the role of redox proteins in this process. Since electron transfer is involved in the light-dependent activation of CRY (Lin et al. 2001; Froy et al. 2002), this step should precede TIM degradation. As expected, all these redox genes also blocked light-dependent TIM degradation (Fig. 3C). RNAi-mediated knockdown of a cytochrome P450 gene, Cyp49a1, also blocked CRY and TIM degradation. In addition, knockdown of a TH gene Pale (Ple), involved in catecholamine biosynthesis and pigmentation of the cuticle, inhibited CRY and TIM degradation, suggesting a novel role for TH in the circadian light entrainment pathway.
We also examined the effects of RNAi against three kinases and two phosphatases identified in the screen. As noted above, tyrosine phosphorylation has been previously implicated in light-dependent TIM degradation. RNAi knockdown of a dual specific phosphatase ssh (Niwa et al. 2002) blocked light-dependent CRY degradation but had little effect on TIM degradation. SSH is involved in actin filament reorganization and is activated by phosphotidylinositol 3′-kinase (PI3′-kinase) in an insulin-dependent manner (Huang et al. 2006). RNAi knockdown of three kinases identified in the screen had a moderate effect in blocking the degradation of CRY/TIM. Nipped-A, a kinase involved in Notch signaling, and Stretchin-MLCK (Strn-Mlck), a myosin light chain kinase, blocked both CRY and TIM degradation. Knockdown of a TGFβ-activated kinase, Tak, and of a peptido-glycan receptor, PGRP-SA, both of which are involved in the IMD immune pathway in Drosophila, moderately inhibited CRY degradation. The effect of knocking down IMD pathway components Tak and PGRP-SA was restricted to CRY degradation; they did not affect TIM, suggesting that components of this pathway may be specifically utilized for the regulation of CRY. In addition to these kinase/phosphatases, dsRNA against an SH2 domain-containing RAS-GTPase, vacuolar peduncle (vap), blocked both CRY and TIM degradation.
We also retested a number of components known to be involved in protein degradation pathways. RNAi against an SCF adaptor gene, skpB, had a strong effect on both CRY and TIM degradation. The role of the SCF complex in light-dependent TIM degradation has been previously described (Koh et al. 2006); the current results indicate that it may also be required for CRY degradation. On the other hand, a putative SUMO protease (CG10107) blocked light-dependent CRY degradation but did not affect TIM degradation (Fig. 3). In follow-up studies, we assayed for SUMOylation of CRY in S2R+ cells but were unable to detect this, suggesting that CRY is not directly regulated by SUMOylation (data not shown). Thus, other components of the CRY degradation complex may be regulated by SUMOylation.
Finally, our list of 22 genes included two HECT domain-containing E3 ligases (CG17735 and CG5604) and a RING domain-containing E3 ligase (CG11321). While neither affected TIM degradation, RNAi against CG17735 in particular had a profound effect on CRY degradation. However, of all the ubiquitin ligases tested in the secondary screen, dsRNA against Bruce (for baculovirus inhibitor of apoptosis repeat [BIR]-containing ubiquitin-conjugating enzyme), a composite E2–E3 ligase, was most effective in blocking CRY degradation (Fig. 3A,B). It also partially blocked TIM degradation. However, the magnitude of the effect on TIM was less than that on CRY. Interestingly, Bruce RNAi also affected baseline levels of CRY when it was fused to a tag other than the one used in Figure 3, suggesting that BRUCE may affect overall CRY levels under some conditions (data not shown).
In vivo validation of candidate genes that affect CRY degradation
In order to validate results of the cell culture screen in vivo, we examined flies mutant for each of three candidate genes: the E2–E3 ubiquitin ligase, Bruce; the dual specific phosphatase, ssh; and the HECT domain-containing E3 ligase CG17735. All these candidates had a strong effect on CRY degradation in the cell culture studies. BRUCE/apollon/Bric6 is a large (528 kD) composite E2–E3 ligase with a BIR domain, a hallmark of inhibitor of apoptosis proteins (IAPs) (Bartke et al. 2004; Hay and Guo 2006). Indeed, BRUCE plays an anti-apoptotic role by inhibiting Reaper- and Grim-mediated apoptosis (Vernooy et al. 2002). Given the strong effect of Bruce RNAi in blocking CRY degradation in S2 cells, we were interested in testing the phenotypes of Bruce mutations in flies.
We focused these studies on a line containing a piggyBac transposon insertion, Brucee00984 (referred to as BruceRB), in the coding region (exon 8) of the Bruce gene (Supplemental Fig. S1A). To assess the effects of this transposon insertion on Bruce mRNA levels, we performed RT–qPCR (Supplemental Fig. 1B). Using a probe 3′ to the insertion site, we could not detect any transcript indicating that the piggyBac insertion results in premature termination of the Bruce transcript (Supplemental Fig. S1). This would result in a truncated protein lacking the critical C-terminal UBC domain.
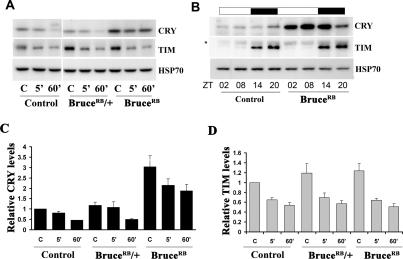
We next evaluated the role of BRUCE in regulating CRY and TIM stability following an acute light pulse at night. In the BruceRB line, we found that CRY levels were elevated under baseline conditions, and although degradation occurred in response to a 5-min or 60-min pulse, relatively high levels remained (Fig. 4A). These results suggested that the major effect of Bruce knockdown is to increase the overall stability of CRY. To further investigate the role of BRUCE in regulating CRY stability, we evaluated CRY levels in a 12:12 light/dark (LD) cycle. In wild-type (control) flies, the light sensitivity of CRY results in an oscillation of protein levels in LD cycles, with higher levels expressed during the night. In BruceRB mutants, CRY levels were elevated in the dark and CRY protein persisted following the transition into the light period (Fig. 4B). Since CRY does not cycle in constant darkness (Emery et al. 1998), indicating that its cyclic expression in LD cycles is driven entirely by its response to light, its incomplete degradation in an LD cycle in Bruce mutants supports the idea that light-dependent degradation of CRY in flies is affected by loss of Bruce. We note that the cell culture assay involves a 6-h exposure to light, and so it may be somewhat analogous to an LD cycle, thereby accounting for the robust effects of Bruce knockdown in this assay. In Bruce flies, levels of CRY were even higher during the day than at night, perhaps because of the higher levels of cry mRNA in the daytime. We infer that normally light-induced CRY degradation negates any influence of cry mRNA cycling on protein expression, but in Bruce mutants, the attenuated response to light unmasks such regulation. The effects of BRUCE on CRY levels were supported by analysis of a fly line carrying an RNAi construct that knocks down expression of Bruce specifically in the eye (data not shown).
Figure 4.
Effect of BRUCE on light-dependent CRY and TIM degradation. (A) Bruce mutants have elevated CRY levels that persist even after prolonged light exposure. A representative Western blot image of light-dependent CRY and TIM degradation in yellow white (control) or BruceRB mutants following an acute light pulse (5 min) or prolonged light pulse (60 min) as compared with the dark control (D). High levels of CRY protein were observed in BruceRB mutants, and CRY was not efficiently degraded even after a prolonged light exposure. (B) Western blot analysis of CRY and TIM levels in control and BruceRB mutants in 12:12 light/dark (LD) cycle. Fly heads collected during the light phase (open bar) or dark phase (dark bar) were probed for TIM, CRY, and HSP-70 (loading control). In control flies, CRY levels are high at late night (ZT20), but in BruceRB mutants, CRY levels are low at this time point. Also the overall CRY levels are elevated by more than fivefold in BruceRB mutants. TIM levels are low during the day and high at night in both wild-type and BruceRB mutant flies. (C,D) Quantification of light-dependent CRY and TIM degradation in Bruce mutants. CRY and TIM levels were quantified in the samples described in A and normalized to levels of the respective protein in the control line maintained in the dark. HSP70 levels were determined to control for loading. BruceRB mutants show significantly elevated CRY levels when compared with wild-type siblings and Bruce heterozygotes (P < 0.008). Light-dependent degradation of CRY was also reduced in Bruce mutants (in the data shown here, levels of CRY were significantly reduced in the heterozygous sibling [P < 0.01] but not in the mutant [P = 0.1]). Statistical analysis was conducted using ANOVA followed by a post-hoc Tukey HSD test. An average of four independent experiments is shown.
The cell culture results suggested that BRUCE may have a minor effect on TIM; however in BruceRB mutants, the regulation of TIM was largely unaffected, either following an acute light pulse or under LD cycles (Fig. 4A,B). Thus both cell culture and fly experiments show that BRUCE specifically regulates CRY stability. Since some degradation of CRY occurs following acute light exposure in Bruce mutants, we suggest that there are redundant mechanisms for regulating light-dependent CRY degradation. In fact, the screen identified a number of E3 ligases, one of which, CG17735, we further characterized (see below). We also assayed behavioral rhythms of BruceRB mutant and found that many of the flies either lacked rhythms altogether or had weak rhythms (data not shown). This effect on rhythms was not accompanied by any major defects in the clock neurons (Supplemental Fig. S2) or in the molecular oscillations of clock proteins in LD cycles or constant darkness (Fig. 4B; data not shown). However, this weak rhythmicity, which may or may not map to Bruce, made it difficult to assess the resetting of behavioral rhythms in these flies by light.
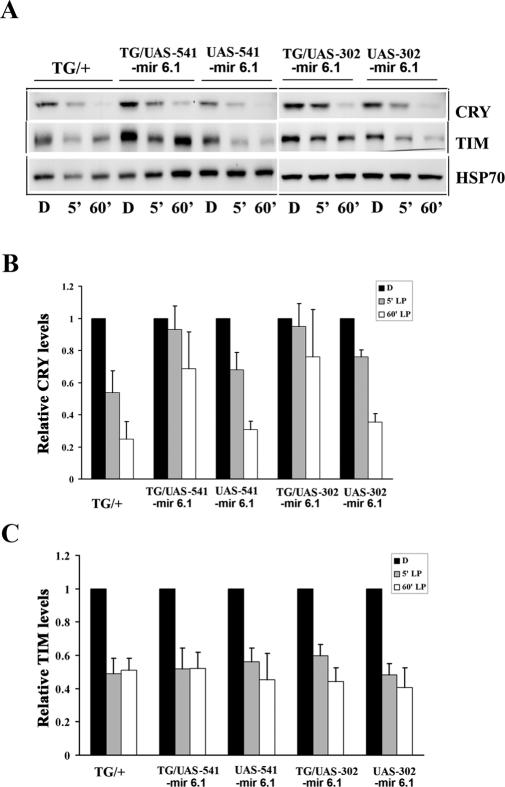
To assess the contribution of the HECT domain-containing novel E3 ligase CG17735 to light-dependent CRY degradation, we generated two independent fly lines (541 and 302) in which CG17735 expression was knocked down through a microRNA (mir6.1)-based RNAi approach described previously (Supplemental Fig. S3A; Chen et al. 2007). We expressed the UAS-mir 6.1-CG17735 construct under the control of a tim-Gal4 driver so as to drive expression in all clock cells and verified through qPCR that levels of CG17735 mRNA were knocked down (Supplemental Fig. S3B). In CG17735 knockdown flies, CRY degradation, but not TIM, was reduced, in response to an acute light pulse (Fig. 5A). We note that it did not completely block CRY degradation by light; this could be either because the knockdown was not complete or because other proteins can also mediate light-dependent CRY degradation. Thus, Bruce may regulate overall CRY stability while CG17735 regulates CRY degradation following acute light exposure.
Figure 5.
CG17735 regulates light-dependent CRY degradation. (A) Western blot analysis showing the effects of mir 6.1-CG17735 on light-dependent CRY and TIM degradation. Two independent UAS-mir 6.1 CG17735 constructs (541 and 302) were expressed in clock cells using a tim-Gal4 (TG) driver. The TG/UAS-mir 6.1-CG17735, or TG/+ (control) or UAS-mir 6.1-CG17735/CyO (sibling control) flies were treated with either a 5-min light pulse (gray bars) or a 60-min light exposure (white bars) at ZT20 or kept in darkness (black bars). All flies were collected at ZT21. CRY (top) and TIM (middle) were visualized by Western blot analysis using CRY and TIM antibodies, respectively. (Bottom) HSP70 levels were determined to control for loading. Although TIM levels were variable in the 541 line and appear to be higher in the blot shown here, they were generally not affected by knockdown of CG17735. (B,C) Quantification of the effects of CG17735 RNAi on CRY and TIM degradation following a 5- or 60-min light pulse. CRY and TIM levels were quantified, and for each genotype they were normalized to the levels of the respective protein in the dark (D) sample. Error bars represent SEM. Data shown were averaged from four independent experiments for all lines except TG, which is based on three experiments. Post-hoc comparisons (Tukey’s test) show that CRY degradation following the 5-min light pulse is significantly less (P < 0.05) in the CG17735 mir 6.1-expressing lines as compared with the TG control.
Interestingly, free-running rhythms of CG17735 knockdown flies had periods that were considerably longer than those of controls: ∼27 h as compared with 24 h in controls (Supplemental Fig. S3C). This difference in period shifted the phase response curve (PRC) of these flies such that a delay, rather than an advance, occurred in response to a light pulse at ZT20 (Supplemental Fig. S3D). However, at ZT22 an advance was seen in both control and experimental flies. Thus, overall the CG17735 knockdown flies did not display major defects in the behavioral response to light.
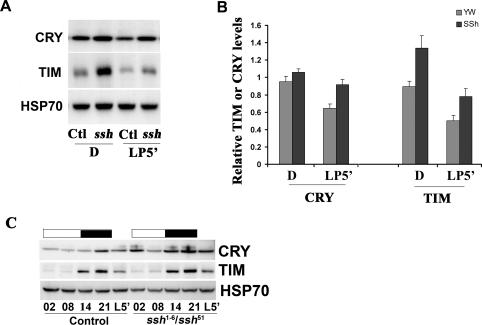
Finally, we determined the effects of a mutation in the dual specific phosphatase, ssh, on light-dependent CRY stability. Null mutations of ssh are lethal (Niwa et al. 2002), and so we characterized a heteroallelic combination of two hypomorphic mutations, ssh1-6 and ssh51, that disrupts ssh expression (Niwa et al. 2002). Following an acute light pulse at ZT20, higher levels of residual CRY were observed in the ssh hypomorphic mutant (Fig. 6A). However after prolonged light exposure (60 min), CRY degradation was equivalent to that in controls (data not shown). The response of TIM to light was normal (Fig. 6). We also examined CRY and TIM levels in a 12:12 LD cycle. As in Bruce mutants CRY levels were elevated in the dark, and increased residual CRY was detected following transition into light at ZT2 (Fig. 6B), indicating that light-dependent degradation is impaired under these conditions. Free-running behavioral rhythms were normal, and their response to light was largely intact in the ssh hypomorphic mutants (Supplemental Fig. 4). These in vivo results support the cell culture data showing that CG17735, ssh and Bruce affect CRY stability in response to light.
Figure 6.
CRY stability is increased in ssh loss-of-function mutants. (A) Western blot analysis of CRY and TIM degradation in response to an acute light pulse. Flies were given an acute light pulse for 5 min at ZT20, then incubated in the dark and collected at ZT21. (B) Quantification of CRY and TIM levels in the dark-maintained (D) and light-pulsed samples (LP5′). The data represent the average of six biologically independent samples. Levels of CRY were reduced in control samples by 32% (P < 0.01, as compared with the dark control). This light-dependent degradation was significantly less (13%) in ssh mutants (P = 0.09 when compared with its dark control). Although TIM levels were somewhat higher in ssh mutants, its degradation by a light pulse was unaffected (degradation of 47% vs. 42% in control and ssh, respectively). (C) CRY levels are increased in the ssh mutants during the early morning (2 h into the daytime, ZT02). In contrast, TIM levels were low in the daytime and high in the nighttime in both the control and ssh flies. Transheterozygotes of ssh1-6 and ssh51 alleles were used, and Student’s t-test was performed for light-dependent degradation within genotypes.
Discussion
Using a luciferase-based CRY degradation assay, we employed a genome-wide RNAi knockdown approach to identify genes required for the CRY light response. A subset of these genes was further validated by secondary assays. To place these genes in the context of the circadian light entrainment pathway, we determined whether they are required for the degradation of the TIM clock protein by light. In addition, we investigated the role of two ubiquitin ligases (CG17735 and BRUCE) and a dual specific phosphatase (SSH) on light-dependent CRY degradation in vivo. Utilizing this unbiased approach, we identified several new genes, including protein kinases, phosphatases, molecules involved in regulating cellular redox, and ubiquitin ligases, none of which were previously identified as regulators of circadian rhythms. Further, our in vivo analysis validates the cell-based screen and demonstrates novel roles for the composite E2–E3 ligase BRUCE, the E3 ligase CG17735, and the phosphatase SSH in regulating CRY stability.
CRY contains a flavin and a pterin-binding pocket, and intramolecular electron transfer has been reported in its activation by light (Lin et al. 2001; Froy et al. 2002). In fact, if the cellular redox system is compromised, light-induced CRY degradation is impaired (Lin et al. 2001). However, the identity of redox molecules that participate in the circadian light response has remained largely unknown. Here we report the identification of three thioredoxin domain-containing redox molecules (GstE7, Txl, and CG11790) and a cytochrome p450 CYP49a1 that modulate light-dependent CRY and TIM degradation. These results suggest that cellular redox status and electron transfer modulate light-dependent activation of CRY, which in turn affects subsequent transmission of the light signal to TIM and the degradation of CRY itself.
The screen also identified a novel role for the TH gene, ple, in regulating light-dependent CRY and TIM degradation. TH catalyzes a rate limiting step in the synthesis of L-Dopamine (DA) and requires the cofactor tetrahydrobiopterin (BH4). S2 cells have been used successfully to demonstrate the role of DA signaling in FGFR receptor endocytosis and cytoskeleton rearrangement (Hsouna et al. 2007); thus, they may express components of the dopamine pathway. The mechanism of action of TH and DA signaling in light-dependent CRY degradation is not clear. However, TH has been implicated in oxidative mechanisms that lead to the production of free radicals, which may underlie its effects on CRY degradation (Adams et al. 1997).
A number of signal transduction molecules involved in the insulin signaling and IMD pathways were also identified by the screen. A phosphatase SSH appears to be of particular importance for the regulation of CRY. Knock down of ssh blocks light-dependent CRY degradation in cultured cells and also affects CRY stability in flies. However, it has little effect on TIM degradation. Thus, dephosphorylation of CRY, or a component required for CRY degradation, by SSH is important for regulating CRY stability. Currently, we presume that SSH plays an indirect role in regulating CRY degradation. Further studies aimed at understanding the phosphorylation sites on CRY or its interacting components are needed to understand the mechanism of SSH action on CRY. We note that the phenotype of ssh mutant flies probably underrepresents its actual role in the regulation of CRY because the mutant we used is a hypomorph rather than a null (a complete knockout of ssh is lethal).
Of the ubiquitin-conjugating enzymes identified in this screen, a composite E2–E3 ligase, BRUCE, and a novel HECT domain-containing E3 ligase CG17735 had the strongest effects on light-dependent CRY degradation. Neither of these ligases had a significant effect on the TIM light response in flies. BRUCE exerts its anti-apoptotic role through its N-terminal IAP domain or by targeting proapoptotic proteins, such as SMAC2, for degradation using its C-terminal E2–E3 ligase activity (Bartke et al. 2004). In flies, apart from its anti-apoptotic activity, it also plays an important role in regulating sperm individualization, a process by which the sperm nuclear syncytium differentiates into individual sperm (Huh et al. 2004). This is similar to the apoptotic pathway proteins, DIAP1, DIAP2, and Dredd, which perform a nonapoptotic role in the IMD immune pathway (Hay and Guo 2006). Thus a nonapoptotic role for Bruce in regulating CRY stability is not surprising. It is interesting to note that Bruce knockdown in S2 cells has a small effect on baseline levels but primarily blocks light-induced CRY degradation, suggesting that CRY is rendered stable and nonresponsive to light in the absence of BRUCE. The genetic evidence from loss-of-function mutations of Bruce in the fly suggest that BRUCE affects the stability of CRY in LD cycles. The same appears to be true for SSH. As noted earlier, this effect on CRY in LD cycles most likely also represents impaired degradation in response to light. We infer that the mechanisms involved in the CRY response to parametric and nonparametric entrainment may be somewhat different, and the cell culture assay may allow detection of molecules required for either mechanism. The fact that all these candidates identified in the cell culture screen have the same general effect on CRY stability in cells and flies demonstrates the power of the screen for identifying novel circadian regulatory molecules.
CG17735 affects the degradation of CRY by light in cell culture as well as in flies. Given that this gene encodes a putative E3 ligase, it is tempting to speculate that this is the E3 ligase that targets CRY for degradation in response to acute light exposure. However, until a null mutant of CG17735 can be obtained, we cannot know the extent to which it is required for the regulation of CRY. Nor can we know the behavioral consequences of completely blocking CRY degradation. It is intriguing that the knockdown of CG17735 is associated with a long period, but at this point it is unclear if this is due to the effect on CRY or on some other, as yet unidentified, clock target.
Although we identified various components of the CRY degradation complex, the importance of light-dependent CRY degradation to circadian resetting behavior is not clear. Bruce mutants demonstrate a strong defect in CRY degradation; however, due to the weak behavioral rhythms in these mutants, the effect on circadian photic sensitivity could not be addressed. Also we note that light-dependent CRY degradation, although reduced, still persists in BruceRB mutants, suggesting that it is regulated by redundant pathways. The other two mutants assayed (ssh and CG17735) did not completely abolish CRY degradation either, but in this case, it may be because neither is a null mutant. Interestingly, TIM degradation was not affected in any of these mutants, suggesting that they specifically affect CRY degradation. TIM is targeted for light-dependent degradation by an F-box-containing protein, JET, which has minimal effect on CRY degradation (Koh et al. 2006). This result again supports the existence of two distinct destruction complexes that regulate CRY and TIM degradation. However, some aspects of the degradation pathways may be similar. The demonstration that SkpB, a component of the SCF complex, is critical for both CRY and TIM degradation suggests that some components are shared between the two destruction complexes. In addition, components required for the activation of CRY are presumably also required for the TIM light response. It is likely that other classes of molecules identified in the screen, such as those that affect redox status, are involved in CRY activation and therefore also affect TIM and the behavioral response. Further analysis of these molecules in clock neurons should address the role of redox and CRY degradation in circadian photosensitivity.
Materials and methods
Expression constructs and cell culture
The pIZ-luc-cry plasmid was generated by fusing the luciferase (luc) gene with cry. The coding region of luc was amplified by PCR using Pfu polymerase (New England Biolabs) from pGL3-Basic (Promega), and cloned in-frame with wild-type cry or cry carrying a point mutation, cryb, as an N-terminal or C-terminal fusion in pIZ-V5 (Lin et al. 2001). In another plasmid, a myc tag sequence was added to the N terminus of cry or cryb cDNA (Busza et al. 2004), and a stop codon was introduced at the end of cry and cloned into pIZ-V5/His A (Invitrogen). Insect cell expression constructs for jet, tim were described previously (Ceriani et al. 1999; Koh et al. 2006). A Renilla luciferase gene expressed under the control of Actin5C or RNA polIII promoter was described previously (Darlington et al. 1998; Nybakken et al. 2005)
S2R+ cells from the Drosophila RNAi Screening Center (DRSC; http://flyrnai.org) were grown in Schneider’s medium containing 10% FBS, split at 1:8 dilution, and maintained at 25°C. Transfection was performed using Effectene (Qiagen) with expression constructs according to the manufacturer’s instructions. Thirty-six hours post-transfection, cells were exposed to bright light in the presence of DMSO or the proteasome inhibitor MG132. All cells were harvested and processed for Western blot analysis or luciferase assay.
RNA-mediated interference
For RNAi experiments performed independently of the DRSC, DNA fragments 600–800 base pairs (bp) in length, containing various coding regions of Prosβ2, morgue, DmS13, or Slimb, were amplified with primers containing T7 promoters at both the 5′ and 3′ end, and dsRNA was generated by T7 transcription and annealed as described previously (Sathyanarayanan et al. 2004). Prosβ2 was amplified using the following primers: 5′-ACTGCGCCAAGATCCATTAC-3′ and 5′-GTTGAGGTTCC GGTCTTGAA-3′; morgue was amplified using the following primers: 5′-AGACATCATGGGCTTTGAGG-3′ and 5′-CAGC CGATACCAGTTTGGAT-3′; and DmS13 was amplified using 5′-CTGCTACGTCTTGGAGGTGCTATGCCACAGG-3′ and 5′-ACAGTGCTCATTGTAGTCGGACAACGTGAGGC-3′. A T7 promoter sequence (GAATTAATACGACTCACTATAGG GAGA) was added to the 5′ end of both forward and reverse primers.
Primary screen and analysis
Two sets of 58 plates (384-well) containing 0.08 μg of dsRNA per well were provided by the DRSC. The primary screen was transfection-based, utilizing a dual luciferase assay. The N-terminal luc fusion construct of cry, pIZ-luc-cry (12 ng per well), was mixed with pAct-Renilla-luc (9 ng per well), and a lipid complex was prepared with Effectene (300 μL per plate) and enhancer (250 μL per plate) in Buffer EC (Qiagen). The DNA–effectene mix was transferred to the screening plate by using multidrop 384 and mixed gently. Cells (2.6 × 104 per well) in complete medium were added to the plate. Plates were wrapped in foil and incubated in dark for 4 d at 25°C in a humidified container. On the fifth day, one set of plates was exposed to white light for 12 h, while the other was wrapped in two layers of foil and placed adjacent to the light-exposed plates. Firefly and Renilla luciferases assays were performed using the Dual-glo luciferase assay (Promega) according to the manufacturer’s instructions. All cell culture medium was aspirated off the cells, an equal volume of Dual-Glo Luciferase reagent was mixed with complete medium, and 40 μL of the mix were dispensed using a MicroFill (BioTek). Luciferase levels were read using a Analyst HT plate reader (Molecular Dynamics). Subsequently, Renilla luciferase activity was measured. Each receptacle plate was assigned a number and a bar code for automatic identification by the plate reader. The transient transfection efficiency was estimated by utilizing a GFP expression construct. About 25%–30% of the cells were GFP-positive at the end of the assay (data not shown).
To identify genes involved in regulating light-dependent CRY-luc degradation, the luciferase activity was normalized to Renilla luciferase values to give a normalized luciferase score. The normalized luciferase scores were compared between light (L) and dark (D) samples, and the relative light-dependent change in luciferase level was calculated (D − L/D). Using the relative light-dependent change in luciferase levels, a Z-score for each well was determined based on the interior-well plate average and interior-well plate standard deviation (as described in Nybakken et al. 2005). Z-scores are a measure of how different a particular value is from the mean in terms of the number of standard deviations from that mean. The interior wells are all wells except those on the outer edges of the plates, which, we found empirically, it was best not to include in these calculations due to occasional edge effects found in some plates. Z-scores from duplicate screens were averaged, and the average Z-score was used to identify hits. The artificial change in normalized luciferase scores arising from a decrease in renilla luciferase levels observed at the edges of the plates or in one of the two replicate plates was eliminated by comparing dark and light levels of luciferase. Most of the Heidelberg collection (plates 35–54) did not target legitimate genes (many target noncoding RNA) and were not included in the second replicate. Due to a jam in the MicroFill, addition of luciferase reagent to most of the wells in plate numbers 45 and 53 was affected, and thus these results were eliminated from analysis. Among the two replicates experiments, we found that the second replicate showed less variability. In order to get maximum coverage, positives (<2 Z-score) from the second replicate were also included in the secondary screen.
From the primary screen, positives corresponding to annotated genes were selected for reconfirmation. Hits that were predicted to target a number of genes (off-targets) were eliminated. The remaining positives from the primary screen were cherry-picked, and DNA templates were amplified by PCR using T7 primers. The length of the amplified product was checked on an agarose gel and purified using Montage PCR cleanup kit (Millipore). dsRNA was generated by T7 transcription (Sathyanarayanan et al. 2004). Reconfirmation of the hits was performed by utilizing the primary screen assay. pIZ-luc-cry and pAct-renilla luciferase constructs and dsRNA were cotransfected into S2R+ cells as described above. dsRNA against gfp was utilized as a negative control, and Prosβ2 was utilized as a positive control. Here the Z-scores were calculated utilizing GFP values as a mean (n = 3).
Western blot analysis
Cells (S2R+) were cotransfected with 320 ng of dsRNA and 25 ng of pIZ-cry-V5 (or pIZ-myc-cry) along with 50 ng of pAct-tim and 50 ng of pFlag-jet in a 12-well plate. Five days post-transfection, one set of sample was exposed to light while the other was kept in the dark for 2 h. Cells were washed with PBS and processed for Western blot analysis as described in Sathyanarayanan et al. (2004). The blots were probed with anti-V5 antibody (Invitrogen), anti-TIM antibody (UPR8), and anti-HSP70 antibody (Sigma). A myc epitope tag mouse monoclonal antibody (SantaCruz Biotechnologies) was utilized to probe for myc-tagged CRY (1:1000). The blots were quantified using a Kodak image station. dsRNA against gfp was used as a negative control. Light-dependent CRY-V5 and TIM degradation with the various RNAi treatments were quantified relative to the degradation seen with gfp RNAi. The experiment was repeated twice, and a degradation score for each dsRNA was calculated relative to gfp. A relative CRY/TIM degradation score was calculated by normalizing CRY and TIM Z-scores.
Western blot analysis for adult flies
In adult flies, the CRY and TIM degradation levels were evaluated by Western blot analysis. Flies were entrained to 12 h:12 h LD cycles for 3–4 d and collected on dry ice at given time points. Ten adult heads were collected for each genotype, and similar Western methods were followed for fly head extracts as described above. Rabbit anti-CRY antibody (Rush et al. 2006) was used to probe endogenous CRY. TIM and HSP-70 levels were determined as described above. After ECL, the blots were visualized with a Kodak Image Station, and images were captured by a CCD camera. Each experiment was repeated four times, and the CRY and TIM levels were normalized and plotted as described in each figure legend.
Fly lines, locomotor activity measurements, and statistics
miRNA-based CG17735 RNAi lines 541 and 302 were generated by cloning a hairpin targeting CG17735 into the pUAST vector containing mir6.1 as described below. The piggyBac insertion line BruceRB (Brucee00984) was obtained from the Bloomington Stock Center. Other fly lines were kindly provided by several researchers (see the Acknowledgments). Flies were entrained to a 12 h:12 h LD cycle at 25°C. Locomotor activity of individual flies was monitored and analyzed as described previously (Yuan et al. 2005). Circadian phase-resetting experiments were performed as described (Yuan et al. 2005). The length of the circadian period, τ, was calculated using χ2-periodogram analysis.
Construction of miRNAs targeting CG17735
The miRNA-based CG17735 RNAi construct was designed based on the mir6.1 precursor as described previously (Chen et al. 2007). The 22-bp sequences corresponding to mature mir6.1 were replaced with sequences perfectly complementary to sequences from the CG17735 gene, generating two new miRNAs: mir6.1-CG17735-1 and mir6.1-CG17735-2 (Supplemental Fig. S3A). Two sites in the CG17735 transcript were targeted to minimize the possibility that mRNA secondary structure would prevent the miRNA-RISC complex from being able to bind and cleave the mRNA. The use of multiple miRNAs targeting a common transcript, but at distinct positions, also provides a method for limiting the possibility that a single mutational event (in either the miRNA or the target sequence) will lead to a loss of effective target knockdown. The sites in CG17735 that are targeted are CG17735-1 (5′-CAACGTCGTCTTAACCATAACT-3′) and CG17735-2 (5′-CAAGCGCGTGTCATACTAAATT-3′). To generate a mir6.1 stem–loop backbone that generates a mature miRNA complementary to these target sites, we annealed pairs of primers. CG17735-1-1 (5′-GGCAGCTTACTTAAACTTAATCACAGCCTTTAATGTCAACGTCGTCTTAACCATACCTTAAGTTAATATACCATATC-3′) and CG17735-1-2 (5′-AATAATGATGTTAGGCACTTTAGGTACCAACGTCGTCTTAACCATAACTTAGATATGGGTATATTAACTTAAGGT-3′) were annealed and filled in using PCR. This product was then amplified using primers Mir6 5′ EcoRI/BglII (5′-GGCGAATTCCGCCAGATCTTTTAAAGTCCACAACTCATCAAGGAAAATGAAAGTCAAAGTTGGCAGCTTACTTAAACTTA-3′) and Mir6 3′ BamHI/NotI (5′-GGCCGCGGCCGCACGGATCCAAAACGGCATGGTTATTCGTGTGCCAAAAAAAAAAAAAATTAAATAATGATGTTAGGCAC3′). These primers add mir6.1 flanking sequences that are thought to promote miRNA processing, as well as several restriction sites. A miRNA that targets CG17735-2 was generated similarly, beginning with primers CG17735-2-1 (5′-GGCAGCTTACTTAAACTTAATCACAGCCTTTAATGTCAAGCGCGTGTCATACTAACTTTAAGTTAATATACCATATC-3′) and CG17735-2-2 (5′AATAATGATGTTAGGCACTTTAGGTACCAAGCGCGTGTCATACTAAATTTAGATATGGTATATTAACTTAAAGT-3′). PCR products were purified with Qiagen PCR purification columns and then digested with appropriate restriction enzymes. For CG17735-1, these were EcoRI and BamHI; for CG17735-2, these were BglII and NotI. Digested products were then ligated into pUAST vector cut with EcoRI and NotI.
Immunohistochemistry
Adult fly brains were collected at ZT2, dissected, and processed as described (Sathyanarayanan et al. 2004). Samples were incubated with primary antibodies diluted as follows: anti-PDF (HH74), 1:1000; anti-PER (UPR34), 1:1000. Eight to 12 fly brain hemispheres were examined per condition. Immunofluorescent images were obtained with a Leica confocal microscope.
Acknowledgments
We thank members of the DRSC, especially Bernard Mathey-Prevot, Susan Armknecht, Matthew Booker, and Norbert Perrimon for providing the genome-wide RNAi library and for help with RNAi screening; Patrick Emery for the anti-CRY antibody; Tadashi Uemura for ssh fly stocks; Yanshan Fang for the pIZ-myc-cry plasmid; Kent Nybakkan for the PolIII-rluc plasmid; members of the Perrimon laboratory for help with the assay set-up; and Kyunghee Koh and Susan Harbison for help with data analysis. S.S. and X.Z. were supported in part by NIH grant NS048471.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1652308.
References
- Adams J.D., Klaidman L.K., Ribeiro P. Tyrosine hydroxylase: Mechanisms of oxygen radical formation. Redox Rep. 1997;3:273–279. doi: 10.1080/13510002.1997.11747123. [DOI] [PubMed] [Google Scholar]
- Ashmore L.J., Sehgal A. A fly’s eye view of circadian entrainment. J. Biol. Rhythms. 2003;18:206–216. doi: 10.1177/0748730403018003003. [DOI] [PubMed] [Google Scholar]
- Bartke T., Pohl C., Pyrowolakis G., Jentsch S. Dual role of BRUCE as an antiapoptotic IAP and a chimeric E2/E3 ubiquitin ligase. Mol. Cell. 2004;14:801–811. doi: 10.1016/j.molcel.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Busza A., Emery-Le M., Rosbash M., Emery P. Roles of the two Drosophila CRYPTOCHROME structural domains in circadian photoreception. Science. 2004;304:1503–1506. doi: 10.1126/science.1096973. [DOI] [PubMed] [Google Scholar]
- Cashmore A.R. Cryptochromes: Enabling plants and animals to determine circadian time. Cell. 2003;114:537–543. [PubMed] [Google Scholar]
- Ceriani M.F., Darlington T.K., Staknis D., Mas P., Petti A.A., Weitz C.J., Kay S.A. Light-dependent sequestration of TIMELESS by CRYPTOCHROME. Science. 1999;285:553–556. doi: 10.1126/science.285.5427.553. [DOI] [PubMed] [Google Scholar]
- Chen C.H., Huang H., Ward C.M., Su J.T., Schaeffer L.V., Guo M., Hay B.A. A synthetic maternal-effect selfish genetic element drives population replacement in Drosophila. Science. 2007;316:597–600. doi: 10.1126/science.1138595. [DOI] [PubMed] [Google Scholar]
- Collins B., Mazzoni E.O., Stanewsky R., Blau J. Drosophila CRYPTOCHROME is a circadian transcriptional repressor. Curr. Biol. 2006;16:441–449. doi: 10.1016/j.cub.2006.01.034. [DOI] [PubMed] [Google Scholar]
- Darlington T.K., Wager-Smith K., Ceriani M.F., Staknis D., Gekakis N., Steeves T.D., Weitz C.J., Takahashi J.S., Kay S.A. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- Emery P., So W.V., Kaneko M., Hall J.C., Rosbash M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell. 1998;95:669–679. doi: 10.1016/s0092-8674(00)81637-2. [DOI] [PubMed] [Google Scholar]
- Emery P., Stanewsky R., Helfrich-Forster C., Emery-Le M., Hall J.C., Rosbash M. Drosophila CRY is a deep brain circadian photoreceptor. Neuron. 2000;26:493–504. doi: 10.1016/s0896-6273(00)81181-2. [DOI] [PubMed] [Google Scholar]
- Froy O., Chang D.C., Reppert S.M. Redox potential: Differential roles in dCRY and mCRY1 functions. Curr. Biol. 2002;12:147–152. doi: 10.1016/s0960-9822(01)00656-x. [DOI] [PubMed] [Google Scholar]
- Grima B., Lamouroux A., Chelot E., Papin C., Limbourg-Bouchon B., Rouyer F. The F-box protein slimb controls the levels of clock proteins period and timeless. Nature. 2002;420:178–182. doi: 10.1038/nature01122. [DOI] [PubMed] [Google Scholar]
- Hardin P.E. Essential and expendable features of the circadian timekeeping mechanism. Curr. Opin. Neurobiol. 2006;16:686–692. doi: 10.1016/j.conb.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Hay B.A., Guo M. Caspase-dependent cell death in Drosophila. Annu. Rev. Cell Dev. Biol. 2006;22:623–650. doi: 10.1146/annurev.cellbio.21.012804.093845. [DOI] [PubMed] [Google Scholar]
- Hosack D.A., Dennis G., Sherman B.T., Lane H.C., Lempicki R.A. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsouna A., Lawal H.O., Izevbaye I., Hsu T., O’Donnell J. Drosophila dopamine synthesis pathway genes regulate tracheal morphogenesis. Dev. Biol. 2007;308:30–43. doi: 10.1016/j.ydbio.2007.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T.Y., DerMardirossian C., Bokoch G.M. Cofilin phosphatases and regulation of actin dynamics. Curr. Opin. Cell Biol. 2006;18:26–31. doi: 10.1016/j.ceb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Huh J.R., Vernooy S.Y., Yu H., Yan N., Shi Y., Guo M., Hay B.A. Multiple apoptotic caspase cascades are required in nonapoptotic roles for Drosophila spermatid individualization. PLoS Biol. 2004;2:E15. doi: 10.1371/journal.pbio.0020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T., Matsumoto A., Kato T., Jr., Togashi S., Ryo H., Ikenaga M., Todo T., Ueda R., Tanimura T. DCRY is a Drosophila photoreceptor protein implicated in light entrainment of circadian rhythm. Genes Cells. 1999;4:57–65. doi: 10.1046/j.1365-2443.1999.00237.x. [DOI] [PubMed] [Google Scholar]
- Ivanchenko M., Stanewsky R., Giebultowicz J.M. Circadian photoreception in Drosophila: Functions of cryptochrome in peripheral and central clocks. J. Biol. Rhythms. 2001;16:205–215. doi: 10.1177/074873040101600303. [DOI] [PubMed] [Google Scholar]
- Ko H.W., Jiang J., Edery I. Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime. Nature. 2002;420:673–678. doi: 10.1038/nature01272. [DOI] [PubMed] [Google Scholar]
- Koh K., Zheng X., Sehgal A. JETLAG resets the Drosophila circadian clock by promoting light-induced degradation of TIMELESS. Science. 2006;312:1809–1812. doi: 10.1126/science.1124951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan B., Levine J.D., Lynch M.K., Dowse H.B., Funes P., Hall J.C., Hardin P.E., Dryer S.E. A new role for cryptochrome in a Drosophila circadian oscillator. Nature. 2001;411:313–317. doi: 10.1038/35077094. [DOI] [PubMed] [Google Scholar]
- Kulkarni M.M., Booker M., Silver S.J., Friedman A., Hong P., Perrimon N., Mathey-Prevot B. Evidence of off-target effects associated with long dsRNAs in Drosophila melanogaster cell-based assays. Nat. Methods. 2006;3:833–838. doi: 10.1038/nmeth935. [DOI] [PubMed] [Google Scholar]
- Kume K., Zylka M.J., Sriram S., Shearman L.P., Weaver D.R., Jin X., Maywood E.S., Hastings M.H., Reppert S.M. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- Lin C., Todo T. The cryptochromes. Genome Biol. 2005;6:220. doi: 10.1186/gb-2005-6-5-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F.J., Song W., Meyer-Bernstein E., Naidoo N., Sehgal A. Photic signaling by cryptochrome in the Drosophila circadian system. Mol. Cell. Biol. 2001;21:7287–7294. doi: 10.1128/MCB.21.21.7287-7294.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher P. Redox control of neural function: Background, mechanisms, and significance. Antioxid. Redox Signal. 2006;8:1941–1970. doi: 10.1089/ars.2006.8.1941. [DOI] [PubMed] [Google Scholar]
- Murad A., Emery-Le M., Emery P. A subset of dorsal neurons modulates circadian behavior and light responses in Drosophila. Neuron. 2007;53:689–701. doi: 10.1016/j.neuron.2007.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidoo N., Song W., Hunter-Ensor M., Sehgal A. A role for the proteasome in the light response of the timeless clock protein. Science. 1999;285:1737–1741. doi: 10.1126/science.285.5434.1737. [DOI] [PubMed] [Google Scholar]
- Niwa R., Nagata-Ohashi K., Takeichi M., Mizuno K., Uemura T. Control of actin reorganization by Slingshot, a family of phosphatases that dephosphorylate ADF/cofilin. Cell. 2002;108:233–246. doi: 10.1016/s0092-8674(01)00638-9. [DOI] [PubMed] [Google Scholar]
- Nybakken K., Vokes S.A., Lin T.Y., McMahon A.P., Perrimon N. A genome-wide RNA interference screen in Drosophila melanogaster cells for new components of the Hh signaling pathway. Nat. Genet. 2005;37:1323–1332. doi: 10.1038/ng1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush B.L., Murad A., Emery P., Giebultowicz J.M. Ectopic CRYPTOCHROME renders TIM light sensitive in the Drosophila ovary. J. Biol. Rhythms. 2006;21:272–278. doi: 10.1177/0748730406290416. [DOI] [PubMed] [Google Scholar]
- Sathyanarayanan S., Zheng X., Xiao R., Sehgal A. Posttranslational regulation of Drosophila PERIOD protein by protein phosphatase 2A. Cell. 2004;116:603–615. doi: 10.1016/s0092-8674(04)00128-x. [DOI] [PubMed] [Google Scholar]
- Shearman L.P., Sriram S., Weaver D.R., Maywood E.S., Chaves I., Zheng B., Kume K., Lee C.C., van der Horst G.T., Hastings M.H., et al. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- Stanewsky R. Genetic analysis of the circadian system in Drosophila melanogaster and mammals. J. Neurobiol. 2003;54:111–147. doi: 10.1002/neu.10164. [DOI] [PubMed] [Google Scholar]
- Stanewsky R., Kaneko M., Emery P., Beretta B., Wager-Smith K., Kay S.A., Rosbash M., Hall J.C. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell. 1998;95:681–692. doi: 10.1016/s0092-8674(00)81638-4. [DOI] [PubMed] [Google Scholar]
- Vernooy S.Y., Chow V., Su J., Verbrugghe K., Yang J., Cole S., Olson M.R., Hay B.A. Drosophila Bruce can potently suppress Rpr- and Grim-dependent but not Hid-dependent cell death. Curr. Biol. 2002;12:1164–1168. doi: 10.1016/s0960-9822(02)00935-1. [DOI] [PubMed] [Google Scholar]
- Yuan Q., Lin F., Zheng X., Sehgal A. Serotonin modulates circadian entrainment in Drosophila. Neuron. 2005;47:115–127. doi: 10.1016/j.neuron.2005.05.027. [DOI] [PubMed] [Google Scholar]
- Zeng H., Qian Z., Myers M.P., Rosbash M. A light-entrainment mechanism for the Drosophila circadian clock. Nature. 1996;380:129–135. doi: 10.1038/380129a0. [DOI] [PubMed] [Google Scholar]